ABSTRACT
Methylation is essential to the physiology of all cells, including the obligate intracellular bacterium Chlamydia. Nevertheless, the methylation cycle is under strong reductive evolutionary pressure in Chlamydia. Only Parachlamydia acanthamoebae and Waddlia chondrophila genome sequences harbor homologs to metK, encoding the S-adenosylmethionine (SAM) synthetase required for synthesis of SAM, and to sahH, which encodes the S-adenosylhomocysteine (SAH) hydrolase required for detoxification of SAH formed after the transfer of the methyl group from SAM to the methylation substrate. Transformation of a conditional-lethal ΔmetK mutant of Escherichia coli with a genomic library of Chlamydia trachomatis L2 identified CTL843 as a putative SAM transporter based on its ability to allow the mutant to survive metK deficiency only in the presence of extracellular SAM. CTL843 belongs to the drug/metabolite superfamily of transporters and allowed E. coli to transport S-adenosyl-l-[methyl-14C]methionine with an apparent Km of 5.9 µM and a Vmax of 32 pmol min−1 mg−1. Moreover, CTL843 conferred a growth advantage to a Δpfs E. coli mutant that lost the ability to detoxify SAH, while competition and back-transport experiments further implied that SAH was an additional substrate for CTL843. We propose that CTL843 acts as a SAM/SAH transporter (SAMHT) serving a dual function by allowing Chlamydia to acquire SAM from the host cell and excrete the toxic by-product SAH. The demonstration of a functional SAMHT provides further insight into the reductive evolution associated with the obligate intracellular lifestyle of Chlamydia and identifies an excellent chemotherapeutic target.
IMPORTANCE
Obligate intracellular parasites like Chlamydia have followed a reductive evolutionary path that has made them almost totally dependent on their host cell for nutrients. In this work, we identify a unique transporter of a metabolite essential for all methylation reactions that potentially bypasses the need for two enzymatic reactions in Chlamydia. The transporter, CTL843, allows Chlamydia trachomatis L2 to steal S-adenosylmethionine (SAM) from the eukaryotic host cytosol and to likely remove the toxic S-adenosylhomocysteine (SAH) formed when SAM loses its methyl group, acting as a SAM/SAH transporter (SAMHT). In addition to reflecting the adaptation of Chlamydia to an obligate intracellular lifestyle, the specific and central roles of SAMHT in Chlamydia metabolism provide a target for the development of therapeutic agents for the treatment of chlamydial infections.
Introduction
Chlamydiae live as obligate intracellular parasites in specialized vacuoles within eukaryotic cells. The vast majority of species within this phylum of Gram-negative bacteria cause diseases in animals and/or humans and belong to the genus Chlamydia, although the list of Chlamydia-like organisms capable of causing disease continues to expand (1). Chlamydia trachomatis causes severe ocular and urogenital infections in humans and was the first Chlamydia species to be fully sequenced (2). Since that time, the sequences of 26 additional chlamydial genomes have been released in public databases (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/microbial_taxtree.html). All chlamydiae show signs of incomplete biosynthetic pathways, similar to other organisms that have adapted to parasitic/symbiotic lifestyles, such as mycoplasmas, phytoplasmas, and alpha- and gammaproteobacteria, including rickettsiae. Although novel enzymes have evolved to bypass some of these metabolic “holes” (3), the reduced genome size of obligate intracellular organisms is possible mostly because many metabolites do not need to be synthesized by the complex pathways characteristic of free-living bacteria but instead are transported from the substrate-rich host cell cytoplasm by novel transport systems absent in free-living bacteria (4). For example, a specific nucleotide transport protein allows Protochlamydia amoebophila to acquire the universal electron carrier NAD (NAD+) from the host in exchange for bacterial ADP, rather than synthesizing this compound (5).
S-Adenosyl-l-methionine (also known as AdoMet or SAM) is an essential intermediate in the physiology of all cells (6). The majority of SAM is used for methyltransferase reactions in which the S-methyl group of SAM is transferred to acceptor substrates, including nucleic acids, proteins, phospholipids, biological amines, and a long list of small molecules. During the transmethylation reaction, SAM is converted to S-adenosylhomocysteine (also known as AdoHcy or SAH) by the removal of the S-methyl group. SAH is a potent inhibitor of SAM-dependent methyltransferases and is therefore rapidly detoxified either by a SAH hydrolase or by a methylthioadenosine (MTA)/SAH nucleosidase (7, 8). Although SAM is not likely to be used as a methyl group donor for DNA in Chlamydia due to the absence of DNA methyltransferases (9), we recently showed that methylation of C. trachomatis 16S rRNA by the bacterial KsgA methylase is critical for bacterial fitness (10), confirming an earlier report that SAM-dependent methylation reactions take place in chlamydiae (11).
In this study, we examined the methylation cycle in Chlamydia. Comparative genome analysis showed that the methylation cycle is under reductive evolutionary pressure in Chlamydia. Only two Chlamydia-like organisms, Parachlamydia acanthamoebae and Waddlia chondrophila, appear to possess both an S-adenosylmethionine synthetase, the conserved but energetically demanding enzyme that catalyzes the only known route of SAM synthesis (12), and a recognized SAH catabolizing enzyme. Due to the lack of genetic tools for manipulating chlamydiae, we used Escherichia coli as a surrogate host to identify the strategy employed by the majority of chlamydiae to acquire SAM and eliminate SAH. By use of C. trachomatis serovar L2 strain 434/Bu as a model, CTL843 was identified through a genetic screen in a SAM synthetase-deficient E. coli strain and shown to mediate transport of SAM and likely SAH, acting as a SAMHT (SAM/SAH transporter). While serving as another example of the specialization of Chlamydia within its eukaryotic cell niche, the discovery of a novel means of Chlamydia host-cell interaction also presents an excellent chemotherapeutic target for the treatment of chlamydial infections.
RESULTS
The methylation cycle is under reductive evolutionary pressure in Chlamydia.
We recently showed that the conserved dimethyltransferase KsgA plays an important role in Chlamydia biology (10). KsgA transfers two methyl groups from the activated methyl donor SAM to the 16S rRNA molecules in ribosomes, generating the toxic by-product SAH. S-Adenosylmethionine synthetase (MAT or AdoMet synthetase [EC 2.5.1.6]) is the enzyme that synthesizes SAM from methionine and ATP. MAT is a highly conserved protein that is well studied at the primary, secondary, and tertiary structural levels and is an essential enzyme in prokaryotes and eukaryotes (12, 13). Comparative analysis of Chlamydia genome sequences identified open reading frames (ORFs) sharing 60% identity with E. coli MAT in P. acanthamoebae and W. chondrophila (i.e., pah_c014o152 and wcw_0127, respectively) and harboring all the conserved amino acid residues required for SAM synthetase activity (see Fig. S1 in the supplemental material). On the other hand, no MAT homolog was found in the other chlamydial genome sequences available (as of March 2011), with the exception of P. amoebophila, where a gene encoding a truncated and likely nonfunctional version of MAT (i.e., Pc1819) was identified.
Because SAH formed from SAM during methylation reactions needs to be eliminated due to its inhibitory effect on methyltransferases, we also looked for the presence of genes encoding putative SAH-detoxifying enzymes in Chlamydia genome sequences. Two different classes of enzymes degrade SAH in living cells. Most microbes, including E. coli, harbor an MTA/SAH nucleosidase (MTAN; EC 3.2.2.9) to metabolize SAH to adenine and S-ribosylhomocysteine, whereas mammals and some microbes employ a specific SAH hydrolase (SahH) (EC 3.3.1.1) to metabolize SAH into homocysteine and adenosine. Although putative MTAN ORFs are identified in the genome annotation of Chlamydia felis (i.e., CF0410), Chlamydia caviae (i.e., CCA0593), and Chlamydia pneumoniae (i.e., Cpn0232 for C. pneumoniae TW-183), sequence alignments revealed that most of the conserved residues required for MTAN activity in Arabidopsis thaliana, E. coli, and Staphylococcus aureus were missing in these chlamydial homologs (Fig. S2). Further examination revealed that the products of these three chlamydial ORFs belong to the PNP_UDP_1 superfamily (PF01048), which includes purine nucleoside phosphorylase (PNP), uridine phosphorylase (UdRPase), and 5′-methylthioadenosine phosphorylase (MTA phosphorylase). Thus, they are unlikely to be involved in SAH hydrolysis. Consistent with the presence of MAT in P. acanthamoebae and W. chondrophila, SahH homologs possessing the conserved motifs representative of active SahH enzymes were identified in these organisms, i.e., pah_c022o240 and wcw_1005, respectively (Fig. S3). Since out of 26 chlamydial genomes analyzed, only two, P. acanthamoebae and W. chondrophila, appear to have the genes for a complete methylation cycle (SAM synthesis, methylation activity, and SAH detoxification), the question is raised as to how the majority of Chlamydia species (which all possess at least one SAM-utilizing enzyme), including C. trachomatis, synthesize SAM and detoxify SAH.
