Abstract
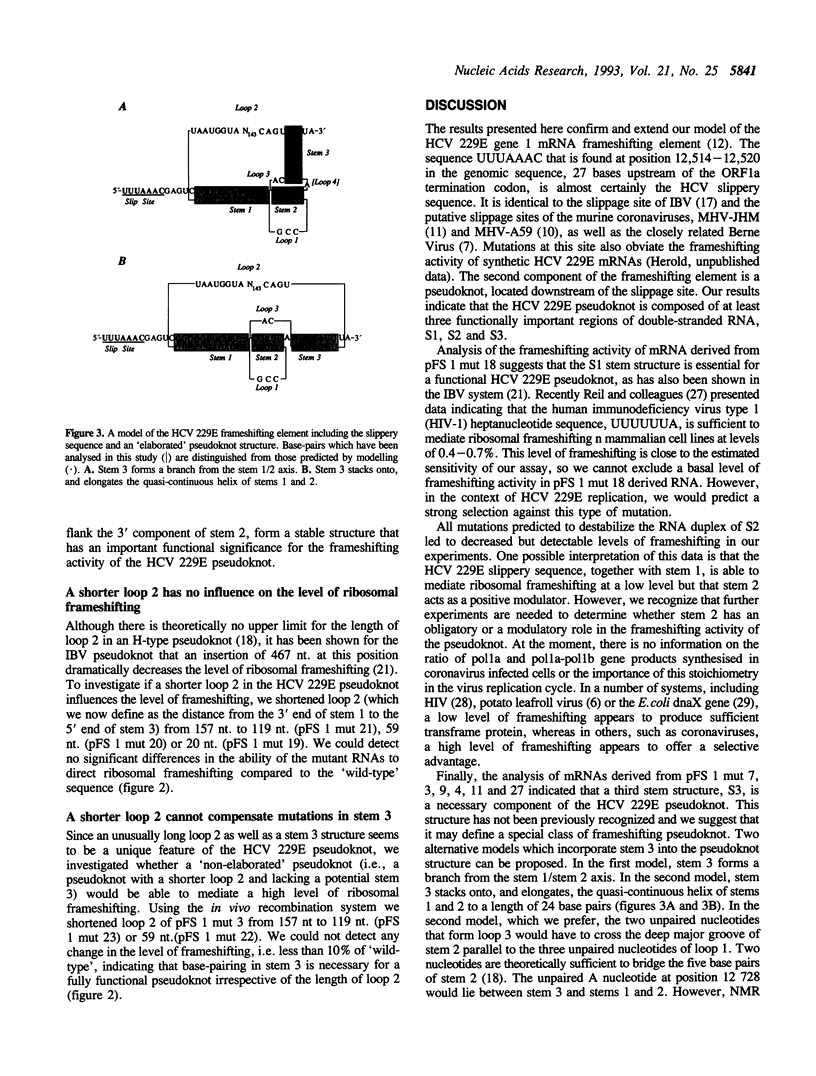
The RNA polymerase gene (gene 1) of the human coronavirus 229E is approximately 20 kb in length and is located at the 5' end of the positive-strand genomic RNA. The coding sequence of gene 1 is divided into two large open reading frames, ORF1a and ORF1b, that overlap by 43 nucleotides. In the region of the ORF1a/ORF1b overlap, the genomic RNA displays two elements that are known to mediate (-1) ribosomal frameshifting. These are the slippery sequence, UUUAAAC, and a 3' pseudoknot structure. By introducing site-specific mutations into synthetic mRNAs, we have analysed the predicted structure of the HCV 229E pseudoknot and shown that besides the well-known stem structures, S1 and S2, a third stem structure, S3, is required for a high frequency of frameshifting. The requirement for an S3 stem is independent of the length of loop 2.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brault V., Miller W. A. Translational frameshifting mediated by a viral sequence in plant cells. Proc Natl Acad Sci U S A. 1992 Mar 15;89(6):2262–2266. doi: 10.1073/pnas.89.6.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbeek P. J., Pachuk C. J., Noten A. F., Charité J., Luytjes W., Weiss S. R., Spaan W. J. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 1990 Apr 11;18(7):1825–1832. doi: 10.1093/nar/18.7.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Boursnell M. E., Binns M. M., Bilimoria B., Blok V. C., Brown T. D., Inglis S. C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987 Dec 1;6(12):3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Jenner A. J., Inglis S. C. Mutational analysis of the "slippery-sequence" component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1992 Sep 20;227(2):463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Rolley N. J., Jenner A. J., Inglis S. C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1991 Aug 20;220(4):889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro M., Parkin N., Varmus H. E. An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):713–717. doi: 10.1073/pnas.89.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J. D., Icho T., Wickner R. B. A -1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P. J. Alternative readings of the genetic code. Cell. 1993 Aug 27;74(4):591–596. doi: 10.1016/0092-8674(93)90507-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D., Oroszlan S. The where, what and how of ribosomal frameshifting in retroviral protein synthesis. Trends Biochem Sci. 1990 May;15(5):186–190. doi: 10.1016/0968-0004(90)90159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Varmus H. E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985 Dec 13;230(4731):1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- Lee H. J., Shieh C. K., Gorbalenya A. E., Koonin E. V., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M. M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991 Feb;180(2):567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R., Dixon M., Smith R., Peters G., Dickson C. Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: two frameshift suppression events are required for translation of gag and pol. J Virol. 1987 Feb;61(2):480–490. doi: 10.1128/jvi.61.2.480-490.