Abstract
Since the discovery by this laboratory of the zinc finger transcription factor, KLF10, a member of the Krüppel-like family of transcription factors, there have been multiple publications regarding its functions and its immediate family members, in numerous cell types. KLF10 has been shown to be rapidly induced by TGFβ1, 2, 3, E2, epidermal growth factor, and bone morphogenetic protein-2. TGFβ inducible early gene-1 activates the TGFβ-Smad signaling pathway via repression of Smad 7 expression and activation of Smad 2 expression and activity. Overall, KLF10 has been implicated in cell differentiation, as a target gene for a variety of signaling pathways, and in serving as a potential marker for human diseases such as breast cancer, cardiac hypertrophy, and osteoporosis. Like other KLF members, KLF10 is expressed in specific cell types in numerous tissues and is known to be involved in repressing cell proliferation and inflammation as well as inducing apoptosis similar to that of TGFβ KLF10 binds to Sp-1-GC rich DNA sequences and can activate or repress the transcription of a number of genes. Overall, KLF10 has been shown to play a major role in the TGFβ inhibition of cell proliferation and inflammation and induction of apoptosis, and its overexpression in human osteoblasts and pancreatic carcinoma cells mimics the actions of TGFβ.
Keywords: KLF10, TIEG1, TGFβ inducible early gene, Krüppel family, estrogen, TGFβ, Smads, apoptosis, differentiation, bone, heart, cancer, immune system
Discovery and overall characterization of KLF10
The KLF10 gene, originally termed TGFβ inducible early gene-1 (TIEG), was initially identified in normal human fetal osteoblasts (hFOB) following TGFβ treatment using differential display PCR. Further studies revealed that KLF10 mRNA increased within 30 min of TGFβ treatment and reached a maximum of approximately 10-fold above control levels at 90 min post-treatment [1]. This induction of KLF10 mRNA was independent of new protein synthesis and very transient, with a rapid return to normal (pretreated) levels within 2 h. The KLF10 protein levels follow closely behind the mRNA levels. As depicted in Fig. 1, the KLF10 gene spans 6.5 kb and contains four exons. It is located on human chromosome 8q22.2. Computer analysis of the 5′-upstream region of KLF10 shows no TATA box or initiator sequence, but does show consensus sequence similarities to binding sites for several transcription factors including Sp1, JunB, and aromatic hydrocarbon/receptor-ligand complexes. Transcriptional analysis of genomic regions containing 5′flanking regions reveals that the KLF10 promoter has significant activity in hFOB cells and pancreatic cancer cells.
Fig. 1.

Model of the chromosomal locale and the genomic and protein structure of KLF10 (TIEG1). A. Genomic structure showing promoter, exons, and introns of KLF10. B. Protein structure of KLF10 showing various protein and DNA binding domains.
The KLF10 gene and the EGRα (early growth response-α) gene, discovered in prostate cancer cells [2], were independently discovered and found to be very homologous [3]. Our laboratory demonstrated that both the KLF10 and EGRα proteins are the products of a single gene [1,3]. It was subsequently shown that KLF10 and EGRα are transcribed from differentially regulated, alternative promoters. Both use common exons for almost all of their coding regions with the exception of the small exons at the 5′-end. Thus, the KLF10 and EGRα genes produce proteins, which differ in sequence only by 12 amino acids at the amino termini [3]. However, the functional consequences of this small difference in structure appear extensive. Northern analysis of mRNA from various human tissues and several cell lines reveal that KLF10 is the predominant transcript expressed and regulated by growth factors and cytokines.
KLF10 protein: Structure, function, and regulation
Also depicted in Fig. 1., sequence analysis indicates that KLF10 mRNA encodes a 480 amino acid protein that contains a three zinc finger DNA binding domain at the C-terminal end and several proline-rich Src homology-3 (SH3) binding domains at the N-terminal end [1]. These SH3 domains bind the Sp1 protein for transcriptional regulation [4]. The KLF10 also contains three unique repression domains (R1-R3) spread throughout the protein structure [1,5,6]. Thus, KLF10 was classified as a member of the Krüppel-like family of transcription factors, all of which bind to GC rich Sp1-like sequences to regulate gene transcription. The regulation of KLF10 expression in human osteoblast (OB) cells is growth factor/cytokine specific with a strong induction by TGFβ and bone morphogenetic protein-2 (BMP-2), BMP-4, and activin, with a moderate induction by epidermal growth factor, and no induction by other growth factors/cytokines (e.g., BMP-6, IGF-1, IGF-2, FGF, TNFα IL-6, IL-1B) [1,7]. Subsequent studies demonstrated that estrogen increased, but testosterone and glucocorticoids decreased, KLF10 in human OB cells in culture [8,9]. Recently our laboratory demonstrated that the estrogen regulation of KLF10 gene transcription occurs via the first intron of the gene [10]. Both growth factors and estrogen rapidly induce the expression of KLF10 (30 min) followed by a return to control values within 2–4 h. The cellular actions of KLF10 ultimately result in the inhibition of cell proliferation and induction of apoptosis [7,11,12].
Using a KLF10 specific polyclonal antibody and immunoprecipitation methods in normal hFOB cells, KLF10 was shown to encode a 72-kDa protein whose levels are rapidly, but transiently, increased within 2 h of TGFβ treatment [13]. Polarized confocal microscopic analysis of hFOB cells revealed that KLF10 protein was localized in the nucleus in untreated cells; however, as expected, the levels of KLF10 protein in the nucleus increased when the cells were treated with TGFβ for 2 h. Similar analyses of untreated human keratinocytes revealed that KLF10 was localized in the cytoplasm but was translocated to the nucleus after H2O2 treatment [13].
Subsequent immunohistochemical studies of human tissues demonstrated that KLF10 protein is expressed in epithelial cells of the placenta, breast, and pancreas, as well as in skeletal and smooth muscle cells, heart cells, glial cells, fibroblasts, pancreatic carcinoma cells, cerebral cortex and cerebellum cells, myeloid cells, OB cells, and select cells in the bone marrow [13–20]. All cells of the kidney displayed negative staining for this protein. Interestingly, as described later, a breast cancer stage specific expression of KLF10 protein was reported by our laboratory [13,21].
Evolutionary conservation of KLF10
The protein sequence of human KLF10 (hKLF10) was shown to be closely related to three mouse sequences [(mGIF, GC-binding protein, and mKLF10)] [5]. Due to the extremely high level of nucleotide sequence identity (more than 96%), these three sequences likely represent homologous genes. In fact, the predicted amino acid sequence of the murine KLF10 protein is 85% identical to the human KLF10. Like human KLF10, the mGIF cDNA was recently characterized as a negative-acting transcription factor that can be induced by another TGFβ family member, Glial cell-Derived Neurotrophic Factor (GDNF) in murine neuroblastoma cells [19].
KLF11 and other related family members
The possibility that there are multiple KLF-related genes in the mouse was suggested by the presence of multiple bands on genomic Southern blots [5,19]. Cook et al (1998) identified a closely related KLF family member, KLF11, in human tissues. KLF11 (TIEG2), which inhibits cell proliferation, is localized in the nucleus, and shares other properties similar to that of KLF10. As with the KLF10 gene, KLF11 is a ubiquitously expressed transcription factor, which also contains the Krüppel-like three zinc finger motif at the C-terminal end of the protein. The KLF11 gene encodes a nuclear protein, which binds GC rich/Sp1-like sequences to regulate gene expression and inhibit cell proliferation. KLF10 and KLF11 can regulate some of the same genes [22]. Like KLF10, KLF11 contains three repressor domains [6], but can activate gene transcription (monoamine oxidase B gene) when bound to Sp1 sites along with the Sp1 protein [23]. Both KLF10 and KLF11 inhibit cell proliferation and induce apoptosis [11,24–27]. The discovery and characterization of another related KLF10 in mice, termed mTIEG3, which is highly homologous to KLF11 (mTIEG2) and to KLF10 (TIEG1), opens the possibility that three mouse and human KLF10 proteins (KLF10, KLF11, and TIEG3) may have evolved [28]. As with KLF10 and KLF11, TIEG3 has been shown to contain three conserved repressor motifs and to be induced by TGFβ to repress target gene transcription. These sites in KLF11 (TIEG2) have been shown to interact with the core-pressor, mSin3A, in repressing gene transcription [29,30]. In general, these mouse genes appear to have different expression patterns depending on the cell/tissue type but likely serve the same functions as their two human counterparts (human KLF10 and KLF11). These common functions are described below.
