ABSTRACT
Natural habitats vary in available nutrients and room for bacteria to grow, but successful colonization can lead to overcrowding and stress. Here we show that competing sibling colonies of Paenibacillus dendritiformis bacteria survive overcrowding by switching between two distinct vegetative phenotypes, motile rods and immotile cocci. Growing colonies of the rod-shaped bacteria produce a toxic protein, Slf, which kills cells of encroaching sibling colonies. However, sublethal concentrations of Slf induce some of the rods to switch to Slf-resistant cocci, which have distinct metabolic and resistance profiles, including resistance to cell wall antibiotics. Unlike dormant spores of P. dendritiformis, the cocci replicate. If cocci encounter conditions that favor rods, they secrete a signaling molecule that induces a switch to rods. Thus, in contrast to persister cells, P. dendritiformis bacteria adapt to changing environmental conditions by inducible and reversible phenotypic switching.
IMPORTANCE
In favorable environments, species may face space and nutrient limits due to overcrowding. Bacteria provide an excellent model for analyzing principles underlying overcrowding and regulation of density in nature, since their population dynamics can be easily and accurately assessed under controlled conditions. We describe a newly discovered mechanism for survival of a bacterial population during overcrowding. When competing with sibling colonies, Paenibacillus dendritiformis produces a lethal protein (Slf) that kills cells at the interface of encroaching colonies. Slf also induces a small proportion of the cells to switch from motile, rod-shaped cells to nonmotile, Slf-resistant, vegetative cocci. When crowding is reduced and nutrients are no longer limiting, the bacteria produce a signal that induces cocci to switch back to motile rods, allowing the population to spread. Genes encoding components of this phenotypic switching pathway are widespread among bacterial species, suggesting that this survival mechanism is not unique to P. dendritiformis.
Introduction
Earth provides a variety of natural habitats for all living organisms, but most successful species will eventually experience overcrowding, which can lead to extreme behaviors (1–4). Bacteria have evolved mechanisms to ensure survival of populations when faced with harsh environmental conditions or nutrient stress (5–8). In some cases, the stress arises when the bacteria are extremely successful in exploiting a niche, and their rapid expansion leads to overcrowding, resulting in resource depletion. To survive such sibling competition, some Gram-positive bacteria transition from a vegetative phase into dormant endospores that can withstand lack of nutrients, extreme heat, UV radiation, or antibacterial compounds (5, 6). Once favorable growth conditions are encountered, spores germinate and resume vegetative growth, restoring the population. To delay transition to a dormant phase, some species exhibit cannibalism, killing siblings and feeding on the released nutrients (6). Competition is not limited to members of a single colony, and nutrient stress is exacerbated when sibling colonies encroach. Paenibacillus dendritiformis (9–12), a Gram-positive, spore-forming bacterial species, produces a toxic protein, termed “sibling lethal factor,” Slf, which reduces population density by killing the swarming cells at the interface between competing colonies (10, 11).
Here we describe a newly discovered mechanism for surviving bacterial overcrowding during sibling colony competition: P. dendritiformis undergoes inducible and reversible switching between rod-shaped and coccoid phenotypes, each of which allows optimal growth under different conditions. Sublethal levels of Slf induce a switch in phenotype from motile rods to Slf-resistant, vegetative cocci. The cocci are nonmotile but are resistant to cell wall antibiotics and osmotic stress. When conditions are favorable for spreading, the cocci secrete an inducer and switch to rods. Thus, a small fraction of the population avoids lethal competition and can maintain the population by multiplying as cocci and switching to motile rods when competition is reduced.