Positive selection of C. trachomatis L2 metK-complementing ORF in E. coli.
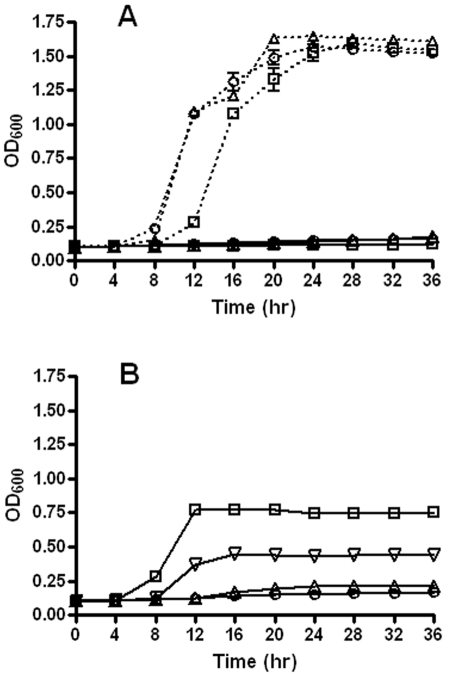
metK is essential in E. coli, and E. coli cells are impermeable to extracellular SAM (14–16). To construct a conditional-lethal ΔmetK mutant of E. coli, the rescuing copy of metK needs to be tightly controlled. Although metK deletion was first obtained in ATM770 (Table 1), this strain was still able to grow in the absence of IPTG (isopropyl-β-d-thiogalactopyranoside), indicating that the lactose promoter controlling metK in pREF71 was still leaky in the absence of inducer (data not shown), as previously encountered with the ara promoter (16). On the other hand, by placement of a copy of the metK gene containing an alternate GUG start codon (to reduce translational efficiency) under the control of the arabinose-inducible, glucose-repressible ara promoter, the growth and/or survival of the ΔmetK ATM778 mutant was made dependent on the presence of arabinose (Fig. 1A). We screened a C. trachomatis serovar L2 genomic DNA library in ATM778 in the presence of glucose and identified several colonies on medium supplemented with 1 mM SAM, while no colonies were detected under the same conditions in the absence of extracellular SAM. Two of 12 independent colonies characterized displayed consistent SAM-dependent growth in the presence of glucose (i.e., repression of E. coli metK expression) (Fig. 1B). Rescuing plasmids were isolated and analyzed by restriction mapping and sequence analyses. All library clones revealed the same 2,528-bp insert harboring C. trachomatis L2 CTL843 and 123 bp of upstream sequence in the same orientation as the lactose promoter of pUC, followed by CTL842 with 7 bp of upstream sequence, in reverse orientation. Subcloning of this ORF into pBluescript (i.e., pRAK368, Table 1) confirmed that CTL843 conferred a SAM-dependent growth phenotype to ATM778 in the presence of glucose and IPTG (Fig. 1B), suggesting that CTL843 functions as a transporter for SAM.
TABLE 1 .
Bacterial strains and plasmids
| Strain or plasmid | Genotype/description | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80 Δ(lacZY-argF)U169 deoR recA1 endA1 phoA hsdR17 supE44 λ− thi-1 gyrA96 relA1 Δ(lacZ)M15 |
54 |
| MG1655 | F− λ− ilvG rfb-50 rph-1 | E. coli Genetic Stock Collection |
| MC4100 | F−
araD139 Δ(argF-lac)U169 rpsL150 relA1 deoC1 rbsR fthD5301 fruA25 λ− |
55 |
| BW25113 | Δ(araD-araB)567 ∆lacZ4787 (::rrnB-3) λ−
rph-1 Δ(rhaD- haB)568 hsdR514 |
56 |
| ATM609 | BW25113 transformed with pKD46 | 56 |
| ATM770 | ∆metK::kan/pREF71 | Allelic exchange mutant of ATM609/pREF71 |
| ATM777 | MC4100/pREF73 | Cmr transformant of MC4100 with pREF73 |
| ATM778 | ∆metK::kan/pREF73 | Kmr transductant of ATM777 with P1 grown on ATM770 |
| NC13 | MC4100 ∆pfs(8-226)::kan | 17 |
| ATM1113 | ∆metK::kan/pREF73/pREF77 | Apr transformant of ATM778 with pREF77 |
| ATM1114 | ∆metK::kan/pREF73/pRAK368 | Apr transformant of ATM778 with pRAK368 |
| ATM1115 | MG1655 ∆pfs::kan | Kmr transductant of MG1655 with P1 grown on NC13 |
| ATM915 | MG1655/pRAK368 | Apr transformant of MG1655 with pRAK368 |
| ATM1116 | ∆pfs::kan/pRAK368 | Kmr transductant of ATM915 with P1 grown on NC13 |
| ATM1117 | ∆metK::kan/pREF73/pUC18 | Apr transformant of ATM778 with pUC |
| Plasmids | ||
| pKD46 | Para gam bet exo oriR101 repA101(Ts); Apr | 56 |
| pAM238 | IPTG-inducible expression vector; Spcr; pSC101 derivative; low copy number |
57 |
| pREF71 | pAM238::AUG-metKEc | This work |
| pBAD33 | Arabinose-inducible expression vector; Cmr; p15A derivative; low copy number |
58 |
| pREF73 | pBAD33::GUG-metKEc | This work |
| pCtL2 | 10-fold coverage of C. trachomatis serovar L2 genome, 2.2-kb average insert size; Apr; pUC19 derivative, 500–700 copies per cell |
3 |
| pREF77 | pUC19::ctl843Ct; isolate 9 | This work |
| pGEMT | PCR cloning vector; Apr; high copy number | Promega |
| pRAK367 | pGEMT::lacIq | This work |
| pBluescript II SK(+) | IPTG-inducible expression vector; Apr; pUC derivative; 300–500 copies per cell |
Stratagene |
| pRAK368 | pBluescript II SK(+)::samhtCt lacIq | This work |
FIG 1 .
Growth of E. coli ∆metK mutant in the presence or absence of extracellular SAM. ATM778 (∆metK) transformed with the indicated plasmids was grown at 37°C in LB supplemented with Ap, Cm, 0.2% glucose (solid lines), or 0.2% arabinose (dotted lines), in the absence (A) or presence (B) of 1 mM SAM. Absorbance (OD600) was measured in a Bioscreen growth curve analyzer and plotted against time (hours). Error bars represent the standard deviations from four replicates. Symbols: ○, pUC-empty vector; □, pREF77-ctl843; ▵, pRAK368-ctl843; ▿, pRAK368-ctl843 with IPTG.
Kinetic analysis of SAM uptake in whole cells expressing recombinant CTL843.
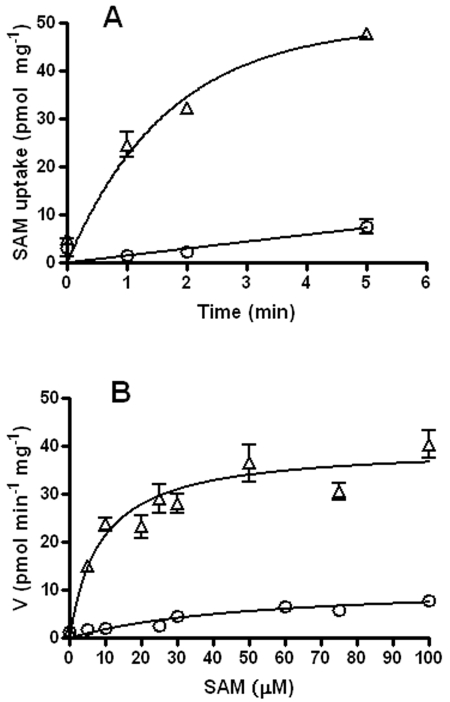
To investigate CTL843 function, uptake of [14C]SAM into intact cells of E. coli was analyzed by a rapid filtration assay. Control cells showed a slow, low linear uptake of SAM (Fig. 2A). In contrast, expression of CTL843 in ATM915 showed a marked increase in the amount of intracellular label over time in a process that was linear during the first minute and then reached saturation indicative of carrier-mediated transport (Fig. 2A). Cell suspensions were also incubated with 5 to 100 µM [14C]SAM, and uptake was measured after 30 s to estimate the kinetic constants of recombinant CTL843 (Fig. 2B). From these data, the calculated apparent Km values (reflecting SAM affinity for CTL843) and Vmax values (reflecting CTL843 activity) were 5.88 ± 0.11 µM and 31.57 ± 0.09 pmol min−1 mg−1, respectively. To assess the role of the proton gradient in CTL843 function, ATM915 was pretreated for 5 minutes with 20 µM carbonyl cyanide m-chlorophenylhydrazone (CCCP), which acts as a channel through the inner membrane to dissipate the H+ gradient. CCCP treatment reduced [14C]SAM intracellular uptake by 50%.