Mechanism of the rapid turnover of KLF10 protein
As mentioned earlier, KLF10 induction is abruptly transient. Using yeast-2-hybrid and co-immunoprecipitation assays, an E3 ubiquitin ligase, Seven in Absentia homologue-1 (SIAH1) protein, was found to interact with KLF10 [22]. KLF10 and SIAH1 have been shown to interact through a conserved SIAH-binding motif in KLF10 (i.e., a degradation site or “degron”) [31]. This site on TIEG1 was shown to be essential for SIAH directed proteasomal degradation of TIEG1 [24]. Co-expression of SIAH1 results in proteasomal degradation of KLF10, but not KLF11. Importantly, co-expression of SIAH1 completely reverses the repression of the Smad7 promoter activity by KLF10 [22]. Furthermore, overexpression of a dominant negative SIAH1 stabilized KLF10 resulting in enhancement of TGFβ/Smad-dependent transcriptional activity. These findings indicate that the ability of TGFβ to modulate gene transcription is regulated by proteasomal degradation of the downstream transcription factor, KLF10, through the SIAH pathway. Verification of this view was supported by our studies showing that proteasomal inhibitors block the transient expression patterns of KLF10 [22]. In this manner, the rapid turnover of KLF10 may serve to limit the duration and/or magnitude of KLF10 responses.
Role of KLF10 in cellular pathways
KLF10 gene expression is regulated by the TGFβ-Smad pathway
The overexpression of KLF10 in human osteosarcoma MG-63 cells caused changes in gene expression and cell proliferation, which mimicked those of the MG-63 cells treated with TGFβ (e.g., increased alkaline phosphatase activity, decreased levels of osteocalcin mRNA and protein, and decreased cell proliferation) [7]. The degree of these changes correlated with the level of KLF10 expression. The fact that KLF10 encodes a three zinc-finger Krüppel-like transcription factor, whose overexpression has been shown to mimic the effects of TGFP in human osteosarcoma and pancreatic carcinoma cells, supports a primary role for KLF10 as an important transcription factor in the TGFβ signaling pathway [6,7,11,26,32]. Studies utilizing transient transfection of KLF10, along with a Smad binding element (SBE) reporter construct into fibroblast cells, displayed an induction of the SBE reporter activity known to be regulated by TGFβ” [33]. Additionally, KLF10 expression also enhanced the induction of the endogenous TGFβ regulated genes p21 and PAI-1 [33]. The ability of KLF10 to enhance TGFβ action was shown to be Smad dependent since KLF10 had no effect on SBE transcription in the absence of Smad4 expression or when an inhibitory Smad protein, Smad7, was overexpressed [33]. Furthermore, KLF10 overexpression enhanced the TGFβ induced Smad2 phosphorylation and increased the transcription of Smad2, but not Smad3 or Smad4 [34]. Lastly, KLF10 bound to and repressed a specific element in the proximal promoter of the inhibitory Smad7 gene. Thus, as outlined in Fig. 2, KLF10 increases the activity of the TGFβ/Smad signal transduction pathway by dual mechanisms, that is, relieving the negative feedback through repression of the inhibitory Smad7 and inducing Smad2 expression and phosphorylation [10]. Figure 2 also depicts the estrogen induction of KLF10 expression [32,35]. This could well explain the estrogen-TGFβ/Smad crosstalk [10]. Interestingly, similar to KLF10, we observed that KLF11 is also able to enhance TGFβ/Smad signaling and repress Smad7 gene expression [22]. However, the mechanisms of KLF10 and KLF11 are distinct, at least in part, because KLF10, but not KLF11, is targeted for degradation by SIAH1 [22].
Fig. 2.
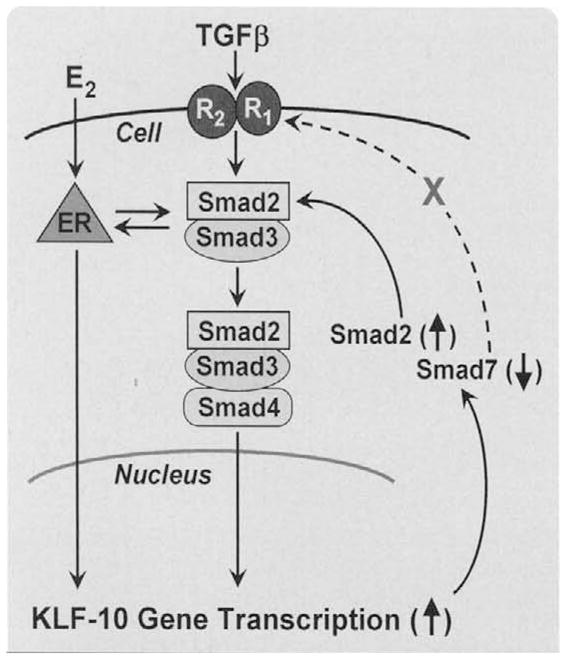
This figure outlines the role of KLF10 protein as a target and as a regulator of the TGFβ-Smad 2,3 signaling pathway. TGFβ binds to its membrane receptors (R2 R1) to activate the Smad signaling pathway. KLF10 (TIEG) is one of the rapid, early response genes whose expression is activated. KLF10 mRNA is rapidly translated into the KLF10 protein, which as a gene transcription repressor inhibits Smad 7 gene expression and, as a gene activator, induces Smad 2 gene expression. The ultimate outcome is a continued activation of the TGFβ-Smad signaling pathway, that is, an extension of the signaling period. The KLF10 mRNA and protein are rapidly turned over, thus the signaling is fine tuned to the KLF10 levels and activity. The steroid, estrogen (E), also induces the expression of KLF10 gene transcription, which is one mechanism of the well documented crosstalk between TGFβ and estrogen signaling.
Role of KLF10 in TGFβ mediated apoptosis
Members of the TGFβ family of peptides are known to exert antiproliferative effects and induce apoptosis in many epithelial cell types. In the exocrine pancreas, these peptides not only regulate normal cell growth, but alterations in these pathways have been associated with neoplastic transformation. Therefore, the identification of molecules that regulate cell proliferation and apoptotic cell death in response to TGFβ is necessary for a better understanding of normal morphogenesis as well as carcinogenesis of the pancreas. Tachibana et al (1997) characterized the expression and function of KLF10 in exocrine pancreatic epithelial cells. The gene was expressed in both acinar and ductular epithelial cell populations from the exocrine pancreas. Overexpression of KLF10 in the TGFβ-sensitive epithelial cell line PANC1 and hepatoma cell lines was sufficient to induce apoptosis [11,27]. This KLF10-induced apoptosis appears to be similar to the P53 induced mitochondrial apoptosis pathway, which occurs in pancreatic epithelial cells [27], as well as other epithelial cells [36], lymphoma cells [37], leukemic cells [38], and retinal cells [39]. Interestingly, Jin et al [25], demonstrated that KLF10 induces apoptosis in leukemic or melanoma cells through the mitochondrial pathway by inducing Bax and Bim upregulation, Bcl-2 and Bcl-XL downregulation, release of cytochrome c from the mitochondria, activation of caspase 3 and the disruption of mitochondrial membrane potential.
