Abstract
The interaction of G protein-coupled receptors (GPCRs) with heterotrimeric G proteins represents one of the most fundamental biological processes. However, the molecular architecture of the GPCR/G protein complex remains poorly defined. In the present study, we applied a comprehensive GPCR/Gα chemical cross-linking strategy to map a receptor/Gα interface, both prior to and after agonist-induced receptor activation. By employing the M3 muscarinic acetylcholine receptor (M3R)/Gαq system as a model system, we examined the ability of ~250 combinations of Cys-substituted M3R and Gαq proteins to undergo cross-link formation. We identified many specific M3R/Gαq contact sites, both in the inactive and the active receptor conformation, allowing us to draw conclusions regarding the basic architecture of the M3R/Gαq interface and the nature of the conformational changes following receptor activation. Since heterotrimeric G proteins as well as most GPCRs share a high degree of structural homology, our findings should be of broad general relevance.
INTRODUCTION
G protein-coupled receptors (GPCRs) are involved in regulating nearly all known physiological functions and represent excellent drug targets1–3. All GPCRs are predicted to contain a transmembrane (TM) core formed by a bundle of seven TM helices (TM1-7) which are connected by three extracellular and three intracellular loops (Fig. 1a). Physiologically, GPCRs are activated by extracellular ligands which enable the receptors to interact with and activate distinct sets of heterotrimeric G proteins (Gαβγ). Specifically, ligand-activated GPCRs catalyze the exchange of GDP for GTP on the Gα subunit. GTP binding to Gα is predicted to trigger the dissociation of the heterotrimeric G protein into Gα-GTP and free βγ which are then able to modulate the activity of a multitude of downstream effector enzymes and ion channels4.
Figure 1.
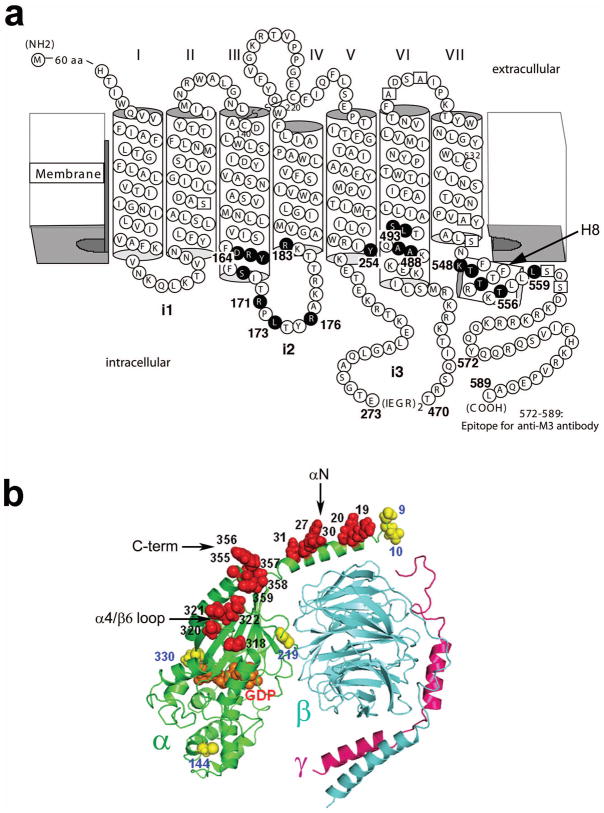
Introduction of Cys substitutions into the M3R and Gαq. (a) Secondary structure of a modified version of the rat M3R (M3′(3C)-Xa). All Cys substitutions were introduced into the M3′ (3C)-Xa construct23 that lacked most endogenous Cys residues and the central portion of the i3 loop (A274–K469; see Methods for details). Endogenous Cys residues were substituted with serine or alanine (open squares), except for C140, C220 and C532 which proved to be essential for proper receptor function23. Amino acids that were replaced with Cys residues are highlighted by black circles. The numbering of M3R residues is based on the sequence of the full-length rat M3R. (b) Model of human Gαq (green) in complex with a βγ dimer (β, cyan; γ, purple). The five endogenous Cys residues present in Gαq are highlighted in yellow. We introduced Cys substitutions into a modified version of Gαq (Gαq-C-less) in which C144, C219, and C330 had been replaced with alanine (C9 and C10 were found to be critical for Gαq function and were therefore left intact). Amino acids in Gαq that were subjected to Cys substitution mutagenesis are highlighted in red. The model of the Gq trimer was generated as described under Methods.
High-resolution X-ray structures have been obtained for several class I GPCRs (‘rhodopsin-like’ GPCRs)5–11 and various G protein heterotrimers (αβγ) and isolated Gα subunits in different functional states (for recent reviews, see refs. 12–14). Combined with biochemical and biophysical data12–14, these structures reveal a surface on Gα that is predicted to face the intracellular side of GPCRs (see below for more details). However, a high-resolution crystal structure of a GPCR/G protein complex is not available at present.
Many studies have shown that multiple GPCR regions participate in receptor/G protein interactions15. These receptor regions include the TM3/i2 loop junction, the i2 loop, the cytoplasmic ends of TM5 and TM6, and helix 8 (H8; Fig 1a). Similarly, various experimental approaches have implicated multiple Gα regions in productive receptor/G protein coupling12–14. The three key regions of Gα that have been implicated by most studies include the N-terminal helix (αN; this helix is only observed in the presence of bound βγ), the α4/β6 loop, and the C-terminal portion11–14,16–21.
Biochemical and biophysical approaches have identified a small number of receptor/Gα interaction sites using specific receptor/Gα combinations11,19–21. However, the molecular architecture of the receptor/Gα interface still remains poorly defined, primarily due to the lack of studies examining receptor/Gα contact sites in a systematic and comprehensive fashion. However, such knowledge is essential for understanding the structural basis underlying one of the most fundamental biological processes.