FIG 2 .
Kinetics of SAM transport in E. coli. (A) Time course of [14C]SAM uptake in E. coli at 37°C as a function of time. The time course assay was carried out in transport buffer supplemented with 10 µM [14C]SAM. Time point assays were done in triplicate. (B) Effect of substrate concentration on [14C]SAM uptake at 37°C. The data represent six independent experiments done in triplicate. The background transport seen with MG1655 (○) was subtracted from the transport seen with ATM915 (MG1655 expressing ctl843 [Δ]) with GraphPad Prism software, and then nonlinear regression analysis was performed to obtain the Km and Vmax values.
SAH is a possible substrate for recombinant CTL843.
The specificity of CTL843 for SAM was determined by measuring the effect of structurally related derivatives on SAM transport. Competition studies were performed in the presence of a 10-fold excess of unlabeled putative competitive inhibitors (Table 2). As expected, addition of excess cold SAM competed for binding and uptake of [14C]SAM in induced ATM915 cells. While we observed a slight increase in SAM uptake in the presence of methionine or homocysteine, only SAH, the molecule formed during the course of SAM-dependent methylation reactions, strongly inhibited SAM uptake. Other structural analogs (adenosine, MTA [formed from SAM during spermidine synthesis], and sinefungin [a synthetic analog of SAH]) had negligible effects on the ability of CTL843 to transport SAM. The uptake of [14C]SAM was measured in the presence of various concentrations of SAH to estimate the apparent Ki value of SAH for CTL843, which was determined to be 4.12 µM.
TABLE 2 .
Effect of putative inhibitors on SAM uptake by E. coli ATM915 expressing CTL843
| Unlabeled SAM analoguesa | Molecular structure | Change (%)b |
|---|---|---|
| S-Adenosyl-l-methionine (SAM) |
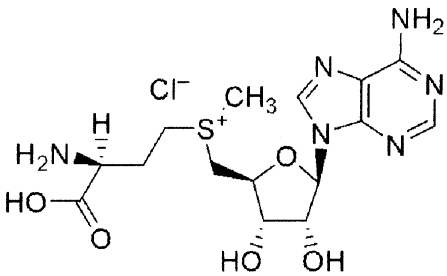
|
−74 ± 8 |
| S-Adenosyl-l-homocysteine (SAH) |
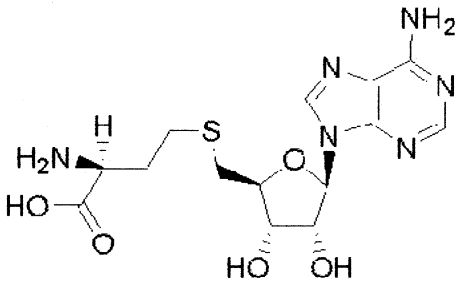
|
−84 ± 10 |
| 5′-Deoxy-5′-(methylthio)adenosine (MTA) |
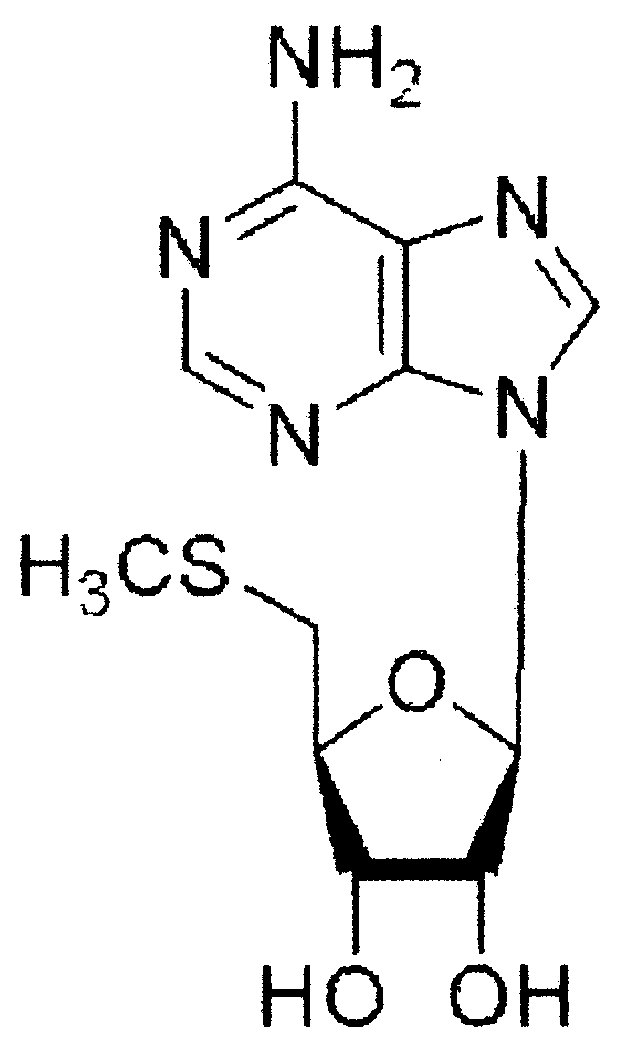
|
15 ± 5 |
| Adenosylornithine (sinefungin) |
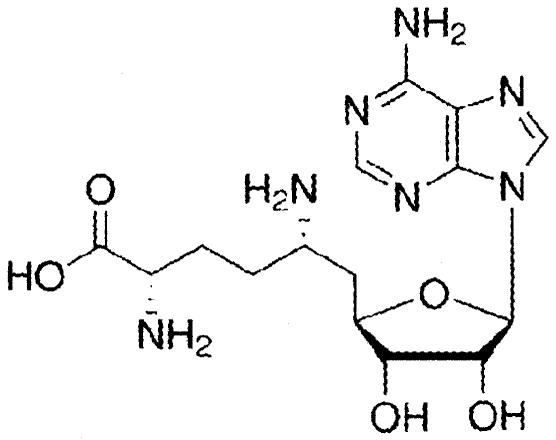
|
−10 ± 13 |
| l-Adenosine |
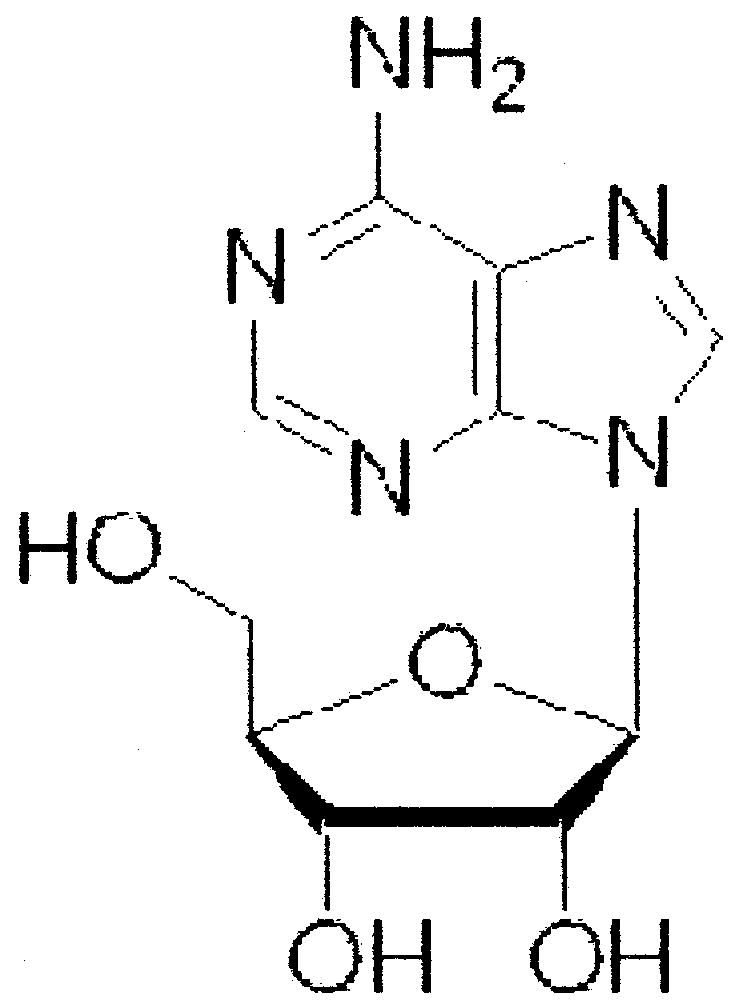
|
6 ± 8 |
| l-Homocysteine |
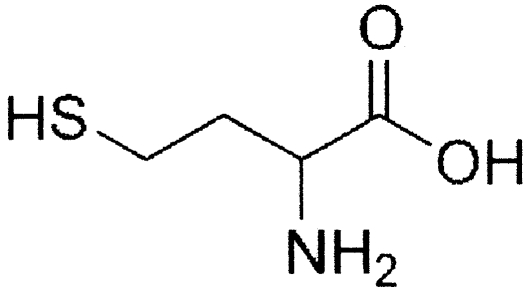
|
23 ± 5 |
| l-Methionine |
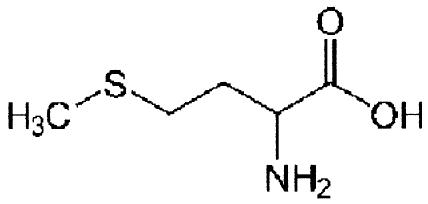
|
32 ± 9 |
One hundred micromolar of the unlabeled compound.