Role of KLF10 in cell proliferation
To determine whether KLF10 plays a central role in eliciting the antiproliferative effects of TGFβ, a tetracycline inducible KLF10 overexpressing breast cancer cell line and KLF10 null mouse embryo fibroblasts (MEFs) were developed [40]. Similar to TGFβ treatment, KLF10 overexpression increases the expression of the cyclin dependent kinase inhibitor p21 and significantly decreases cellular proliferation. Interestingly, while cellular proliferation of wild-type MEFs is inhibited by TGFβ, the cell proliferation of KLF10 null MEFs is stimulated by TGFβ Furthermore, KLF10 null MEF cells display a decrease in Smad dependent transcription with a concomitant prolonged increase in Smad7 expression compared with wild-type cells. These data strongly support that KLF10 plays a central role in the antiproliferative response via the TGFβ-Smad pathway. Similar to known tumor suppressor genes, the reduced levels of KLF10 expression correlate with the more advanced stages/development of cancer [13,21]. Together, these results support a role for KLF10 in linking TGFβ-mediated signaling cascades to the regulation of cell growth, antiproliferation, and apoptosis.
Phenotypic and disease implications of KLF10
Skeletal disease: Skeletal phenotype of the KLF10 knockout mouse
Osteoporosis
Figure 3 summarizes the important bone related genes in which KLF10 has been shown to regulate either by repression or activation of gene transcription. Overall, the expression of KLF10, like TGFβ treatment, in osteoblasts enhances osteoblast and osteoclast differentiation, bone turnover, and bone formation [41]. Recent studies by Hopwood et al (2009) examined gene expression profiles in a dozen osteoporotic patients and identified KLF10 as one of 150 genes whose expression in skeletal tissues differed markedly between osteoporotic and nonosteoporotic patients. Further, Yerges et al (2009) examined 4608 SNPs in 383 bone related genes in 2000 caucasion men over age 65. Using quantitative computerized tomography, KLF10 along with four other genes were “robustly” associated with volumetric cortical bone mineral density [42].
Fig. 3.
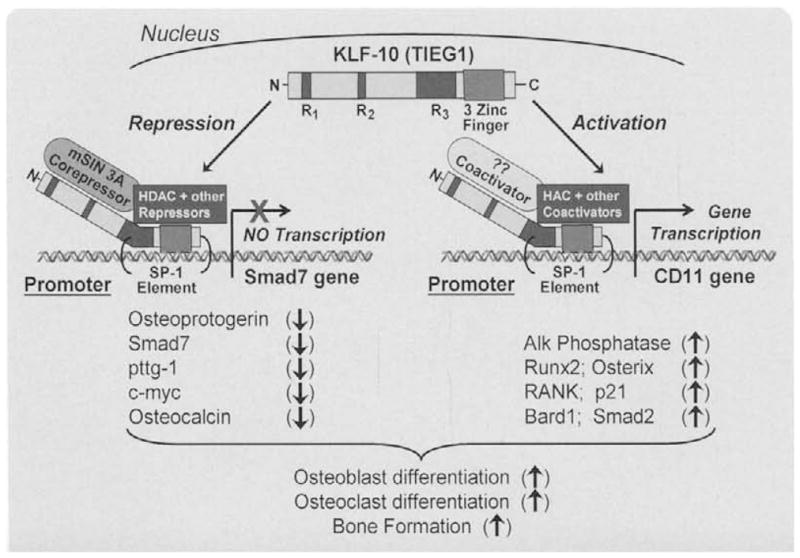
Model depicting the KLF10 co-activation and compression of known target gene expression in bone forming osteoblast (OB) cells. The result of this gene regulation is the induction of osteoblast and osteoclast differentiation and encouragement of bone formation and remodeling.
KLF10 knockout mice
To elucidate the functions of KLF10 in skeletal development, maintenance, and disease, we generated KLF10 knockout (KO) mice. Three-point bending tests on mixed background, knockout animals indicated that the femurs of female KLF10 KO mice were significantly weaker than those of wild-type animals [43]. The femurs of male knockout animals were not affected. pQCT analyses of female KLF10 KO tibias revealed marked decreases in multiple bone parameters in both the diaphyseal and metaphyseal regions, compared with wild-type mice. Micro-CT analysis of the femoral head and vertebrae revealed increases in femoral head trabecular separation and decreases in cortical bone thickness and vertebral bone volume in female KO mice relative to wild-type controls. In addition, electron microscopy indicated a significant decrease in osteocyte number in the femurs of female KO mice, suggesting that defects in osteoblast differentiation might exist. Interestingly, no changes in the bones of male KO mice were observed using any of these techniques. Figure 4 outlines the effects of knocking out TIEG on the skeleton (and other tissues) of mice. The resulting changes in expression of key osteoblast genes are also listed. The bones of KLF10 KO female, but not male, mice display an osteopenic gender-specific phenotype with significantly weaker bones and reduced amounts of cortical and trabecular bone suggesting an important role for KLF10 in skeletal development and/or homeostasis. The gender specificity of the osteopenia due to the TIEG gene ablation is speculated to be due to reproductive hormone differences. This aspect is currently under investigation.
Fig. 4.
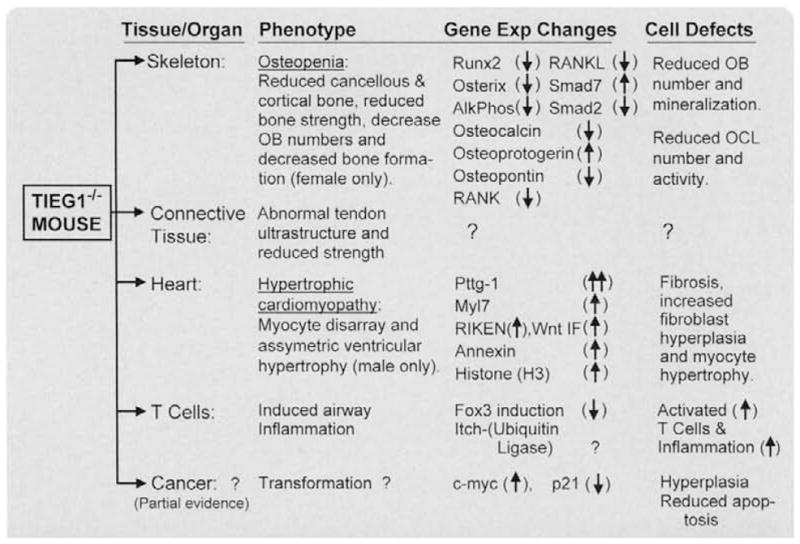
Reported defects/diseases of the KLF10 KO mouse. This table summarizes the cell and organ defects, which occur in KLF10 KO mice. Also listed are some of the genes whose expression are significantly altered in these cells/ organs. See text for referenced resources.
KLF10 regulation of osteoblast and osteoclast differentiation
As depicted in Fig. 4, the bone forming osteoblast from KLF10 KO mice displays reduced expression of important OB differentiation markers, including Runx2, osterix, and osteocalcin. The cells also displayed a reduced capacity to mineralize bone [35,44]. As mentioned above, the data of the KLF10 KO mice suggest an important role for KLF10 in osteoblast differentiation and bone formation [43]. A recent study analyzing osteocytes of KLF10 KO mice revealed changes in osteocyte surface and density as well as hypomineralized bone matrix surrounding the osteocytes [45].
The KLF10 KO mice also display deficiencies in the bone resorbing osteoclast functions [44]. When OBs from KLF10 KO and wild-type mice were cultured with marrow and spleen cells from wild-type mice, significantly fewer osteoclasts developed in the KLF10 KO OB cocultures compared with the osteoclasts that developed in the wild-type OB co-cultures. As depicted in Fig. 5, examination of gene expression in the KLF10 KO OBs revealed that decreased RANKL, a required inducer of osteoclastogenesis, and increased OPG levels, an inhibitor of osteoclastogenesis, relative to wild-type OBs, are likely responsible for the reduced ability of these osteoblasts to support osteoclast differentiation [41,44]. Thus, as shown in Fig. 5, it appears from these studies that KLF10 expression in OBs is critical for both osteoblast differentiation and mineralization as well as OB support of osteoclast differentiation.