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S. H., Copeland T. D., Hatanaka M., Oroszlan S. Characterization of ribosomal frameshifting for expression of pol gene products of human T-cell leukemia virus type I. J Virol. 1993 Jan;67(1):196–203. doi: 10.1128/jvi.67.1.196-203.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin N. T., Chamorro M., Varmus H. E. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J Virol. 1992 Aug;66(8):5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij C. W. Pseudoknots: a new motif in the RNA game. Trends Biochem Sci. 1990 Apr;15(4):143–147. doi: 10.1016/0968-0004(90)90214-v. [DOI] [PubMed] [Google Scholar]
- Pleij C. W., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985 Mar 11;13(5):1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer D., Tacke E., Schmitz J., Kull B., Kaufmann A., Rohde W. Ribosomal frameshifting in plants: a novel signal directs the -1 frameshift in the synthesis of the putative viral replicase of potato leafroll luteovirus. EMBO J. 1992 Mar;11(3):1111–1117. doi: 10.1002/j.1460-2075.1992.tb05151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J. D., Wyatt J. R., Tinoco I., Jr Conformation of an RNA pseudoknot. J Mol Biol. 1990 Jul 20;214(2):437–453. doi: 10.1016/0022-2836(90)90192-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reil H., Kollmus H., Weidle U. H., Hauser H. A heptanucleotide sequence mediates ribosomal frameshifting in mammalian cells. J Virol. 1993 Sep;67(9):5579–5584. doi: 10.1128/jvi.67.9.5579-5584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddell S. Coronavirus JHM: coding assignments of subgenomic mRNAs. J Gen Virol. 1983 Jan;64(Pt 1):113–125. doi: 10.1099/0022-1317-64-1-113. [DOI] [PubMed] [Google Scholar]
- Snijder E. J., den Boon J. A., Bredenbeek P. J., Horzinek M. C., Rijnbrand R., Spaan W. J. The carboxyl-terminal part of the putative Berne virus polymerase is expressed by ribosomal frameshifting and contains sequence motifs which indicate that toro- and coronaviruses are evolutionarily related. Nucleic Acids Res. 1990 Aug 11;18(15):4535–4542. doi: 10.1093/nar/18.15.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C. K., Draper D. E. Unusual mRNA pseudoknot structure is recognized by a protein translational repressor. Cell. 1989 May 19;57(4):531–536. doi: 10.1016/0092-8674(89)90123-2. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z. Translational frameshifting in the Escherichia coli dnaX gene in vitro. Nucleic Acids Res. 1991 May 11;19(9):2457–2462. doi: 10.1093/nar/19.9.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C., Tzeng T. H., Bruenn J. A. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Jones D. H., Grose C. Site-directed mutagenesis of herpesvirus glycoprotein phosphorylation sites by recombination polymerase chain reaction. PCR Methods Appl. 1992 Feb;1(3):205–207. doi: 10.1101/gr.1.3.205. [DOI] [PubMed] [Google Scholar]
- den Boon J. A., Snijder E. J., Chirnside E. D., de Vries A. A., Horzinek M. C., Spaan W. J. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J Virol. 1991 Jun;65(6):2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam E. B., Pleij C. W., Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990 Jul;4(2):121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]