RESULTS
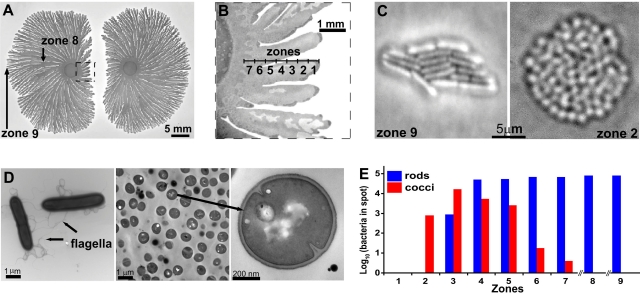
To examine the survival of P. dendritiformis during colonial competition, we inoculated two colonies onto low-nutrient peptone agar and allowed them to grow at 30°C until inhibition and killing of bacteria by Slf at the interface were evident (Fig. 1A and 1B, zone 1). Microscopy of cells within the zone of inhibition (Fig. 1A and 1B, zones 2 to 7) revealed the presence of small (0.7-µm-diameter) vegetative cocci that lack flagella (Fig. 1C and 1D). Comparison of the DNA sequences of the 16S rRNA genes of the cocci and rods showed that they were identical, ruling out contaminants (data not shown). Only cocci were recovered from the area closest to the competing colony (Fig. 1A and 1B, zone 2). No spores were seen in the zone of inhibition or in zones 2 and 3 (spore observation was done as previously described [11]). The proportion of rods increased with increasing distance from the competing colony (Fig. 1E). The correlation between proximity to the competing colony and proportion of cocci suggested that their presence was related to the concentration of the lethal factor Slf (11). To test this, we placed purified Slf next to a single growing colony, producing a zone of killing (Fig. 2A). Colonies of cocci could be seen in the inhibited region (Fig. 2A to 2C, zones 2 to 7) after approximately 2 weeks. No spores were observed in zones 1 to 3, where only cocci were found, and few spores were observed in zones 4 and 5. Spore density in zones 6 and 7 was the same as that in colonies not exposed to Slf (see Fig. S5 in reference 11).
FIG 1 .
The switch from rods to cocci. (A and B) Low (A) and high (B) magnifications of a competing colony. (C) Colonies formed from a single bacterium taken from zone 9 (rods) and from zone 2 (cocci). (D) Transmission electron microscopy of P. dendritiformis rod-shaped, motile cells (left). Cocci (cross-section image) are shown at the same magnification as are the rods (middle panel) and at a higher magnification, revealing incipient cell division (right). (E) The number of bacteria recovered from each zone (250- by 250-µm area) indicated in panels A and B. No bacteria were recovered from zone 1, and only cocci grew from zone 2. The proportion of rod-shaped bacteria increased with increasing distance from the inhibited interface, and only rods were recovered from zones 8 and 9.
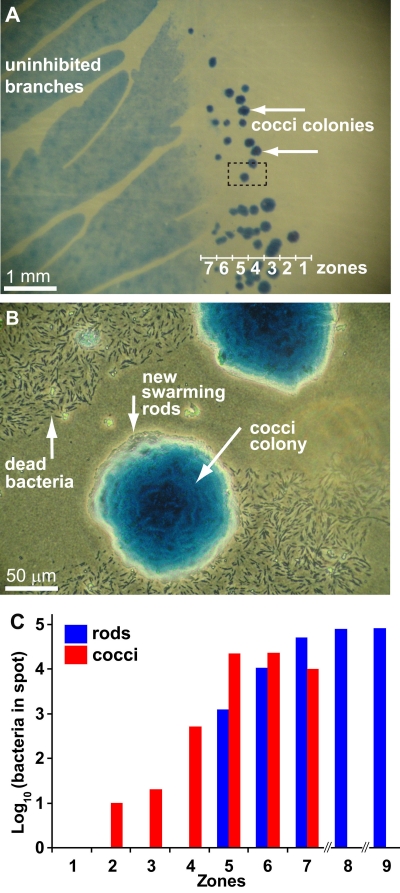
FIG 2 .
Slf induces the switch from rods to cocci in a single growing colony. Purified Slf was placed 1 cm from the edge of the colony 4 days after inoculation. (A) Coccus colonies (arrows) in the inhibited regions 4 weeks after Slf introduction. (B) Higher magnification of the marked rectangle in panel A. Dead bacteria are visible in the inhibited region near coccus colonies. Rods are swarming from the edges of the coccus colonies. (C) The number of bacteria in each zone, 3 days after introduction of Slf, was determined as described for Fig. 1E.