Fig. 5.

Models depicting (A) normal/wild-type, and (B) KLF10 knockout mouse processes of osteoblast (OB) support of osteoclast (OCL) differentiation. In the wild-type mouse (A), the OBs are shown to produce high levels of RANKL and M-CSF required for OCL differentiation, as well as low levels of OPG, the inhibitor of OCL differentiation. The result is enhanced OCL differentiation, high bone turnover, and an increase in bone formation. In (B) the KLF10 ablated OB cells produce low levels of RANKL, high levels of OPG, which retards OCL differentiation. The result is a reduction in bone turnover and a loss of bone (osteopenia).
Connective tissue disease: Studies of the KLF10 knockout mouse
Also summarized in Fig. 4, studies were performed to examine the structure, function, and healing potential of flexor tendons in KLF10 KO mice, and to further examine what role the KLF10 pathway plays in flexor tendon repair [46]. Wild-type and KLF10 KO mice showed healing of lacerated tendons, but the chronologic expression pattern of TGFβ was found to be different. The KLF10 KO tendons had delayed expression of TGFβ and healing when compared with wild-type tendons. The collagen mRNA expression pattern was similar between both groups, but the expression levels were different, with KLF10 KO tendons having a lower expression of collagen type I mRNA. Additional studies focused on the age-dependent changes in the architecture and mechanical properties of tendons isolated from KLF10 KO mice [47]. The results revealed that the fascicles of the KLF10 KO mice at 3 months of age exhibit decreased fast and static stresses compared with those of wild-type mice. Electron microscopy revealed an increase in fibril size in KLF10 KO mouse tendons relative to wild-type controls. These data indicate a structural/functional role for KLF10 in tendon microarchitecture and strength in adult mice. The mouse model described in this study provides a novel means for further understanding of the tendon healing process through isolated deletion of specific factors.
Heart disease: The KLF10 knockout mouse story
Also outlined in Fig. 4, a novel finding was reported by our laboratory in the KLF10 KO mouse implicating an important role for KLF10 in cardiac hypertrophy [48]. KLF10 KO male mice, 4–16 months of age, were analyzed by echocardiography, transcript profiling by gene microarray, and immunohistochemistry. The male, but not female, KLF10 KO mice develop features of cardiac hypertrophy including asymmetric septal hypertrophy, increased ventricular size at 16 months of age, increased heart weight to body weight ratio, increased fibrosis, and increased wall thickness relative to wild-type animals. Interestingly, female mice did not develop cardiac hypertrophy or fibrosis. Masson Trichrome staining demonstrated evidence of myocyte disarray and myofibroblast fibrosis. Interestingly, cardiomyocyte specific Smad4 KO mice also develop cardiac hypertrophy [49]. Additional insights into the molecular mechanism by which KLF10 regulates heart development were elucidated by microarray analysis of the left ventricles which demonstrated that KLF10 KO heart tissues display a 14-fold increase in pituitary tumor-transforming gene-1 (Pttg1) (Fig. 4). Increases in Pttg1 and histone H3 protein levels were confirmed in the KLF10 KO mouse hearts by RT-PCR. These data implicate KLF10 and possibly one of its target genes, Pttg1, in the development of cardiac hypertrophy in the male KLF10 KO mouse.
Immunological system: The knockout mouse story in Treg cells
Recent collaborative studies involving this laboratory with the Feinberg laboratory have also shown that the lack of KLF10 in the knockout mice results in atherosclerosis via activation of the immune system [50]. In other collaborative studies, KLF10 was shown to interact with the ubiquitin ligase, Itch, to ubiquinate KLF10 [51]. This ubiquitination results in induction of both Foxp3 transcription factor levels and activity leading to activation of Treg cells and induction of airway inflammation. Related studies by the Feinberg laboratory [50], also showed that KLF10 regulated Treg cell suppressor function and CD4+ CD25 T cell activation through regulating TGFβ and Foxp3 gene transcription resulting in T regulation of T cell activity. The KLF10 KO mice displayed enhanced CD4+ CD25 T cell activity, which promoted inflammation, atherosclerosis, and accumulation of peripheral proinflammatory cytokines [50].
Role of KLF10 in breast cancer
The earlier description of the actions of KLF10 in inhibiting cell proliferation and inducing apoptosis in pancreatic and other (prostate) cancer cells indicates a role for KLF10 as a tumor suppressor [11–13,26,27,37,40,52–57]. The correlation between the levels of KLF10 protein and the stage of human breast cancer, its prime location on human chromosome 8q22.2, along with certain other proto-oncogens and tumor suppressor genes [58], and past studies revealing its role in pancreatic carcinoma, further supports a major role for KLF10 as a tumor suppressor in a variety of cancers [7,12,13,21,26,40]. The fact that KLF10 plays an important role in cell proliferation and apoptosis, and its levels inversely correlated with breast cancer stages, further supports a tumor suppressor function for KLF10.
To summarize the breast cancer studies, Reinholz et al (2004) measured the mRNA levels of KLF10 and its target genes, Smad7 (inhibitor of Smad signaling), Smad2 (inducer of Smad signaling), and BARD1 (binds and ubiquinates BRCA-1 to block DNA repair), using real-time PCR in 14 normal human breast, as well as 5 noninvasive, 57 invasive (including 29 with outcome data), and 5 metastatic human breast tumor tissues (Fig. 6) [21]. KLF10 and Smad7 mRNA levels were lower in all noninvasive tumors compared with normal breast tissues but Smad7 mRNA levels increased in more advanced stages of cancer. KLF10, BARD1, and Smad2 mRNA levels were lower in invasive cancers compared with normal breast tissues. In addition, KLF10, Smad2, and BARD1, provided discriminatory ability to distinguish between normal and tumor samples, N− and N+ tumors, and N-/good outcome (no recurrence for at least 5 years) and N-/bad outcome (recurrence within 3 years) breast cancer patients. As outlined in Fig. 6, these studies support the hypothesis that the differential gene expression of KLF10 and its induced target genes, Smad2, and BARD1 (both of which are tumor suppressor genes), as well as Smad7, play a significant role in the proliferation of breast cancer cells [21]. The altered expression of KLF10 in breast cancer tissues compared with normal tissues, further supports a role of KLF10 in breast cancer development/maintenance [21,25,53.55,59].
Fig. 6.
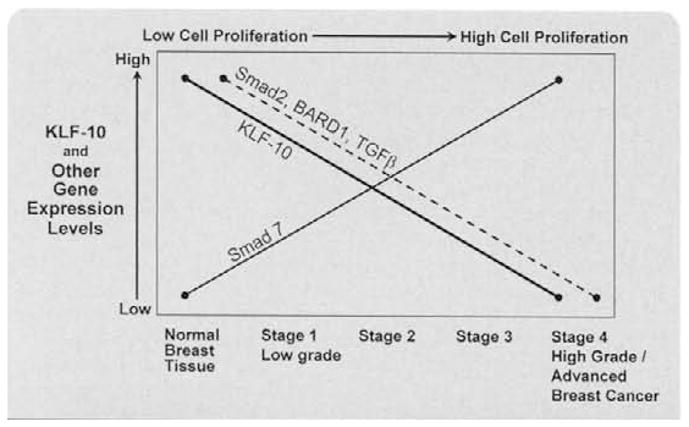
Graph depicting the expression levels of KLF10 and its target genes (Smad 2, Bard 1, Smad 7) in human breast cancers at various stages. (Reproduced with permission from Reinholz et al. Breast Cancer Res. Treat., 86 (2004), 75–88).