Change in radioactivity observed in presence of unlabeled compound compared to control with only 10 µM [14C] SAM.
In the absence of a commercially available source of radioactive SAH to follow the direct transport of SAH by CTL843 (see the supplemental material), we utilized a genetic assay instead. Elevated levels of SAH are toxic for cells, and loss of pfs (which encodes the MTA/SAH nucleosidase responsible for detoxification of both MTA and SAH in E. coli) leads to diminished bacterial growth (7, 17). Although this Δpfs phenotype was relatively unstable, based on the high-frequency (i.e., ~105), spontaneous appearance of mutants with fitness comparable to that of the wild-type strain, first-passage colonies formed when the Δpfs::kan mutation was transduced into MG1655 (see Text S1 in the supplemental material) were always about 60% larger in diameter in the strain expressing CTL843 (ATM988, Table 1), indicating that the chlamydial transporter offered an immediate fitness advantage to the pfs mutant (Fig. S4). The partial complementation of the growth phenotype in ATM988 likely reflects the ability of CTL843 to recognize and export SAH out of the cells and the inability of CTL843 to recognize MTA (Table 2) and is consistent with the data indicating that SAH is a competitive inhibitor of SAM transport.
Recombinant CTL843 mediates specific counterexchange of SAM with SAM and SAH.
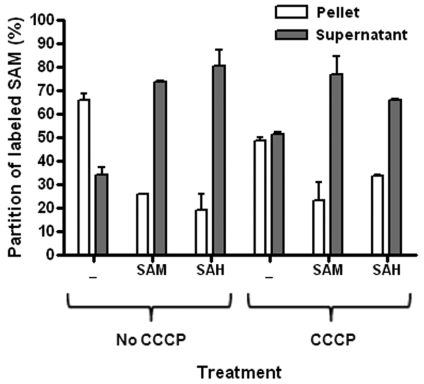
In order to further characterize CTL843 as an SAM/SAH antiporter, back-exchange studies were performed. Cultures of ATM915 expressing CTL843 were preloaded with labeled SAM, washed to remove external radioactivity, and resuspended in M9 minimal salts supplemented with putative counterexchange substrates at a 10-fold excess over labeled SAM and incubated at 37°C. After 10 min, the cells were centrifuged and radioactivity was counted in the cells and in the supernatant. Quantification of exported radioactivity allows differentiation between counterexchange and unidirectional transport. E. coli expressing CTL843 and preloaded with [14C]SAM released significant amounts of internal label (~80% of the initial amount) after resuspension in buffer medium supplemented with unlabeled SAM (thus, against the SAM concentration gradient) or SAH. CCCP treatment did not affect the counterexchange of labeled SAM with SAM or SAH (Fig. 3), indicating that exchange of internal SAM with external SAM or SAH in excess was energy independent. On the other hand, maintenance of the cellular SAM gradient observed in the absence of “competitors” required an intact proton motive force, as SAM concentration reached equilibrium across the membranes in the presence of CCCP (Fig. 3). Thus, CTL843 is active in both directions and can function as an active symporter or energy-independent antiporter (SAM uptake coupled to labeled SAM exit). The similitude between SAM and SAH in the ability to promote the efflux of labeled SAM from the cells strongly suggests that SAH is also being transported into the cells in exchange for SAM. Consequently, we propose that CTL843 be renamed C. trachomatis L2 SAMHT for SAM/SAH transporter.
FIG 3 .
Efflux of [14C]SAM from E. coli cells expressing CTL843. Preloading of labeled SAM (10 µM) in ATM915 was performed initially for 1 min at 37°C and then stopped by dilution and washes at 4°C. Samples were resuspended in duplicate in M9 minimal salts with or without 100 µM cold SAM or cold SAH and incubated at 37°C in the presence or absence of 20 µM CCCP. After 10 min, the partition of labeled SAM was determined by counting radioactivity in the supernatants and in the pellets after centrifugation at 13,000 rpm for 90 s.
Comparative analysis of SAMHT.
An ORF similar to CTL843 was found in all genome sequences available for the order Chlamydiales, with amino acid identity of >90% for Chlamydia homologs, 60 to 67% for Chlamydophila homologs, and 34 to 38% for the Chlamydia-like homologs. All CTL843 homologs contain a duplication of the evolutionarily conserved domain EamA, also known as DUF6 (PF00892; http://pfam.sanger.ac.uk/family/PF00892). While EamA is a signature for transporters belonging to the drug and metabolite transporter (DMT) superfamily, the presence of two EamA domains further subclassifies these chlamydial ORFs in the drug/metabolite exporter (DME) family exhibiting 10 alpha-helical transmembrane spanners (TMSs) (http://www.tcdb.org). Interestingly, this family of transporters also contains the SAM transporters belonging to Rickettsieae, another group of obligate intracellular bacteria. However, Rickettsia prowazekii RP076 (15) shares only about 20% identity with C. trachomatis L2 CTL843 (Fig. 4). Additional phylogenetic analysis failed to support any evolutionary relationship between the rickettsial and the chlamydial SAM transporters (Fig. S5).
FIG 4 .
Sequence alignments of C. trachomatis L2 CTL843 and R. prowazekii RP076 SAM transporter. Amino acid sequences were first aligned on a web interface of MAFFT (53) and were subsequently aligned via Clustal W2 to illustrate the similarity. Identical residues are denoted with an asterisk and are highlighted in black boxes. Strongly conserved residues and weakly conserved residues are marked with a colon and a period, respectively, and are displayed in gray boxes.
DISCUSSION
Bacterial evolution toward obligate intracellular parasitism in a eukaryotic host is thought to be associated with loss of genetic information, especially for genes that become redundant within the host niche (18). These losses are not deleterious for the organism provided that the missing genes, such as those encoding biosynthetic functions, can be compensated for by an increased repertoire of transport functions that allow the organism access to essential nutrients in the intracellular environment. In this work, we used genetic and biochemical strategies to identify a C. trachomatis transporter, CTL843, for the metabolites SAM and likely SAH. We propose that CTL843 and its respective Chlamydia homologs be named SAMHT to clarify their role as a SAM and possible SAH transporter.
Our attempts to genetically complement an E. coli ΔmetK mutant, ATM778, failed to identify a gene in C. trachomatis L2 that encodes SAM synthetase activity. Complementation of ATM778 with C. trachomatis L2 CTL843 ORF was dependent on the presence of extracellular SAM and was related to the expected level of expression in the cells (compared to pRAK368, pREF77 is expected to yield 200 extra copies per cell during replication). CTL843 allows SAM to enter cells, therefore bypassing the E. coli membrane permeability barrier for SAM. A similar effect has been reported previously for the Rickettsia prowazekii SAM transporter (14, 15). However, this transporter shares only ~20% amino acid homology with CTL843. In addition, we showed that C. trachomatis L2 CTL843 expression partially complemented the growth defect of an E. coli Δpfs mutant lacking MTA/SAH nucleosidase activity, suggesting that recombinant CTL843 was able to reduce the internal buildup of toxic SAH in the mutant cells. Whole-cell transport assays further supported CTL843 as a transporter for SAM and SAH: (i) uptake of [14C]SAM into recombinant E. coli cells was saturable, exhibiting kinetic constants indicative of a high-affinity carrier with a Km value below 6 µM; (ii) uptake of [14C]SAM was specifically inhibited by SAH at an equivalently high affinity; and (iii) internal [14C]SAM was expelled out of recombinant cells in the presence of extracellular SAM or SAH following a countertransport mechanism. Although we were unable to monitor direct transport of SAH in E. coli cells expressing CTL843 due to the lack of commercially available labeled SAH and to the limit of detection of SAH by high-performance liquid chromatography (HPLC) (see Text S1 in the supplemental material), we believe that our genetic and biochemical data strongly support the idea that both SAH and SAM are substrates for C. trachomatis L2 CTL843.