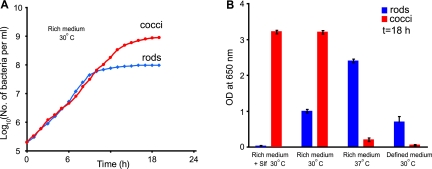
To determine differences between the two phenotypes, the growth and metabolism of cocci and rods under different conditions were compared. Cocci and rods were restreaked multiple times for isolation and maintained as separate stocks. Cocci and rods produced pure cultures in rich (LB) broth and grew at the same rates during exponential phase (Fig. 3A). At 30°C in LB broth, cocci reached a higher density than did the rods, but the rods grew to higher density at 37°C. The rods also outgrew the cocci in chemically defined medium (rich defined medium [RDM] [13]) (Fig. 3B). Addition of purified Slf to cultures confirmed that cocci but not rods were resistant to killing by Slf (Fig. 3B). A more detailed metabolic profiling using Biolog Phenotype MicroArrays (see Fig. S1 in the supplemental material) showed differences in carbon, nitrogen, phosphorus, and sulfur source utilization and in resistance to environmental stresses and antibiotics. In particular, cocci were much more resistant than rods were to osmotic stress and penicillins, indicating differences in cell wall and perhaps membrane structures. Thus, the cocci and rods exhibited striking differences in their abilities to survive and replicate under certain environmental and nutrient conditions.
FIG 3 .
Growth of P. dendritiformis rods and cocci under different conditions. Rods and cocci were cultured separately. (A) Growth of rods and cocci in LB broth at 30°C with aeration. (B) Density of cultures of rods and cocci after 18 h of growth under different conditions. Rich medium, LB broth; defined medium, RDM. Error bars are the standard deviations for 10 cases.
The absence of cocci in areas of colonies not exposed to Slf suggested that the cocci were not preexisting in the population but were induced by exposure to Slf. To confirm this hypothesis, cultures of rods were treated with ampicillin, which kills rods but not cocci, and grown at 30°C in LB to permit outgrowth of any preexisting cocci in the population. No cocci were recovered from the cultures, whereas they were readily detected after similar treatment with Slf (data not shown).
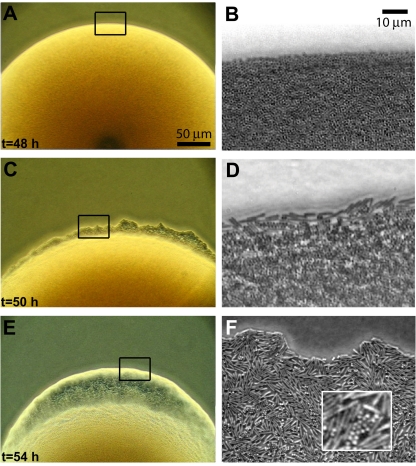
If the switch were an adaptive response to overcrowding, it would be likely that cocci could switch back to the rod morphology under conditions that favor motile rods. Therefore, individual cocci were inoculated on LB swarm plates (1% agar) and observed for the appearance of motile rods. The colonies expanded slowly for the first 48 h, during which time only cocci were detected (Fig. 4A and 4B; see Movie S1 in the supplemental material). After 50 h, rod-shaped, motile bacteria were observed at the edge of the colony (Fig. 4C and 4D; Movie S2), and within 4 h after the initial appearance of rods, the motile rods began to swarm in a thin liquid layer (Fig. 4E and 4F; Movies S3 and S4). The rods multiplied and 2 h later began to swarm in multiple layers, similar to colonies initiated from single rods (Movie S5).
FIG 4 .
P. dendritiformis switch from cocci to rods. Low (A, C, and E)- and high (B, D, and F)-resolution pictures of a colony grown from a single coccus on LB agar. The colony contains only cocci 48 h after inoculation (A and B). At 50 h (C and D), rods, swimming individually, appear at the colony edge. Four hours later (E and F), the rods are swarming. Rods appeared nearly simultaneously (within ~10 min) at multiple locations along the edge of the same colony and other colonies grown on the same plate.