Role of KLF10 in other cancers
As outlined in Fig. 7, KLF10 also appears to play a role in other cancers, including human prostate cancer (as EGRα) [52,60,61], pancreatic cancers [26,53,59,62], human colo-rectal cancer [63], lymphoma cells [37,64], non-small cell carcinoma [65], renal cell carcinoma [66], multiple myeloma [67], and finally, brain cancers [57]. The above studies strongly support that KLF10 plays an important role in inhibiting cell proliferation and inducing apoptosis. These studies also support that KLF10 has the properties of a tumor suppressor [7,11]. KLF11 appears to have similar actions. See a recent review by Ellenreider (2008) describing role of KLF10 and KLF11 in cancer [54].
Fig. 7.
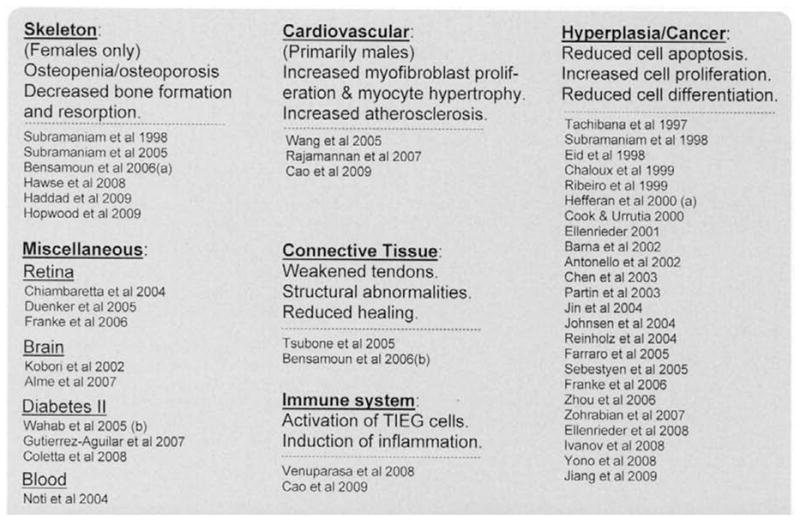
List of abnormal tissue structure and function which correlates with altered TIEG expression. (References to these relationships are also listed.)
Role of KLF10 in other diseases
Due to the limited space for this review, we will only mention that KLF10 and other KLF family members appear to play key regulatory roles in cell differentiation, apoptosis, inflammation, and differentiation in other tissues. References of articles describing the relationships of altered KLF10 expression with various organ/tissue abnormalities and diseases are listed in Fig. 7. Examples include the retina [39,68,69], and bone marrow [15]. In addition, the expression of KLF10 is altered in the brain following injury [14], and in response to stress [70]. KLF10 expression is induced in response to the action of brain derived neurotrophic factor [70], in response to GDNF (glial cell line-derived neurotrophic factor) [19,71], and to nerve growth factor [72].
KLF10 regulates cell differentiation and apoptosis in brain cells [73,74] and may play a significant role in the development of type 2 diabetes [75]. Insulin has been shown to increase KLF10 expression in muscle tissues [76,77]. Finally, KLF10 gene variants even show a moderate correlation with the development of diabetes [78]. In general, KLF10 plays a role in activating the inflammatory response, including stress-induced inflammation, leading to increases in cardiovascular, pulmonary, and other organ diseases, autoimmune abnormalities, and diabetes.
Overview and conclusions
KLF10 was originally discovered in our laboratory as an immediate early response gene following TGFβ treatment of human osteoblasts [1]. We further characterized this gene as a member of the Krüppel-like family of transcription factors (KLF10). We have shown that KLF10 plays a major role in mediating the effects of TGFβ through regulation of the Smad signaling pathway. As shown in Figs. 3 and 4, KLF10 is known to induce and repress the expression of multiple genes in many cell types by binding to Sp1 sites on the DNA and interactions with other regulatory transcription factors. It functions as a regulator of gene transcription, inhibitor of cell proliferation, and an inducer of apoptosis, resulting in enhanced immune system and inflammatory processes, and thus enhancing cardiovascular disease (atherosclerosis, hypertrophic cardiomyopathy), diabetes, and cancer. References to these studies are listed in Fig. 7. It is not surprising that KLF10 expression is inversely correlated with the severity and stage of breast cancer, implying an important role for KLF10 as a tumor suppressor. Thus, KLF10 appears to play an important role in other types of cancer. Further experiments, including long-term studies in which the KLF10 KO mice are crossed with various mouse models of tumor formation will be necessary to definitively establish the role of KLF10 in cancer. To date, the ablation of the KLF10 gene in mice is known to result in multiple phenotypes including defects in the skeleton, heart, and connective tissues. KLF10 KO mice display a severe osteopenic phenotype, but only in female animals, suggesting a potential role for KLF10 in mediating estrogen signaling in bone. Conversely, male, but not female, KLF10 KO mice develop cardiac hypertrophy nearly identical to the human disease, hypertrophic cardiomyopathy. KLF10 KO mice have also been shown to have defects in the mechanical properties and healing potential of tendons. The known phenotypes of KLF10 KO mice are summarized in Fig. 4. Taken together these data implicate an important role for KLF10 in multiple biological processes and disease states. The redundancy of KLF10, KLF11, and TIEG3, and the specific roles of these proteins in a variety of physiological processes, remains unknown. Ongoing studies by other investigators are likely to identify novel functions of the KLF10 family of proteins and uncover consequences of abnormalities in the expression, structure, and/or function of this important class of transcription factors in other diseases.
Acknowledgments
NIH grant numbers R01 DE-14036 and AR52004; and The Breast Cancer Research Foundation
References
- 1.Subramaniam M, Harris SA, Oursler MJ, Rasmussen K, Riggs BL, Spelsberg TC. Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 1995;23:4907–4912. doi: 10.1093/nar/23.23.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blok LJ, Grossmann ME, Perry JE, Tindall DJ. Characterization of an early growth response gene, which encodes a zinc finger transcription factor, potentially involved in cell cycle regulation. Mol Endocrinol. 1995;9:1610–1620. doi: 10.1210/mend.9.11.8584037. [DOI] [PubMed] [Google Scholar]
- 3.Fautsch MP, Vrabel A, Subramaniam M, Hefferen TE, Spelsberg TC, Wieben ED. TGFbeta-inducible early gene (TIEG) also codes for early growth response alpha (EGRalpha): evidence of multiple transcripts from alternate promoters. Genomics. 1998;51:408–416. doi: 10.1006/geno.1998.5388. [DOI] [PubMed] [Google Scholar]
- 4.Gunther M, Laithier M, Brison O. A set of proteins interacting with transcription factor Sp1 identified in a two-hybrid screening. Mol Cell Biochem. 2000;210:131–142. doi: 10.1023/a:1007177623283. [DOI] [PubMed] [Google Scholar]
- 5.Fautsch MP, Vrabel A, Rickard D, Subramaniam M, Spelsberg TC, Wieben ED. Characterization of the mouse TGFbeta-inducible early gene (TIEG): conservation of exon and transcriptional regulatory sequences with evidence of additional transcripts. Mamm Genome. 1998;9:838–842. doi: 10.1007/s003359900878. [DOI] [PubMed] [Google Scholar]
- 6.Cook T, Gebelein B, Belal M, Mesa K, Urrutia R. Three conserved transcriptional repressor domains are a defining feature of the TIEG subfamily of Sp1-like zinc finger proteins. J Biol Chem. 1999;274:29500–29504. doi: 10.1074/jbc.274.41.29500. [DOI] [PubMed] [Google Scholar]