Unlike rickettsiae, chlamydiae multiply in the cytoplasm of the host cell inside a specialized compartment termed an inclusion. Because the inclusion membrane is permeable to molecules smaller than 520 Da (19), SAH (384.4 Da) and SAM (399.4 Da) are expected to diffuse freely between the host cytoplasm and the bacteria expressing SAMHT. The pI value of SAM is 7.24, and SAH is neutral between pI values of 3.5 and 9.2 (20). Consequently, both molecules are neutral in the mammalian cell cytoplasm and in the chlamydial inclusion, where the pH values are calculated to be 7.29 ± 0.07 and 7.25 ± 0.19, respectively (21). Although the glucose present in the transport assay was probably partially compensating for the deleterious effect of CCCP on membrane potential (22), we still observed inhibition of SAM import in E. coli expressing SAMHT in the presence of CCCP. This shows that C. trachomatis L2 SAMHT acts as a secondary active symporter for SAM, similarly to the rickettsial SAM transporter (15). Conversely, exchange of internal SAM with external SAM or SAH is not affected by CCCP and is therefore energy independent (Fig. 3). This mechanism of antiport transport driven by the inward substrate (SAM) gradient and the outward product (SAH) gradient is most favorable for the chlamydiae, which are energy parasites (23). A similar transport mechanism has been described for the E. coli putrescine-ornithine antiporter PotE, which can excrete putrescine as the result of the antiport activity between putrescine and ornithine, in an energy-independent manner, and can also catalyze putrescine uptake in a process that is energy dependent without excretion of ornithine (24).
Although SAM transporters have been found so far in four evolutionarily diverse transporter families, identification of SAM carriers using sequence similarity is of limited value because their degree of homology inside a family is generally not much higher than that between the different members of the same family. In Saccharomyces cerevisiae (25), humans (26), and plants (27, 28), members of the well-characterized mitochondrial carrier protein (MCP) family (29) transport external SAM in counterexchange with SAM or SAH with less affinity. In Saccharomyces cerevisiae (30) and Leishmania (31), transporters belonging to the amino acid permease superfamily or to the folate biopterin transporter family, respectively, seem more specific to SAM, with affinities in the nanomolar ranges. Like the rickettsial SAM transporter (15), the chlamydial SAMHT homologs to CTL843 belong to the drug/metabolite exporter (DME) family (32) and appear to transport both SAM and SAH with affinities in the micromolar range. Note that we hypothesize that SAH is also a substrate for the rickettsial SAM transporters, based on the inhibition of SAM uptake observed in the presence of SAH (15) and the apparent lack of SAH hydrolase homologs in their genomes (data not shown). Although the DME family was recognized 10 years ago and now has >500 sequenced members in bacteria and archaea (33), only three additional transporters from E. coli have been functionally characterized to date (34–36), and the molecular mechanisms driving the activity of DME transporters are not known. The growth dependence of the E. coli ΔmetK mutant on the activity of SAMHT in the presence of SAM might be the ideal platform to screen for antimicrobial compounds targeting this transport system. In addition to improving our knowledge on the mode of action of DME transporters, SAMHT inhibitors may well lead to the development of a new antichlamydial specific therapy.
The 40 chlamydial species, identified mostly by 16S rRNA sequencing, are classified into seven families (37, 38), among which the Chlamydiaceae (including C. trachomatis) are the most studied due to their importance in human and veterinary medicine. Nevertheless, “Chlamydia-like” organisms have been receiving more attention lately, in particular those residing in free-living amoebae, since an intraprotozoal lifestyle has likely contributed significantly to the adaptation of intracellular bacterial pathogens to higher eukaryotes (39–42). Escobar-Páramo et al. (43) suggested that “ancient” genes will not persist if they do not carry a “lasting adaptive value” to populations. The presence of a CTL843 SAMHT homolog in the nine Chlamydiaceae representatives and the three “Chlamydia-like” organisms that have been sequenced suggests that SAMHT confers adaptive functions that allow the exploration of new niches such as mammalian cells. Since an obligate intracellular lifestyle limits the chance of gene acquisition by horizontal gene transfer, SAMHT may have initially been acquired by an ancestral, facultative intracellular form of Chlamydia. Subsequently, the constant supply of metabolites (i.e., SAM) from the host relaxed the selective pressure to maintain the metabolic pathways involved in SAM synthesis (MAT) and SAH degradation (MTAN or SahH) and resulted in the complete loss or pseudogenization of these genes in all Chlamydiaceae and in P. amoebophila UWE25, respectively.
The isolated asexual reproductive cycle of Chlamydia and the evolutionary bottleneck observed at each transmission or passage to a new host are expected to favor the process of genome degradation (44, 45). Moreover, considering that MAT requires ATP and that chlamydiae parasitize their host for ATP (46), loss of MAT is expected to make the microbe more fit in its interaction with the host and MAT could therefore be under strong pathoadaptive pressure (47). Consequently, it is somewhat surprising that the two emerging pathogens (18, 48) W. chondrophila and P. acanthamoebae have maintained apparently functional MAT and SahH enzymes. In the absence of a genetic system to test the contribution of the three genes to bacterial fitness, we do not know if this reflects a greater need for metabolic versatility, a more effective selection for the maintenance of weakly beneficial genes, or simply a less advanced stage in the process of the reductive evolution of their genomes (49). The answers to these questions will add to a better understanding of the adaptive evolution of Chlamydia towards pathogenicity.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli strains and plasmids used in this study are listed in Table 1, and their construction is described in the supplemental material. E. coli strain DH5α was used for cloning. Strains were grown in Luria-Bertani (LB) broth with aeration or on LB agar, unless indicated otherwise. Medium was supplemented with ampicillin (Ap; 50 µg/ml), kanamycin (Km; 50 µg/ml), chloramphenicol (Cm; 10 µg/ml), spectinomycin (Spc; 100 µg/ml), arabinose (0.2%), glucose (0.5%), isopropyl-β-d-1-thiogalactopyranoside (IPTG; 1 mM), and mannitol (0.5%) as needed.
Whole-cell transport assays. (i) SAM import.
Whole-cell transport assays were performed as described in references 14 and 50 with slight modifications. Overnight LB cultures of MG1655 or ATM915 (Table 1) were subcultured 1:100 into 20 ml LB supplemented with IPTG to induce expression of CTL843 (plus appropriate selective agents as needed) and grown at 37°C with shaking to mid-log phase (optical density at 600 nm [OD600] of ~0.7). Cultures were standardized to an OD600 of 0.5 and concentrated to 2 × 109 bacteria/ml. Cells were centrifuged and washed with M9 minimal salts (51). Washed cells were suspended in cold M9 minimal salts plus 0.5% glucose (i.e., transport buffer), yielding approximately 0.6 to 0.8 mg/ml total protein and about 1 × 108 bacteria per 50-μl reaction mixture. A 10 μM concentration of S-adenosyl-l-[methyl-14C]methionine ([14C]SAM) (GE Healthcare UK Limited) was added to the bacterial suspension for a final volume of 60 µl per reaction mixture, and uptake was initiated at 37°C and followed up to 5 min. Samples were periodically removed, transferred to 20 ml ice-cold M9 minimal salts to stop transport, filtered through a 0.45-µm Durapore membrane filter (Millipore Corp.), and washed once with the same buffer. Filters were placed in scintillation vials, and radioactivity was measured using 5 ml of ReadySafe liquid scintillation cocktail (Beckman Coulter). Disintegration-per-minute values determined by sample counting were normalized to the activity of [14C]SAM (55 mCi/mmol) and to the protein concentration of the sample to express counts as pmol SAM ⋅ minute−1 ⋅ mg total protein−1.
Kinetic analysis was accomplished by incubating cell suspensions with increasing concentrations of [14C]SAM for 30 s. Data were plotted in Prism 4 (GraphPad Software), and apparent Km and Vmax values were determined using the included Michaelis-Menten linear regression template. To examine the dependence of SAM transport on membrane potential, samples were pretreated with 20 µM carbonyl cyanide m-chlorophenylhydrazone (CCCP) for 5 minutes prior to the start of the transport assay. All uptake data are the results of experiments performed in triplicate unless otherwise indicated.