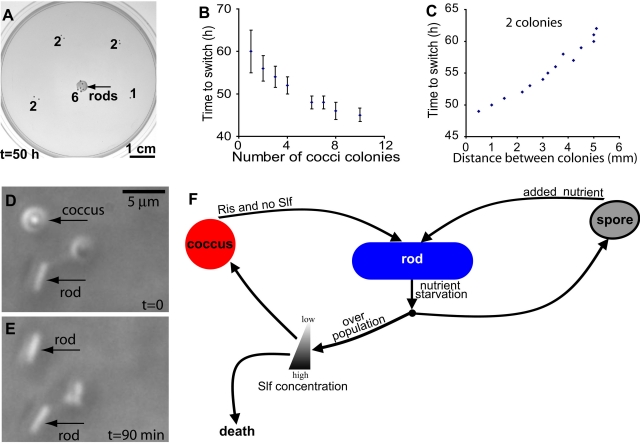
When multiple colonies were present within a spot on the plate, the length of time required for cocci in each colony to switch to rods was proportional to the number of colonies initially present in the spot (Fig. 5A and 5B) and to the proximity of colonies to each other (Fig. 5C). This suggests that the switch is not random but requires a secreted signal which is present in a larger quantity when there are more colonies and diffuses more rapidly when colonies are closer to each other. Such a signaling molecule should be present in culture supernatants. To test for the presence of a secreted inducing signal, cocci were grown in LB broth for 18 h at 30°C, and sterile supernatant from this culture was added to an equal volume of fresh medium prior to inoculation with cocci. In this culture, the switch to rods began at 18 h, whereas switching did not occur in the absence of supernatant until 22 h. This supports the hypothesis that a secreted factor, designated Ris (rod-inducing signal), induces the switch from cocci to rods. Rods grown in rich medium (LB) were also assessed for Ris production. Addition of rod supernatant to a culture of cocci, as described for the coccus supernatant, also induced switching from cocci to rods by 18 h but triggered the switch to the rod phenotype among more than 50% of the population by 18 h, compared to 3% in the culture treated with coccus supernatant. The simplest explanation is that Ris is secreted in greater amounts by the rods. Thus, there may be a positive-feedback loop: cells that switch to the rod phenotype secrete the inducer in larger amounts, accelerating the process of switching among the remaining cocci and ensuring that the transition is complete.
FIG 5 .
Rod-inducing signal (Ris). (A) Six colonies in close proximity show switching of cocci to rods sooner than does one or two colonies. Numbers indicate the number of colonies in a spot. (B) The time required for switch from cocci to rods depends on the number of coccus colonies in close proximity (as in panel A). Error bars are the standard deviations for 10 cases. (C) The time required for the switch to rods is proportional to the distance between two colonies. (D and E) Addition of Ris causes a single coccus to switch to a rod. (F) Circuit model showing the transitions between various cell forms of P. dendritiformis.
Ris was isolated from the culture supernatants of both rods and cocci by performing reverse-phase high-performance liquid chromatography (HPLC) and testing individual fractions for the ability to induce switching. Activity was associated with the fraction eluting at 42 min in both rod and coccus supernatants. The fraction contained a single peak with maximum absorption at 214 nm (see Fig. S2 in the supplemental material). The isolated compound placed near a single coccus caused switching to the rod phenotype in less than 2 h (Fig. 5D and 5E; see also Fig. S2), verifying that a specific secreted signal molecule induces the switch from coccus to rod. Ris did not produce ninhydrin-positive spots on thin-layer chromatography (TLC) plates and did not absorb UV light at 280 nm, suggesting that it is not a peptide.
DISCUSSION
Our results indicate that P. dendritiformis has at least two mechanisms to deal with changing environmental conditions and enable long-term survival of the species (Fig. 5F). First, it has the ability to form spores that are highly resistant to harsh conditions. The second mechanism, the formation of cocci, which are less resistant than spores but able to replicate even in the presence of Slf, offers cells near the leading edge of the colony the opportunity to continuously monitor the level of competition and the environment and to respond to the presence of sufficient nutrients for colony expansion. P. dendritiformis switching between rods and cocci requires specific secreted bacterial signals, Slf and Ris, which induce the switches in response to environmental cues. Thus, the population can be maintained as either rods or cocci under the appropriate conditions. In a culture consisting of all rods, there should be high levels of Ris, maintaining the population in the rod state. Under normal growth conditions, no Slf would be made and no transition to cocci would occur. When there is sudden overcrowding, as in the case of encroaching colonies, Slf is produced, killing most of the rods at the leading edge. This leads to a decrease in Ris production, enabling the transition to cocci in response to low levels of Slf. This is apparent at the edges of the colonies on solid media, where cocci are found in areas of dead rods (Fig. 1E and 2C).
The ability to replicate and maintain the coccoid form in an inducible way distinguishes this phenotype switching from other phenotypic changes such as persistence (14), sporulation (which leads to a dormant state) (5, 6), or formation of cocci in stationary phase (15–19). This ability allows P. dendritiformis to adapt to changing environmental conditions.