- 7.Hefferan TE, Reinholz GG, Rickard DJ, Johnsen SA, Waters KM, Subramaniam M, Spelsberg TC. Overexpression of a nuclear protein. TIEG, mimics transforming growth factor-beta action in human osteoblast cells. J Biol Chem. 2000;275:20255–20259. doi: 10.1074/jbc.C000135200. [DOI] [PubMed] [Google Scholar]
- 8.Leclerc N, Luppen CA, Ho VV, Nagpal S, Hacia JG, Smith E, Frenkel B. Gene expression profiling of glucocorticoid-inhibited osteoblasts. J Mol Endocrinol. 2004;33:175–193. doi: 10.1677/jme.0.0330175. [DOI] [PubMed] [Google Scholar]
- 9.Hofbauer LC, Hicok KC, Khosla S. Effects of gonadal and adrenal androgens in a novel androgen-responsive human osteoblastic cell line. J Cell Biochem. 1998;71:96–108. [PubMed] [Google Scholar]
- 10.Hawse JR, Subramaniam M, Monroe DG, Hemmingsen AH, Ingle JN, Khosla S, Oursler MJ, Spelsberg TC. Estrogen receptor beta isoform-specific induction of transforming growth factor beta-inducible early gene-1 in human osteoblast cells: an essential role for the activation function 1 domain. Mol Endocrinol. 2008;22:1579–1595. doi: 10.1210/me.2007-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tachibana I, Imoto M, Adjei PN, Gores GJ, Subramaniam M, Spelsberg TC, Urrutia R. Overexpression of the TGFbeta-regulated zinc finger encoding gene, TIEG, induces apoptosis in pancreatic epithelial cells. J Clin Invest. 1997;99:2365–2374. doi: 10.1172/JCI119418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cook T, Gebelein B, Mesa K, Mladek A, Urrutia R. Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-beta-inducible Sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J Biol Chem. 1998;273:25929–25936. doi: 10.1074/jbc.273.40.25929. [DOI] [PubMed] [Google Scholar]
- 13.Subramaniam M, Hefferan TE, Tau K, Peus D, Pittelkow M, Jalal S, Riggs BL, Roche P, Spelsberg TC. Tissue, cell type, and breast cancer stage-specific expression of a TGF-beta inducible early transcription factor gene. J Cell Biochem. 1998;68:226–236. [PubMed] [Google Scholar]
- 14.Kobori N, Clifton GL, Dash P. Altered expression of novel genes in the cerebral cortex following experimental brain injury. Brain Res Mol Brain Res. 2002;104:148–158. doi: 10.1016/s0169-328x(02)00331-5. [DOI] [PubMed] [Google Scholar]
- 15.Noti JD, Johnson AK, Dillon JD. The zinc finger transcription factor transforming growth factor beta-inducible early gene-1 confers myeloid-specific activation of the leukocyte integrin CD11d promoter. J Biol Chem. 2004;279:26948–26958. doi: 10.1074/jbc.M310634200. [DOI] [PubMed] [Google Scholar]
- 16.Noti JD, Johnson AK, Dillon JD. The leukocyte integrin gene CD11d is repressed by gut-enriched Kruppel-like factor 4 in myeloid cells. J Biol Chem. 2005;280:3449–3457. doi: 10.1074/jbc.M412627200. [DOI] [PubMed] [Google Scholar]
- 17.Luo X, Ding L, Xu J, Chegini N. Gene expression profiling of leiomyoma and myometrial smooth muscle cells in response to transforming growth factor-beta. Endocrinology. 2005;146:1097–1118. doi: 10.1210/en.2004-1377. [DOI] [PubMed] [Google Scholar]
- 18.Mitsumoto M, Mitsumoto A, Demple B. Nitric oxide-mediated upregulation of the TGF-beta-inducible early response gene-1 (TIEG1) in human fibroblasts by mRNA stabilization independent of TGFbeta. Free Radic Biol Med. 2003;34:1607–1613. doi: 10.1016/s0891-5849(03)00211-9. [DOI] [PubMed] [Google Scholar]
- 19.Yajima S, Lammers CH, Lee SH, Hara Y, Mizuno K, Mouradian MM. Cloning and characterization of murine glial cell-derived neurotrophic factor inducible transcription factor (MGIF) J Neurosci. 1997;17:8657–8666. doi: 10.1523/JNEUROSCI.17-22-08657.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bender H, Wang Z, Schuster N, Krieglstein K. TIEG1 facilitates transforming growth factor-beta-mediated apoptosis in the oligodendroglial cell line OLI-neu. J Neurosci Res. 2004;75:344–352. doi: 10.1002/jnr.10856. [DOI] [PubMed] [Google Scholar]
- 21.Reinholz MM, An MW, Johnsen SA, Subramaniam M, Suman VJ, Ingle JN, Roche PC, Spelsberg TC. Differential gene expression of TGF beta inducible early gene (TIEG). Smad7, Smad2 and Bard1 in normal and malignant breast tissue. Breast Cancer Res Treat. 2004;86:75–88. doi: 10.1023/B:BREA.0000032926.74216.7d. [DOI] [PubMed] [Google Scholar]
- 22.Johnsen SA, Subramaniam M, Monroe DG, Janknecht R, Spelsberg TC. Modulation of transforming growth factor beta (TGFbeta)/Smad transcriptional responses through targeted degradation of TGF beta-inducible early gene-1 by human seven in absentia homologue. J Biol Chem. 2002;277:30754–30759. doi: 10.1074/jbc.M204812200. [DOI] [PubMed] [Google Scholar]
- 23.Ou XM, Chen K, Shih JC. Dual functions of transcription factors, transforming growth factor-beta-inducible early gene (TIEG)2 and Sp3, are mediated by CACCC element and Sp1 sites of human monoamine oxidase (MAO) B gene. J Biol Chem. 2004;279:21021–21028. doi: 10.1074/jbc.M312638200. [DOI] [PubMed] [Google Scholar]
- 24.House CM, Hancock NC, Moller A, Cromer BA, Fedorov V, Bowtell DD, Parker MW, Polekhina G. Elucidation of the substrate binding site of Siah ubiquitin ligase. Structure. 2006;14:695–701. doi: 10.1016/j.str.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 25.Jin W, Di G, Li J, Chen Y, Li W, Wu J, Cheng T, Yao M, Shao Z. TIEG1 induces apoptosis through mitochondrial apoptotic pathway and promotes apoptosis induced by homoharringtonine and velcade. FEBS Lett. 2007;581:3826–3832. doi: 10.1016/j.febslet.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Cook T, Urrutia R. TIEG proteins join the Smads as TGF-beta-regulated transcription factors that control pancreatic cell growth. Am J Physiol Gastrointest Liver Physiol. 2000;278:G513–521. doi: 10.1152/ajpgi.2000.278.4.G513. [DOI] [PubMed] [Google Scholar]
- 27.Ribeiro A, Bronk SF, Roberts PJ, Urrutia R, Gores GJ. The transforming growth factor beta(1)-inducible transcription factor TIEG1. mediates apoptosis through oxidative stress. Hepatology. 1999;30:1490–1497. doi: 10.1002/hep.510300620. [DOI] [PubMed] [Google Scholar]
- 28.Wang Z, Peters B, Klussmann S, Bender H, Herb A, Krieglstein K. Gene structure and evolution of Tieg3, a new member of the Tieg family of proteins. Gene. 2004;325:25–34. doi: 10.1016/j.gene.2003.09.045. [DOI] [PubMed] [Google Scholar]
- 29.Zhang JS, Moncrieffe MC, Kaczynski J, Ellenrieder V, Prendergast FG, Urrutia R. A conserved alpha-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol Cell Biol. 2001;21:5041–5049. doi: 10.1128/MCB.21.15.5041-5049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellenrieder V, Zhang JS, Kaczynski J, Urrutia R. Signaling disrupts mSin3A binding to the Mad1-like Sin3-interacting domain of TIEG2, an Sp1-like repressor. EMBO J. 2002;21:2451–2460. doi: 10.1093/emboj/21.10.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.House CM, Frew IJ, Huang HL, Wiche G, Traficante N, Nice E, Catimel B, Bowtell DD. A binding motif for Siah ubiquitin ligase. Proc Natl Acad Sci USA. 2003;100:3101–3106. doi: 10.1073/pnas.0534783100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tau KR, Hefferan TE, Waters KM, Robinson JA, Subramaniam M, Riggs BL, Spelsberg TC. Estrogen regulation of a transforming growth factor-beta inducible early gene that inhibits deoxyribonucleic acid synthesis in human osteoblasts. Endocrinology. 1998;139:1346–1353. doi: 10.1210/endo.139.3.5830. [DOI] [PubMed] [Google Scholar]