(ii) SAMHTCt specificity.
The substrate specificity of recombinant CTL843 was assessed by measuring the capacity of nonradioactive effectors structurally related to SAM to inhibit the uptake of 10 µM [14C]SAM by E. coli strain ATM915. Values obtained from 5 min of preincubation with 100 µM effectors were expressed as percent inhibition relative to the absence of inhibitor (15, 50). Additionally, the dissociation constant for the binding of SAH to CTL843 (i.e., Ki value) was determined by two methods using Prism 4 software. First, the kinetics of SAM uptake were determined in the presence of 5 and 20 µM cold SAH. Second, the Ki value of SAH was calculated from the half-maximal inhibitory concentration (IC50) estimated by iterative curve fitting for sigmoidal equations describing SAM uptake velocity in the presence of 0 to 100 µM SAH, using the equation of Cheng and Prusoff (52).
(iii) Reversibility of SAM transport.
To determine the capacity of recombinant CTL843 to transport SAM in two directions, induced cells were incubated with 10 µM [14C]SAM for 1 min at 37°C to allow for uptake of labeled SAM and then washed twice in 25 ml cold M9 minimal salts and resuspended in transport buffer to a density of 1 × 109 bacteria/100 µl of bacterial suspension. Nonradioactive SAM or SAH was added to 100 µl of bacteria to a final concentration of 100 µM and incubated at 37°C. Reactions were stopped after 10 min by centrifugation at 13,000 rpm for 90 s, and radioactivity (disintegration-per-minute values) was measured in the supernatants and the pellets. To examine the dependence of SAM exchange on membrane potential, samples were pretreated with 20 µM CCCP for 5 minutes prior to addition of the cold competitive inhibitors. The two independent experiments performed showed the same trend.
SUPPLEMENTAL MATERIAL
Supplemental information. Download Text S1, DOC file, 0.043 MB.
Structure-based sequence alignment of P. acanthamoebae strain Hall’s coccus and W. chondrophila predicted methionine adenosyl transferase (MAT) amino acid sequences (i.e. Pah-co14o152 and wcw-0127, respectively) with those of other organisms. The alignment was generated using the Tcoffee expresso Web server using Arabidopsis thaliana MAT1 sequence (P23686) and the tertiary sequence of rat MAT1 (PDB ID 1O9T), E. coli MAT (PDB ID 1XRC) and Burkholderia pseudomallei MAT (PDB ID 3IML). Identical residues are in black boxes, conserved residues are displayed in gray boxes. The seven conserved motifs described by Sanchez-Perez et al. (J. Mol. Biol. 335:693-706, 2004) are boxed. Download Figure S1, PPT file, 0.105 MB.
Structure-based sequence alignment of C. caviae, C. felis, and C. pneumoniae predicted MTA/SAH nucleosidase (MTAN) amino acid sequences (i.e. CCA 593, CF410, and Cpn232, respectively) with those of other organisms. The alignment was generated using the Tcoffee expresso Web server using the MTAN structure of Staphylococcus aureus (PDB ID 3BL6), Streptococcus pneumoniae (PDB ID 3MMS) and E. coli (PDB ID 3O4V). Residues that are strictly conserved and conservatively substituted in the alignment of 31 pathogenic bacterial species (Siu KK, et al., Acta Crystallogr. F Struct. Biol. Cryst. Commun. 64:343-350, 2008) are displayed as bold white characters in a black box and black character in a gray box, respectively. Residues in the active site are marked with a bold asterisk on top of the alignment. Download Figure S2, DOC file, 0.083 MB.
Structure-based sequence alignment of P. acanthamoebae strain Hall’s coccus and W. chondrophila predicted S-adenosylhomocysteine hydrolases (SAHH) amino acid sequences (i.e. pah_c022o240 and wcw_1005, respectively) with those of other organisms. The alignment was generated using the Tcoffee expresso Web server with the SAHH amino acid sequence from Arabidopsis thaliana (gb∣CAB10173.1∣), Trypanosoma cruzi (gb∣AAP45630.1∣), and Caenorhabditis elegans (gb∣AAB25906.1∣), and the SAHH structures of Mycobacterium tuberculosis (PDB ID 3CE6), Homo sapiens [human] (PDB ID 3MTG), Brucella melitensis (PDB ID 3N58), Rattus norvegicus [rat] (PDB ID 1B3R), Plasmodium falciparum (PDB ID 1V8B), Leishmania major (PDB ID 3G1U) and Burkholderia pseudomallei (PDB ID 3G1U). Residues that are strictly conserved and conservatively substituted are displayed in black boxes and gray boxes, respectively. The catalytic and cofactor (NAD)-binding domains are separated by two helical segments composed of helices αA1-αA2 and αX-αY, respectively (Stepkowski T, et al., Mol. Phylogenet. Evol. 34:15-28, 2005). Residues involved in enzyme catalysis, substrate binding, or binding of cofactor NAD+ are marked with a bold asterisk at the bottom of the alignment. NBS indicates the GXGXXG dinucleotide binding motif. Vertical arrows numbered 1, 2, and 4 relate to lineage-specific indels (Stepkowski T, et al., Mol. Phylogenet. Evol. 34:15-28, 2005). Download Figure S3, PPT file, 0.125 MB.
Effect of CTL843 expression on growth of a wild-type and a pfs mutant strain of E. coli. Strains were grown to exponential phase in LB, washed twice with PBS, and plated on M9 minimal medium with methionine (10 μg/ml). Plates were photographed after 42 hours of incubation at 37°C. The sizes of the small and large colonies in panels B and D were measured to determine the size range for these strains. pRAK368 = ctl843 (SAM/SAH transporter) cloned into pBluescriptII. Download Figure S4, PPT file, 0.310 MB.
Phylogenetic relationships of SAMHTs homologs. The phylogenetic tree is based on the translated nucleotide sequence of the coding regions of the following Chlamydiae and Rickettsiae representatives: C. trachomatis L2 434/Bu [gi∣166153973:971272-972255]; C. muridarum Nigg [gi∣29337300:1005855-1006811]; C. abortus S26/3 [gi∣62184647:1079746-1080648]; C. caviae GPIC [gi∣29839769:1108533-1109438]; C. felis Fe/C-56 [gi∣89897807:63828-64736]; C. pneumoniae AR39 [gi∣58021288:1156561-1157478]; “Candidatus Protochlamydia amoebophila” UWE25 [gi∣46445634:2216282-2217214]; Parachlamydia sp. Halls coccus; Waddlia chondrophila WSU 86-1044 [gi∣297620246:136536-137450]; R. prowazekii strain Madrid E [gi∣15603881:85783-86667]; R. typhi strain Wilmington [gi∣51473215:74987-75871]; R. massiliae MTU5 [gi∣157964072:105400-106290]; R. felis URRWXCal2 [gi∣67458392:62487-63371]; R. conorii str. Malish 7 [gi∣15891923:100945-101829]; R. akari strain Hartford [gi∣157825125:97178-98062]; R. bellii RML369-C [gi∣91204815:19663-20538]; Wolbachia endosymbiont of Drosophila melanogaster [gi∣42519920:605993-606907]; Orientia tsutsugamushi strain Ikeda [gi∣189182907:1812167-1813039], and “Candidatus Amoebophilus asiaticus” 5a2 [gi∣189501470:1534992-1535849]. Translated amino acid sequences were aligned using MAFFT (Katoh K, et al., Nucleic Acids Res. 30:3059-3066, 2002). Subsequently, the evolutionary history was inferred using the neighbor-joining method (Saitou N, Nei M, Mol. Biol. Evol. 4:406-425, 1987). The bootstrap consensus tree inferred from 1,000 replicates (Felsenstein J, Annu. Rev. Genet. 22:521-565, 1988) is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches (Felsenstein J, Annu. Rev. Genet. 22:521-565, 1988). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method (Tamura K, et al., Proc. Natl. Acad. Sci. U. S. A. 101:11030-11035, 2004) and are in the units of the number of base substitutions per site. Codon positions included were 1st+2nd. All positions containing gaps and missing data were eliminated from the data set (Complete deletion option). There were a total of 434 positions in the final data set. Phylogenetic analyses were conducted in MEGA4 (Tamura K, et al., Mol. Biol. Evol. 24:1596-1599, 2007). Download Figure S5, PPT file, 0.056 MB.