Although this form of phenotypic switching has not been described previously, genes of unknown functions with homology to the P. dendritiformis gene encoding Slf are widespread in bacteria and yeasts (20). Therefore, the lethal response to competition and associated phenotypic switching that we have observed in P. dendritiformis may be a common but previously unrecognized mechanism for regulation of population growth in nature.
MATERIALS AND METHODS
Strain and growth media.
Paenibacillus dendritiformis (T morphotype) (10) was maintained at −80°C in LB broth (Sigma) with 20% glycerol. The strain was routinely grown in LB broth at 30°C, with shaking (200 rpm). Low-nutrient peptone medium consisted of Bacto peptone (Difco), 2 g/liter; NaCl, 5 g/liter; and K2HPO4, 5 g/liter (pH adjusted to 7.0). For colony competition assays (10), 1.5% Difco agar (Becton Dickinson) was added to low-nutrient peptone and 12 ml was poured into 8.8-cm-diameter petri plates. The plates were dried for 4 days at 25°C and 50% humidity until the weight decreased by 1 g. EZ-RDM, a modification of the medium developed by Neidhardt et al. (13) (http://www.genome.wisc.edu/resources/protocols/ezmedium.htm) was used for growth in defined medium. The plates were inoculated by placing 5-µl drops of an overnight culture (optical density at 650 nm [OD650] of 1.0) on the surface. For intercolony competition experiments, 2 droplets were inoculated equidistant from the center, along a line through the plate’s center, as described previously (10).
Isolation of cocci.
Bacteria collected from zone 2 (Fig. 1B) were diluted and inoculated onto LB hard (1.6%) agar and grown at 30°C for 12 h. A single coccus colony was picked, diluted, and inoculated on a similar plate. This was repeated 10 times to ensure that only cocci were present in the culture. Each time, Slf was introduced on the colony to eliminate any potential growth of rod-shaped bacteria. After the 10th time, the colony was grown in LB broth for 12 h and stored at −80°C in LB with 20% glycerol.
Biolog Phenotype MicroArrays.
Metabolic activity and sensitivity patterns of the rods and cocci were compared using the Biolog Phenotype MicroArrays (Biolog, Hayward, CA), as described by the manufacturer. Cultures of rods and cocci were grown overnight in LB, and the cells were harvested by centrifugation at 7,000 rpm for 7 minutes in a Sorvall SS-34 rotor (5,800 × g). Bacteria were washed once in normal saline and then diluted to target concentrations of 1 × 107 CFU/ml for rods and 3.2 × 107 CFU/ml for cocci in inoculating fluid IF-0a. Plates PM1 to -20 (Biolog, Hayward, CA) were inoculated with 0.1 ml of diluted bacterial suspensions as described by the manufacturer’s protocol for Bacillus spp. Reduction of the dye was measured over 24 h at 30°C in an OmniLog instrument (Biolog, Hayward, CA).
Slf and Ris isolation.
Slf was isolated and purified from the agar medium as described previously (10, 11). Sodium dodecyl sulfate-polyacrylamide (10%) gel electrophoresis verified that the preparation contained a single protein of 12 kDa. Ris was isolated from cultures grown in 3 ml EZ-RDM at 30°C and 200 rpm for 48 h. Cells were removed from the culture by filtration through a 0.2-µm filter, and the supernatant was analyzed by high-performance liquid chromatography (reverse-phase C18 column eluted with a 1% to 60% acetonitrile gradient; Beckman System Gold 126B, Karat analysis software). Absorption was measured at 214 nm and 280 nm, and each peak was isolated and tested for the ability to trigger the switch from cocci to rods.
Optical microscopy.
Bacteria were imaged using an optical microscope (Olympus IX50) equipped with an LD 60x phase-contrast (PH2) objective lens. Images were captured using a charge-coupled device (CCD) camera with a spatial resolution of 1,004 by 997 pixels over a field of view of 120 by 120 µm2.
TEM.