- 33.Johnsen SA, Subramaniam M, Janknecht R, Spelsberg TC. TGFbeta inducible early gene enhances TGFbeta/Smad-dependent transcriptional responses. Oncogene. 2002;21:5783–5790. doi: 10.1038/sj.onc.1205681. [DOI] [PubMed] [Google Scholar]
- 34.Johnsen SA, Subramaniam M, Katagiri T, Janknecht R, Spelsberg TC. Transcriptional regulation of Smad2 is required for enhancement of TGFbeta/Smad signaling by TGFbeta inducible early gene. J Cell Biochem. 2002;87:233–241. doi: 10.1002/jcb.10299. [DOI] [PubMed] [Google Scholar]
- 35.Hawse JR, Iwaniec UT, Bensamoun SF, Monroe DG, Peters KD, Ilharreborde B, Rajamannan NM, Oursler MJ, Turner RT, Spelsberg TC, Subramaniam M. TIEG-null mice display an osteopenic gender-specific phenotype. Bone. 2008;42:1025–1031. doi: 10.1016/j.bone.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chalaux E, Lopez-Rovira T, Rosa JL, Pons G, Boxer LM, Bartrons R, Ventura F. A zinc-finger transcription factor induced by TGF-beta promotes apoptotic cell death in epithelial Mv1Lu cells. FEBS Lett. 1999;457:478–482. doi: 10.1016/s0014-5793(99)01051-0. [DOI] [PubMed] [Google Scholar]
- 37.Sebestyen A, Barna G, Nagy K, Janosi J, Paku S, Kohut E, Berczi L, Mihalik R, Kopper L. Smad signal and TGFbeta induced apoptosis in human lymphoma cells. Cytokine. 2005;30:228–235. doi: 10.1016/j.cyto.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 38.Jin W, Qu LF, Min P, Chen S, Li H, Lu H, Hou YT. Identification of genes responsive to apoptosis in HL-60 cells. Acta Pharmacol Sin. 2004;25:319–326. [PubMed] [Google Scholar]
- 39.Franke AG, Gubbe C, Beier M, Duenker N. Transforming growth factor-beta and bone morphogenetic proteins: cooperative players in chick and murine programmed retinal cell death. J Comp Neurol. 2006;495:263–278. doi: 10.1002/cne.20869. [DOI] [PubMed] [Google Scholar]
- 40.Johnsen SA, Subramaniam M, Effenberger KE, Spelsberg TC. The TGF-beta inducible early gene plays a central role in the anti-proliferative response to TGF-beta. Signal Transduct. 2004;4:29–35. [Google Scholar]
- 41.Subramaniam M, Hawse JR, Johnsen SA, Spelsberg TC. Role of TIEG1 in biological processes and disease states. J Cell Biochem. 2007;102:539–548. doi: 10.1002/jcb.21492. [DOI] [PubMed] [Google Scholar]
- 42.Yerges LM, Klei L, Cauley JA, Roeder K, Kammerer CM, Ensrud KE, Nestlerode CS, Lewis C, Lang TF, Barrett-Connor E, Moffett SP, Hoffman AR, Ferrell RE, Orwoll ES, Zmuda JM. Candidate gene analysis of femoral neck trabecular and cortical volumetric bone mineral density in older men. J Bone Miner Res. doi: 10.1359/jbmr.090729. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bensamoun SF, Hawse JR, Subramaniam M, Ilharreborde B, Bassillais A, Benhamou CL, Fraser DG, Oursler MJ, Amadio PC, An KN, Spelsberg TC. TGFbeta inducible early gene-1 knockout mice display defects in bone strength and microarchitecture. Bone. 2006a;39:1244–1251. doi: 10.1016/j.bone.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 44.Subramaniam M, Gorny G, Johnsen SA, Monroe DG, Evans GL, Fraser DG, Rickard DJ, Rasmussen K, van Deursen JM, Turner RT, Oursler MJ, Spelsberg TC. TIEG1 null mouse-derived osteoblasts are defective in mineralization and in support of osteoclast differentiation in vitro. Mol Cell Biol. 2005;25:1191–1199. doi: 10.1128/MCB.25.3.1191-1199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haddad O, Hawse JR, Subramaniam M, Spelsberg TC, Bensamoun SF. TIEG1-null osteocytes display defects in their morphology, density and surrounding bone matrix. J Musculoskelet Res. doi: 10.80/10255840903081164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsubone T, Moran SL, Subramaniam M, Amadio PC, Spelsberg TC, An KN. Effect of TGF-beta inducible early gene deficiency on flexor tendon healing. J Orthop Res. 2006;24:569–575. doi: 10.1002/jor.20101. [DOI] [PubMed] [Google Scholar]
- 47.Bensamoun SF, Tsubone T, Subramaniam M, Hawse JR, Boumediene E, Spelsberg TC, An KN, Amadio PC. Age-dependent changes in the mechanical properties of tail tendons in TGF-beta inducible early gene-1 knockout mice. J Appl Physiol, tot. 2006b;101:1419–1424. doi: 10.1152/japplphysiol.00800.2005. [DOI] [PubMed] [Google Scholar]
- 48.Rajamannan NM, Subramaniam M, Abraham TP, Vasile VC, Ackerman MJ, Monroe DG, Chew TL, Spelsberg TC. TGFbeta inducible early gene-1 (TIEG1) and cardiac hypertrophy: discovery and characterization of a novel signaling pathway. J Cell Biochem. 2007;100:315–325. doi: 10.1002/jcb.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J, Xu N, Feng X, Hou N, Zhang J, Cheng X, Chen Y, Zhang Y, Yang X. Targeted disruption of Smad4 in cardiomyocytes results in cardiac hypertrophy and heart failure. Circ Res. 2005;97:821–828. doi: 10.1161/01.RES.0000185833.42544.06. [DOI] [PubMed] [Google Scholar]
- 50.Cao Z, Wara AK, Icli R, Sun X, Packard RR, Esen F, Stapleton CJ, Subramaniam M, Kretschmer K, Apostolou I, von Boehmer H, Hansson GK, Spelsberg TC, Libby P, Feinberg MW. Kruppel-{beta}1 to regulate CD4+CD25− T cells and T regulatory cells. J Biol Chem. doi: 10.74/jbc.M109.000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venuprasad K, Huang H, Harada Y, Elly C, Subramaniam M, Spelsberg T, Su J, Liu YC. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat Immunol. 2008;9:245–253. doi: 10.1038/niXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eid MA, Kumar MV, Iczkowski KA, Bostwick DG, Tindall DJ. Expression of early growth response genes in human prostate cancer. Cancer Res. 1998;58:2461–2468. [PubMed] [Google Scholar]
- 53.Ellenrieder V, Fernandez-Zapico ME, Urrutia R. TGF-beta mediated signaling and transcriptional regulation in pancreatic development and cancer. Curr Opin Gastroenterol. 2001;17:434–440. doi: 10.1097/00001574-200109000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ellenrieder V. TGFbeta regulated gene expression by Smads and Sp1/KLF-like transcription factors in cancer. Anticancer Res. 2008;28:1531–1539. [PubMed] [Google Scholar]
- 55.Sorbello V, Fuso L, Sfiligoi C, Scafoglio C, Ponzone R, Biglia N, Weisz A, Sismondi P, De Bortoli M. Quantitative real-time RT-PCR analysis of eight novel estrogen-regulated genes in breast cancer. Int J Biol Markers. 2003;18:123–129. doi: 10.1177/172460080301800205. [DOI] [PubMed] [Google Scholar]