ACKNOWLEDGMENTS
This work was supported by grants AI44033 and F32 AI078655-01 (D.J.F.) from the National Institute of Allergy and Infectious Diseases, NIH.
Footnotes
Citation Binet R, Fernandez RE, Fisher DJ, Maurelli AT. 2011. Identification and characterization of the Chlamydia trachomatis L2 S-adenosylmethionine transporter. mBio 2(3):e00051-11. doi:10.1128/mBio.00051-11.
REFERENCES
- 1. Corsaro D, Greub G. 2006. Pathogenic potential of novel Chlamydiae and diagnostic approaches to infections due to these obligate intracellular bacteria. Clin. Microbiol. Rev. 19:283–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stephens RS, et al. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759 [DOI] [PubMed] [Google Scholar]
- 3. McCoy AJ, et al. 2006. L-diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proc. Natl. Acad. Sci. U. S. A. 103:17909–17914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andersson JO, Andersson SG. 1999. Insights into the evolutionary process of genome degradation. Curr. Opin. Genet. Dev. 9:664–671 [DOI] [PubMed] [Google Scholar]
- 5. Haferkamp I, et al. 2004. A candidate NAD+ transporter in an intracellular bacterial symbiont related to Chlamydiae. Nature 432:622–625 [DOI] [PubMed] [Google Scholar]
- 6. Grillo MA, Colombatto S. 2008. S-adenosylmethionine and its products. Amino Acids 34:187–193 [DOI] [PubMed] [Google Scholar]
- 7. Parveen N, Cornell KA. 2011. Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism. Mol. Microbiol. 79:7–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stepkowski T, Brzeziński K, Legocki AB, Jaskólski M, Béna G. 2005. Bayesian phylogenetic analysis reveals two-domain topology of S-adenosylhomocysteine hydrolase protein sequences. Mol. Phylogenet. Evol. 34:15–28 [DOI] [PubMed] [Google Scholar]
- 9. Binet R, Maurelli AT. 2009. Transformation and isolation of allelic exchange mutants of Chlamydia psittaci using recombinant DNA introduced by electroporation. Proc. Natl. Acad. Sci. U. S. A. 106:292–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Binet R, Maurelli AT. 2009. The chlamydial functional homolog of KsgA confers kasugamycin sensitivity to Chlamydia trachomatis and impacts bacterial fitness. BMC Microbiol. 9:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pannekoek Y, et al. 2005. The N5-glutamine S-adenosyl-L-methionine-dependent methyltransferase PrmC/HemK in Chlamydia trachomatis methylates class 1 release factors. J. Bacteriol. 187:507–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sánchez-Pérez GF, Bautista JM, Pajares MA. 2004. Methionine adenosyltransferase as a useful molecular systematics tool revealed by phylogenetic and structural analyses. J. Mol. Biol. 335:693–706 [DOI] [PubMed] [Google Scholar]
- 13. Markham GD, Pajares MA. 2009. Structure-function relationships in methionine adenosyltransferases. Cell. Mol. Life Sci. 66:636–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Driskell LO, Tucker AM, Winkler HH, Wood DO. 2005. Rickettsial metK-encoded methionine adenosyltransferase expression in an Escherichia coli metK deletion strain. J. Bacteriol. 187:5719–5722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tucker AM, Winkler HH, Driskell LO, Wood DO. 2003. S-adenosylmethionine transport in Rickettsia prowazekii. J. Bacteriol. 185:3031–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wei Y, Newman EB. 2002. Studies on the role of the metK gene product of Escherichia coli K-12. Mol. Microbiol. 43:1651–1656 [DOI] [PubMed] [Google Scholar]
- 17. Cadieux N, et al. 2002. Identification of the periplasmic cobalamin-binding protein BtuF of Escherichia coli. J. Bacteriol. 184:706–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baud D, Thomas V, Arafa A, Regan L, Greub G. 2007. Waddlia chondrophila, a potential agent of human fetal death. Emerg. Infect. Dis. 13:1239–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heinzen RA, Hackstadt T. 1997. The Chlamydia trachomatis parasitophorous vacuolar membrane is not passively permeable to low-molecular-weight compounds. Infect. Immun. 65:1088–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Farooqui J, Kim S, Paik WK. 1983. Measurement of isoelectric point of S-adenosyl-L-methionine and its metabolic products by an isoelectric-focusing technique. Electrophoresis 4:261–265 [Google Scholar]
- 21. Grieshaber S, Swanson JA, Hackstadt T. 2002. Determination of the physical environment within the Chlamydia trachomatis inclusion using ion-selective ratiometric probes. Cell. Microbiol. 4:273–283 [DOI] [PubMed] [Google Scholar]
- 22. Zeller V, Janoir C, Kitzis MD, Gutmann L, Moreau NJ. 1997. Active efflux as a mechanism of resistance to ciprofloxacin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:1973–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tjaden J, et al. 1999. Two nucleotide transport proteins in Chlamydia trachomatis, one for net nucleoside triphosphate uptake and the other for transport of energy. J. Bacteriol. 181:1196–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kashiwagi K, Miyamoto S, Suzuki F, Kobayashi H, Igarashi K. 1992. Excretion of putrescine by the putrescine-ornithine antiporter encoded by the potE gene of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 89:4529–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marobbio CM, Agrimi G, Lasorsa FM, Palmieri F. 2003. Identification and functional reconstitution of yeast mitochondrial carrier for S-adenosylmethionine. EMBO J. 22:5975–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Agrimi G, et al. 2004. Identification of the human mitochondrial S-adenosylmethionine transporter: bacterial expression, reconstitution, functional characterization and tissue distribution. Biochem. J. 379:183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bouvier F, et al. 2006. Arabidopsis SAMT1 defines a plastid transporter regulating plastid biogenesis and plant development. Plant Cell 18:3088–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Palmieri L, et al. 2006. Molecular identification of an Arabidopsis S-adenosylmethionine transporter. Analysis of organ distribution, bacterial expression, reconstitution into liposomes, and functional characterization. Plant Physiol. 142:855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haferkamp I. 2007. The diverse members of the mitochondrial carrier family in plants. FEBS Lett. 581:2375–2379 [DOI] [PubMed] [Google Scholar]
- 30. Rouillon A, Surdin-Kerjan Y, Thomas D. 1999. Transport of sulfonium compounds. Characterization of the s-adenosylmethionine and s-methylmethionine permeases from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274:28096–28105 [DOI] [PubMed] [Google Scholar]
- 31. Dridi L, Ahmed OA, Ouellette M. 2010. High affinity S-adenosylmethionine plasma membrane transporter of Leishmania is a member of the folate biopterin transporter (FBT) family. J. Biol. Chem. 285:19767–19775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jack DL, Yang NM, Saier MH., Jr. 2001. The drug/metabolite transporter superfamily. Eur. J. Biochem. 268:3620–3639 [DOI] [PubMed] [Google Scholar]
- 33. Yen MR, Chen JS, Marquez JL, Sun EI, Saier MH. 2010. Multidrug resistance: phylogenetic characterization of superfamilies of secondary carriers that include drug exporters. Methods Mol. Biol. 637:47–64 [DOI] [PubMed] [Google Scholar]
- 34. Doroshenko V, et al. 2007. YddG from Escherichia coli promotes export of aromatic amino acids. FEMS Microbiol. Lett. 275:312–318 [DOI] [PubMed] [Google Scholar]
- 35. Livshits VA, Zakataeva NP, Aleshin VV, Vitushkina MV. 2003. Identification and characterization of the new gene rhtA involved in threonine and homoserine efflux in Escherichia coli. Res. Microbiol. 154:123–135 [DOI] [PubMed] [Google Scholar]
- 36. Ohtsu I, et al. 2010. The L-cysteine/L-cystine shuttle system provides reducing equivalents to the periplasm in Escherichia coli. J. Biol. Chem. 285:17479–17487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Corsaro D, Michel R, Walochnik J, Müller KD, Greub G. 2010. Saccamoeba lacustris, sp. nov. (Amoebozoa: Lobosea: Hartmannellidae), a new lobose amoeba, parasitized by the novel chlamydia “Candidatus Metachlamydia lacustris” (Chlamydiae: Parachlamydiaceae). Eur. J. Protistol. 46:86–95 [DOI] [PubMed] [Google Scholar]
- 38. Horn M. 2008. Chlamydiae as symbionts in eukaryotes. Annu. Rev. Microbiol. 62:113–131 [DOI] [PubMed] [Google Scholar]
- 39. Greub G, Raoult D. 2004. Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17:413–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moliner C, Fournier PE, Raoult D. 2010. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol. Rev. 34:281–294 [DOI] [PubMed] [Google Scholar]
- 41. Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. 2005. Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71:20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Toft C, Andersson SG. 2010. Evolutionary microbial genomics: insights into bacterial host adaptation. Nat. Rev. Genet. 11:465–475 [DOI] [PubMed] [Google Scholar]
- 43. Escobar-Páramo P, Faivre N, Buckling A, Gougat-Barbera C, Hochberg ME. 2009. Persistence of costly novel genes in the absence of positive selection. J. Evol. Biol. 22:536–543 [DOI] [PubMed] [Google Scholar]
- 44. Moran NA. 1996. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. U. S. A. 93:2873–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pettersson ME, Berg OG. 2007. Muller’s ratchet in symbiont populations. Genetica 130:199–211 [DOI] [PubMed] [Google Scholar]
- 46. Schmitz-Esser S, et al. 2004. ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to Chlamydiae and Rickettsiae. J. Bacteriol. 186:683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maurelli AT. 2007. Black holes, antivirulence genes, and gene inactivation in the evolution of bacterial pathogens. FEMS Microbiol. Lett. 267:1–8 [DOI] [PubMed] [Google Scholar]
- 48. Greub G. 2009. Parachlamydia acanthamoebae, an emerging agent of pneumonia. Clin. Microbiol. Infect. 15:18–28 [DOI] [PubMed] [Google Scholar]
- 49. Mira A, Ochman H, Moran NA. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17:589–596 [DOI] [PubMed] [Google Scholar]
- 50. Anfora AT, Welch RA. 2006. DsdX is the second D-serine transporter in uropathogenic Escherichia coli clinical isolate CFT073. J. Bacteriol. 188:6622–6628 188/18/6622;10.1128/JB.00634-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Miller JM. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 52. Cheng Y, Prusoff WH. 1973. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22:3099–3108 [DOI] [PubMed] [Google Scholar]
- 53. Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 55. Casadaban MJ, Cohen SN. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. U. S. A. 76:4530–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Binet R, Wandersman C. 1995. Protein secretion by hybrid bacterial ABC-transporters: specific functions of the membrane ATPase and the membrane fusion protein. EMBO J. 14:2298–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental information. Download Text S1, DOC file, 0.043 MB.