We used an FEI Tecnai transmission electron microscope (TEM), operated at 80 kV, and 400-mesh copper carbon grids (from Electron Microscopy Sciences). Rod-shaped cells were collected from the agar, placed on the grids, and then stained for 10 s using 0.5% uranyl acetate. Because there was not sufficient contrast to discern the internal structure of cocci stained by this procedure, the cocci were fixed in formaldehyde (2%) and osmium tetroxide (2%). They were embedded in 3% (wt/vol) agarose and dehydrated with 50, 75, and 100% ethanol and 100% acetone. The sample was infiltrated with resin using five 24-h rinses (25, 50, and 75% and 2 times with 100%). The sample was polymerized in fresh 100% resin at 60°C for 2 days. It was sectioned (60- to 70-nm sections) with a glass knife and examined under a TEM.
DNA sequences of the 16S rRNA.
Oligonucleotide primers for PCR amplification of 16S rRNA, 5′ AGAGTTTGATCCTGGCTCAG 3′ and 5′ TACGGCTACCTTGTTACGACTT 3′, were purchased from IDT (Coralville, IA). PCR was performed using Taq polymerase (New England Biolabs) according to the manufacturer’s instructions. Overnight cultures were used as the templates. DNA sequencing of the amplified fragments was performed at the University of Texas Institute for Cellular and Molecular Biology DNA Core Facility using an ABI Prism 3700 DNA sequencer.
TLC.
The HPLC fraction containing biologically active Ris was analyzed for the presence of peptides by TLC as described in reference 21. The material was spotted onto silica gel 60 Å, 250-µm-thick plates (Whatman). The solvent was 1-butanol–acetic acid–H2O (4:1:5). Ninhydrin spray (Acros Organics) was used for amino acid detection, and 50× minimal essential medium (MEM) amino acid solution (Gibco) was chromatographed as a control.
SUPPLEMENTAL MATERIAL
Comparison of P. dendritiformis rods and cocci by Biolog Phenotype MicroArrays. Graphs show reduction of the tetrazolium dye, indicating metabolic activity, for 24 h at 30°C in the presence of various substrates and inhibitors: data for rods are shown in red and data for cocci are shown in green. Yellow indicates the overlap between the two. Plate 2 shows utilization of carbon sources tested (A), plate 9 contains osmolytes (B), and plate 12 contains antibiotics and inhibitors (C). Rod signal (red) is reduced over time (mainly in the last 3 rows of panel B and most of panel C, probably due to sporulation). Download Figure S1, EPS file, 2.521 MB.
Switch from cocci to rods induced by extracellular signal molecule Ris (rod-inducing signal). Cocci were sparsely (1 bacterium per 10-µm by 10-µm area) inoculated and grown on LB agar at room temperature. (A) A sample containing the signaling molecule (F) was introduced near the cocci at time zero. (B to D) Cocci switched to rods within 90 min, gradually increasing their lengths. (E) In contrast, spore germination required 48 h under these conditions, and the spores had a distinctly different appearance. (F) Separation and isolation of the signaling molecule Ris (rod-inducing signal). Rods were grown in 3 ml defined medium (RDM) (13) at 30°C and 200 rpm for 48 h. RDM was used to facilitate isolation of Ris by HPLC, but an active peak with the same HPLC elution time was also obtained from both cocci and rods grown in LB. Cells were removed from the culture by filtration through a 0.2-µm filter, and the supernatant was analyzed by high-performance liquid chromatography (reversed-phase HPLC; C18 column eluted with a 1% to 60% acetonitrile gradient; Beckman System Gold 126B, Karat analysis software). Twelve peaks were detected. Each peak was isolated and tested for the ability to trigger the switch from cocci to rods. Only the material eluting at 42 min, after separation from other peaks, triggered the switch. Absorption was measured at 214 nm and 280 nm. Download Figure S2, EPS file, 2.674 MB.
Colonies of Paenibacillus dendritiformis (40 h after inoculation), each grown from a single coccus on LB agar. No motile bacteria are evident. The movie was taken using phase-contrast microscopy. Frame dimensions are 288 µm by 216 µm. Download Movie S1, AVI file, 3.117 MB.
Early stages of the switch from cocci to rods (50 h after inoculation). High-resolution, phase-contrast real-time microscopy of motile rods at the edge of a coccus colony shown in Movie 1. Frame dimensions are 96 µm by 72 µm. Download Movie S2, AVI file, 1.585 MB.