- 56.Subramaniam M, Oursler MJ, Rasmussen K, Riggs BL, Spelsberg TC. TGF-beta regulation of nuclear proto-oncogenes and TGF-beta gene expression in normal human osteoblast-like cells. J Cell Biochem. 1995;57:52–61. doi: 10.1002/jcb.240570107. [DOI] [PubMed] [Google Scholar]
- 57.Zohrabian VM, Nandu H, Gulati N, Khitrov G, Zhao C, Mohan A, Demattia J, Braun A, Das K, Murali R, Jhanwar-Uniyal M. Gene expression profiling of metastatic brain cancer. Oncol Rep. 2007;18:321–328. [PubMed] [Google Scholar]
- 58.Hu G, Chong RA, Yang Q, Wei Y, Blanco MA, Li F, Reiss M, Au JL, Haffty BG, Kang Y. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15:9–20. doi: 10.1016/j.ccr.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang L, Chen Y, Chan CY, Wang X, Lin L, He ML, Lin MC, Yew DT, Sung JJ, Li JC, Kung HF. Down-regulation of stathmin is required for TGF-beta inducible early gene 1 induced growth inhibition of pancreatic cancer cells. Cancer Lett. 2009;274:101–108. doi: 10.1016/j.canlet.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 60.Partin JV, Anglin IE, Kyprianou N. Quinazoline-based alpha 1-adrenoceptor antagonists induce prostate cancer cell apoptosis via TGF-beta signalling and I kappa B alpha induction. Br J Cancer. 2003;88:1615–1621. doi: 10.1038/sj.bjc.6600961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yono M, Mane SM, Lin A, Weiss RM, Latifpour J. Differential effects of diabetes induced by streptozotocin and that develops spontaneously on prostate growth in Bio Breeding (BB) rats. Life Sci. 2008;83:192–197. doi: 10.1016/j.lfs.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 62.Antonello D, Moore PS, Zamboni G, Falconi M, Scarpa A. Absence of mutations in the transforming growth factor-beta inducible early gene 1, TIEG1. in pancreatic cancer. Cancer Lett. 2002;183:179–183. doi: 10.1016/s0304-3835(01)00802-3. [DOI] [PubMed] [Google Scholar]
- 63.Chen GG, Xu H, Lee JF, Subramaniam M, Leung KL, Wang SH, Chan UP, Spelsberg TC. 15-hydroxy-eicosatetraenoic acid arrests growth of colorectal cancer cells via a peroxisome proliferator-activated receptor gamma-dependent pathway. Int J Cancer. 2003;107:837–843. doi: 10.1002/ijc.11447. [DOI] [PubMed] [Google Scholar]
- 64.Barna G, Sebestyen A, Chinopoulos CC, Nagy K, Mihalik R, Paku S, Kopper L. TGF beta 1 kills lymphoma cells using mitochondrial apoptotic pathway with the help of caspase-8. Anticancer Res. 2002;22:3867–3872. [PubMed] [Google Scholar]
- 65.Ferraro B, Bepler G, Sharma S, Cantor A, Haura EB. EGR1 predicts PTEN and survival in patients with non-small-cell lung cancer. J Clin Oncol. 2005;23:1921–1926. doi: 10.1200/JCO.2005.08.127. [DOI] [PubMed] [Google Scholar]
- 66.Ivanov SV, Ivanova AV, Salnikow K, Timofeeva O, Subramaniam M, Lerman MI. Two novel VHL targets, TGFBI (BIGH3) and its transactivator KLF10, are up-regulated in renal clear cell carcinoma and other tumors. Biochem Biophys Res Commun. 2008;370:536–540. doi: 10.1016/j.bbrc.2008.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou FL, Zhang WG, Chen G, Zhao WH, Cao XM, Chen YX, Tian W, Liu J, Liu SH. Serological identification and bio-informatics analysis of immunogenic antigens in multiple myeloma. Cancer Immunol Immunother. 2006;55:910–917. doi: 10.1007/s00262-005-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chiambaretta F, De Graeve F, Turet G, Marceau G, Gain P, Dastugue B, Rigal D, Sapin V. Cell and tissue specific expression of human Kruppel-like transcription factors in human ocular surface. Mol Vis. 2004;10:901–909. [PubMed] [Google Scholar]
- 69.Duenker N, Valenciano AI, Franke A, Hernandez-Sanchez C, Dressel R, Behrendt M, De Pablo F, Krieglstein K, de la Rosa EJ. Balance of pro-apoptotic transforming growth factor-beta and anti-apoptotic insulin effects in the control of cell death in the postnatal mouse retina. Eur J Neurosci. 2005;22:28–38. doi: 10.1111/j.1460-9568.2005.04183.x. [DOI] [PubMed] [Google Scholar]
- 70.Alme MN, Wibrand K, Dagestad G, Bramham CR. Chronic fluoxetine treatment induces brain region-specific upregulation of genes associated with BDNF-induced long-term potentiation. Neurol Plast. 2007:26496. doi: 10.1155/2007/26496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Consales C, Volpicelli F, Greco D, Leone L, Colucci-D’Amato L, Perrone-Capano C, di Porzio U. GDNF signaling in embryonic midbrain neurons in vitro. Brain Res. 2007;1159:28–39. doi: 10.1016/j.brainres.2007.04.071. [DOI] [PubMed] [Google Scholar]
- 72.Dijkmans TF, van Hooijdonk LW, Schouten TG, Kamphorst JT, Fitzsimons CP, Vreugdenhil E. Identification of new nerve growth factor-responsive immediate-early genes. Brain Res. 2009;1249:19–33. doi: 10.1016/j.brainres.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 73.Alvarez-Rodriguez R, Barzi M, Berenguer J, Pons S. Bone morphogenetic protein 2 opposes Shh-mediated proliferation in cerebellar granule cells through a TIEG-1-based regulation of Nmyc. J Biol Chem. 2007;282:37170–37180. doi: 10.1074/jbc.M705414200. [DOI] [PubMed] [Google Scholar]
- 74.Gohla G, Krieglstein K, Spittau B. Tieg3/Klf11 induces apoptosis in OLI-neu cells and enhances the TGF-beta signaling pathway by transcriptional repression of Smad7. J Cell Biochem. 2008;104:850–861. doi: 10.1002/jcb.21669. [DOI] [PubMed] [Google Scholar]
- 75.Wahab NA, Weston BS, Mason RM. Modulation of the TGFbeta/Smad signaling pathway in mesangial cells by CTGF/CCN2. Exp Cell Res. 2005;307:305–314. doi: 10.1016/j.yexcr.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 76.Coletta DK, Balas B, Chavez AO, Baig M, Abdul-Ghani M, Kashyap SR, Folli F, Tripathy D, Mandarino LJ, Cornell JE, Defronzo RA, Jenkinson CP. Effect of acute physiological hyperinsulinemia on gene expression in human skeletal muscle in vivo. Am J Physiol. 2008;294:E910–E917. doi: 10.1152/ajpendo.00607.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hansen L, Gaster M, Oakeley EJ, Brusgaard K, Damsgaard Nielsen EM, Beck-Nielsen H, Pedersen O, Hemmings BA. Expression profiling of insulin action in human myotubes: induction of inflammatory and pro-angiogenic pathways in relationship with glycogen synthesis and type 2 diabetes. Biochem Biophys Res Commun. 2004;323:685–695. doi: 10.1016/j.bbrc.2004.08.146. [DOI] [PubMed] [Google Scholar]
- 78.Gutierrez-Aguilar R, Benmezroua Y, Balkau B, Marre M, Helbecque N, Charpentier G, Polychronakos C, Sladek R, Froguel P, Neve B. Minor contribution of 5MAD7 and KLF10 variants to genetic susceptibility of type 2 diabetes. Diabetes Metab. 2007;33:372–378. doi: 10.1016/j.diabet.2007.06.002. [DOI] [PubMed] [Google Scholar]