Structure-based sequence alignment of P. acanthamoebae strain Hall’s coccus and W. chondrophila predicted methionine adenosyl transferase (MAT) amino acid sequences (i.e. Pah-co14o152 and wcw-0127, respectively) with those of other organisms. The alignment was generated using the Tcoffee expresso Web server using Arabidopsis thaliana MAT1 sequence (P23686) and the tertiary sequence of rat MAT1 (PDB ID 1O9T), E. coli MAT (PDB ID 1XRC) and Burkholderia pseudomallei MAT (PDB ID 3IML). Identical residues are in black boxes, conserved residues are displayed in gray boxes. The seven conserved motifs described by Sanchez-Perez et al. (J. Mol. Biol. 335:693-706, 2004) are boxed. Download Figure S1, PPT file, 0.105 MB.
Structure-based sequence alignment of C. caviae, C. felis, and C. pneumoniae predicted MTA/SAH nucleosidase (MTAN) amino acid sequences (i.e. CCA 593, CF410, and Cpn232, respectively) with those of other organisms. The alignment was generated using the Tcoffee expresso Web server using the MTAN structure of Staphylococcus aureus (PDB ID 3BL6), Streptococcus pneumoniae (PDB ID 3MMS) and E. coli (PDB ID 3O4V). Residues that are strictly conserved and conservatively substituted in the alignment of 31 pathogenic bacterial species (Siu KK, et al., Acta Crystallogr. F Struct. Biol. Cryst. Commun. 64:343-350, 2008) are displayed as bold white characters in a black box and black character in a gray box, respectively. Residues in the active site are marked with a bold asterisk on top of the alignment. Download Figure S2, DOC file, 0.083 MB.
Structure-based sequence alignment of P. acanthamoebae strain Hall’s coccus and W. chondrophila predicted S-adenosylhomocysteine hydrolases (SAHH) amino acid sequences (i.e. pah_c022o240 and wcw_1005, respectively) with those of other organisms. The alignment was generated using the Tcoffee expresso Web server with the SAHH amino acid sequence from Arabidopsis thaliana (gb∣CAB10173.1∣), Trypanosoma cruzi (gb∣AAP45630.1∣), and Caenorhabditis elegans (gb∣AAB25906.1∣), and the SAHH structures of Mycobacterium tuberculosis (PDB ID 3CE6), Homo sapiens [human] (PDB ID 3MTG), Brucella melitensis (PDB ID 3N58), Rattus norvegicus [rat] (PDB ID 1B3R), Plasmodium falciparum (PDB ID 1V8B), Leishmania major (PDB ID 3G1U) and Burkholderia pseudomallei (PDB ID 3G1U). Residues that are strictly conserved and conservatively substituted are displayed in black boxes and gray boxes, respectively. The catalytic and cofactor (NAD)-binding domains are separated by two helical segments composed of helices αA1-αA2 and αX-αY, respectively (Stepkowski T, et al., Mol. Phylogenet. Evol. 34:15-28, 2005). Residues involved in enzyme catalysis, substrate binding, or binding of cofactor NAD+ are marked with a bold asterisk at the bottom of the alignment. NBS indicates the GXGXXG dinucleotide binding motif. Vertical arrows numbered 1, 2, and 4 relate to lineage-specific indels (Stepkowski T, et al., Mol. Phylogenet. Evol. 34:15-28, 2005). Download Figure S3, PPT file, 0.125 MB.
Effect of CTL843 expression on growth of a wild-type and a pfs mutant strain of E. coli. Strains were grown to exponential phase in LB, washed twice with PBS, and plated on M9 minimal medium with methionine (10 μg/ml). Plates were photographed after 42 hours of incubation at 37°C. The sizes of the small and large colonies in panels B and D were measured to determine the size range for these strains. pRAK368 = ctl843 (SAM/SAH transporter) cloned into pBluescriptII. Download Figure S4, PPT file, 0.310 MB.
Phylogenetic relationships of SAMHTs homologs. The phylogenetic tree is based on the translated nucleotide sequence of the coding regions of the following Chlamydiae and Rickettsiae representatives: C. trachomatis L2 434/Bu [gi∣166153973:971272-972255]; C. muridarum Nigg [gi∣29337300:1005855-1006811]; C. abortus S26/3 [gi∣62184647:1079746-1080648]; C. caviae GPIC [gi∣29839769:1108533-1109438]; C. felis Fe/C-56 [gi∣89897807:63828-64736]; C. pneumoniae AR39 [gi∣58021288:1156561-1157478]; “Candidatus Protochlamydia amoebophila” UWE25 [gi∣46445634:2216282-2217214]; Parachlamydia sp. Halls coccus; Waddlia chondrophila WSU 86-1044 [gi∣297620246:136536-137450]; R. prowazekii strain Madrid E [gi∣15603881:85783-86667]; R. typhi strain Wilmington [gi∣51473215:74987-75871]; R. massiliae MTU5 [gi∣157964072:105400-106290]; R. felis URRWXCal2 [gi∣67458392:62487-63371]; R. conorii str. Malish 7 [gi∣15891923:100945-101829]; R. akari strain Hartford [gi∣157825125:97178-98062]; R. bellii RML369-C [gi∣91204815:19663-20538]; Wolbachia endosymbiont of Drosophila melanogaster [gi∣42519920:605993-606907]; Orientia tsutsugamushi strain Ikeda [gi∣189182907:1812167-1813039], and “Candidatus Amoebophilus asiaticus” 5a2 [gi∣189501470:1534992-1535849]. Translated amino acid sequences were aligned using MAFFT (Katoh K, et al., Nucleic Acids Res. 30:3059-3066, 2002). Subsequently, the evolutionary history was inferred using the neighbor-joining method (Saitou N, Nei M, Mol. Biol. Evol. 4:406-425, 1987). The bootstrap consensus tree inferred from 1,000 replicates (Felsenstein J, Annu. Rev. Genet. 22:521-565, 1988) is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches (Felsenstein J, Annu. Rev. Genet. 22:521-565, 1988). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method (Tamura K, et al., Proc. Natl. Acad. Sci. U. S. A. 101:11030-11035, 2004) and are in the units of the number of base substitutions per site. Codon positions included were 1st+2nd. All positions containing gaps and missing data were eliminated from the data set (Complete deletion option). There were a total of 434 positions in the final data set. Phylogenetic analyses were conducted in MEGA4 (Tamura K, et al., Mol. Biol. Evol. 24:1596-1599, 2007). Download Figure S5, PPT file, 0.056 MB.