Intermediate stages of the switch from cocci to rods (54 h after inoculation). The movie shows the motility of rods at the edges of the coccus colonies shown in Movie 1. The switch to motile rods occurs almost simultaneously in all the colonies. The movie was taken using phase-contrast microscopy. Frame dimensions are 288 µm by 216 µm. Download Movie S3, AVI file, 10.844 MB.
Intermediate stages of the switch from cocci to rods (54 h after inoculation). High-resolution, phase-contrast real-time microscopy of swarming rods at the edge of a colony of cocci shown in Movie 1. Frame dimensions are 96 µm by 72 µm. Cocci migrate within a single layer of swarming rods. Download Movie S4, AVI file, 8.957 MB.
Late stages of the switch from cocci to rods (56 h after inoculation). High-resolution, real-time microscopy of P. dendritiformis rods arising from the cocci colonies shown in Movie 1. Frame dimensions are 96 µm by 72 µm. Bacteria near the interface swarm in multiple layers. Download Movie S5, AVI file, 9.488 MB.
ACKNOWLEDGMENTS
E.-L.F. acknowledges support by the Robert A. Welch Foundation (F-1573), H.L.S. acknowledges support by the Sid W. Richardson Foundation, and S.M.P. acknowledges support by NIH grant AI16935.
We thank Eshel Ben-Jacob and H. P. Zhang for fruitful discussions; Rasika M. Harshey, Marvin Whiteley, and George Shubeita for their insightful comments on the manuscript; Inna Brainis for providing the bacterial strain; Rachel S. Smith for assistance with TEM; and Alexandra R. Mey for determining 16S rRNA gene sequence.
Footnotes
Citation Be’er A, Florin E-L, Fisher CR, Swinney HL, Payne SM. 2011. Surviving bacterial sibling rivalry: inducible and reversible phenotypic switching in Paenibacillus dendritiformis. mBio 2(3):e00069-11. doi:10.1128/mBio.00069-11.
REFERENCES
- 1. Dobler R, Kolliker M. 2010. Kin-selected siblicide and cannibalism in the European earwig. Behav. Ecol. 21:257–263 [Google Scholar]
- 2. vom Saal FS, Howard LS. 1982. The regulation of infanticide and parental behavior: implications for reproductive success in male mice. Science 215:1270–1272 [DOI] [PubMed] [Google Scholar]
- 3. Belovsky GE, Mellison C, Larson C, Van Zandt PA. 1999. Experimental studies of extinction dynamics. Science 286:1175–1177 [DOI] [PubMed] [Google Scholar]
- 4. Lister BC. 1980. Resource variation and the structure of British bird communities. Proc. Natl. Acad. Sci. U. S. A. 77:4185–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Errington J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1:117–126 [DOI] [PubMed] [Google Scholar]
- 6. Gonzalez-Pastor JE, Hobbs EC, Losick R. 2003. Cannibalism by sporulating bacteria. Science 301:510–513 [DOI] [PubMed] [Google Scholar]
- 7. Claverys JP, Havarstein LS. 2007. Cannibalism and fratricide: mechanisms and raisons d’etre. Nature 5:219–229 [DOI] [PubMed] [Google Scholar]
- 8. Gibbs KA, Urbanowski ML, Greenberg EP. 2008. Genetic determinants of self identity and social recognition in bacteria. Science 321:256–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ben-Jacob E, Cohen I, Gutnick DL. 1998. Cooperative organization of bacterial colonies: from genotype to morphotype. Annu. Rev. Microbiol. 52:779–806 [DOI] [PubMed] [Google Scholar]
- 10. Be’er A, et al. 2009. Deadly competition between sibling bacterial colonies. Proc. Natl. Acad. Sci. U. S. A. 106:428–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Be’er A, et al. 2010. Lethal protein produced in response to competition between sibling bacterial colonies. Proc. Natl. Acad. Sci. U. S. A. 107:6258–6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Be’er A, et al. 2009. Paenibacillus dendritiformis bacterial colony growth depends on surfactant but not on bacterial motion. J. Bacteriol. 191:5758–5764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625 [DOI] [PubMed] [Google Scholar]
- 15. Young KD. 2006. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 70:660–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. James GA, Korber DR, Caldwell DE, Costerton JW. 1995. Digital image analysis of growth and starvation responses of a surface-colonizing Acinetobacter sp. J. Bacteriol. 177:907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lange R, Hengge-Aronis R. 1991. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor sigma S. J. Bacteriol. 173:4474–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luscombe B, Gray T. 1974. Characteristics of Arthrobacter grown in continuous culture. J. Gen. Microbiol. 82:213–222 [Google Scholar]
- 19. Fuhrmann C, Soedarmanto I, Lammler C. 1997. Studies on the rod-coccus life cycle of Rhodococcus equi. J. Veterinarmed. B 44:287–294 [DOI] [PubMed] [Google Scholar]
- 20. Alvaro D, Lisby M, Rothstein R. 2007. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 3:2439–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hadley ME, Hruby VJ, Sharma SD. March 1998. Peptides having potent antagonist and agonist bioactivities at melanocortin receptors. U.S. patent 5731408
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of P. dendritiformis rods and cocci by Biolog Phenotype MicroArrays. Graphs show reduction of the tetrazolium dye, indicating metabolic activity, for 24 h at 30°C in the presence of various substrates and inhibitors: data for rods are shown in red and data for cocci are shown in green. Yellow indicates the overlap between the two. Plate 2 shows utilization of carbon sources tested (A), plate 9 contains osmolytes (B), and plate 12 contains antibiotics and inhibitors (C). Rod signal (red) is reduced over time (mainly in the last 3 rows of panel B and most of panel C, probably due to sporulation). Download Figure S1, EPS file, 2.521 MB.
Switch from cocci to rods induced by extracellular signal molecule Ris (rod-inducing signal). Cocci were sparsely (1 bacterium per 10-µm by 10-µm area) inoculated and grown on LB agar at room temperature. (A) A sample containing the signaling molecule (F) was introduced near the cocci at time zero. (B to D) Cocci switched to rods within 90 min, gradually increasing their lengths. (E) In contrast, spore germination required 48 h under these conditions, and the spores had a distinctly different appearance. (F) Separation and isolation of the signaling molecule Ris (rod-inducing signal). Rods were grown in 3 ml defined medium (RDM) (13) at 30°C and 200 rpm for 48 h. RDM was used to facilitate isolation of Ris by HPLC, but an active peak with the same HPLC elution time was also obtained from both cocci and rods grown in LB. Cells were removed from the culture by filtration through a 0.2-µm filter, and the supernatant was analyzed by high-performance liquid chromatography (reversed-phase HPLC; C18 column eluted with a 1% to 60% acetonitrile gradient; Beckman System Gold 126B, Karat analysis software). Twelve peaks were detected. Each peak was isolated and tested for the ability to trigger the switch from cocci to rods. Only the material eluting at 42 min, after separation from other peaks, triggered the switch. Absorption was measured at 214 nm and 280 nm. Download Figure S2, EPS file, 2.674 MB.
Colonies of Paenibacillus dendritiformis (40 h after inoculation), each grown from a single coccus on LB agar. No motile bacteria are evident. The movie was taken using phase-contrast microscopy. Frame dimensions are 288 µm by 216 µm. Download Movie S1, AVI file, 3.117 MB.
Early stages of the switch from cocci to rods (50 h after inoculation). High-resolution, phase-contrast real-time microscopy of motile rods at the edge of a coccus colony shown in Movie 1. Frame dimensions are 96 µm by 72 µm. Download Movie S2, AVI file, 1.585 MB.
Intermediate stages of the switch from cocci to rods (54 h after inoculation). The movie shows the motility of rods at the edges of the coccus colonies shown in Movie 1. The switch to motile rods occurs almost simultaneously in all the colonies. The movie was taken using phase-contrast microscopy. Frame dimensions are 288 µm by 216 µm. Download Movie S3, AVI file, 10.844 MB.
Intermediate stages of the switch from cocci to rods (54 h after inoculation). High-resolution, phase-contrast real-time microscopy of swarming rods at the edge of a colony of cocci shown in Movie 1. Frame dimensions are 96 µm by 72 µm. Cocci migrate within a single layer of swarming rods. Download Movie S4, AVI file, 8.957 MB.
Late stages of the switch from cocci to rods (56 h after inoculation). High-resolution, real-time microscopy of P. dendritiformis rods arising from the cocci colonies shown in Movie 1. Frame dimensions are 96 µm by 72 µm. Bacteria near the interface swarm in multiple layers. Download Movie S5, AVI file, 9.488 MB.