To address this question, we used the rat M3 muscarinic acetylcholine receptor (M3R), a prototypic Gq-coupled receptor22, as a model system. We carried out systematic cross-linking experiments using Cys-substituted M3R and Gαq constructs, both in the absence and presence of an activating ligand. To facilitate the interpretation of cross-linking data, we employed a modified version of the rat M3R lacking most native Cys residues (M3′(3C)-Xa; Fig. 1a)23. The M3′(3C)-Xa construct lacked the central portion of the third intracellular loop (i3 loop; A274–K469), resulting in the removal of three Cys residues contained within this region23. However, since this construct still retained the functionally critical N- and C-terminal portions of the i3 loop (Fig. 1a)22, it can couple to Gq-type G protein with similar efficacy as the wild-type M3R23. Moreover, seven of the ten remaining endogenous M3R Cys residues were substituted with serine or alanine (Fig. 1a)23. A modified version of human Gαq (‘Gαq-C-less’) in which we had replaced three of the five endogenous Cys residues with alanine served as a template for Cys substitution mutagenesis (Fig. 1b). In total, we examined cross-link formation between 18 mutant M3Rs and 14 mutant Gαq subunits (all constructs contained single Cys substitutions), in all possible combinations. These studies yielded many specific M3R/Gαq contact sites, both in the absence and presence of activating ligands, providing novel insights into the molecular architecture of the M3R/Gq interface and the potential mechanisms governing receptor-mediated G protein activation.
RESULTS
Generation of Cys-substituted mutant M3Rs
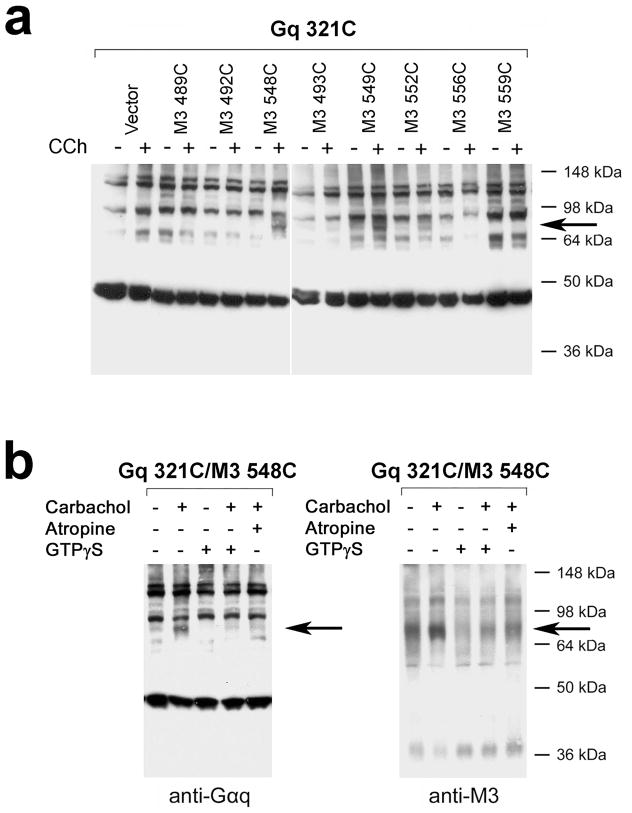
We introduced Cys substitutions into regions/sites of the M3′(3C)-Xa receptor known to be critical for M3R/Gq coupling (Fig. 1a)22. Recent studies carried out with several class I GPCRs strongly suggest that the cytoplasmic side of H8 is also critically involved in receptor/G protein interactions15,24–27. For this reason, we also targeted the corresponding M3R residues (K548, T549, T552, T556, and L559) by Cys substitution mutagenesis (Fig. 1a).
Pharmacological properties of Cys-substituted mutant M3Rs
We first examined whether the various Cys-substituted mutant M3Rs retained the ability to bind muscarinic ligands with high affinity. Radioligand binding studies with membranes prepared from receptor-expressing COS-7 cells showed that all mutant M3Rs receptors were able to bind the muscarinic antagonist, [3H]N-methylscopolamine ([3H]NMS), with high affinity (range of [3H]NMS KD values: 163–877 pM; Supplementary Table 1). All mutant M3Rs were expressed at high density (range of [3H]NMS Bmax values: 5.4–16.4 pmol/mg), except for the D164C and Y166C receptors which were expressed at relatively low levels (Bmax~ 1 pmol/mg; Supplementary Table 1). Moreover, [3H]NMS/carbachol inhibition binding studies demonstrated that the different mutant M3Rs were able to bind the muscarinic agonist, carbachol, with affinities that were similar to or even higher than that observed with the M3′(3C)-Xa receptor from which all mutant receptors were derived (range of carbachol Ki values: 2.5–44.5 μM; Supplementary Table 1).
To examine whether the Cys-substituted mutant M3Rs retained the ability to couple to Gq-type G proteins, we studied their ability to mediate carbachol-induced increases in inositol phosphate (IP) production. Carbachol stimulation of receptor-expressing COS-7 cells led to pronounced increases in IP accumulation in all mutant receptors studied (Supplementary Table 1). However, several of the mutant receptors, including D164C, R165C, L173C, and Y254C, showed pronounced decreases in carbachol potency (increase in EC50) and/or efficacy (decrease in Emax), as compared to the M3′ (3C)-Xa construct (Supplementary Table 1). This observation is consistent with the key roles of these residues in modulating the efficiency of receptor/G protein interactions22.
Generation of Cys-substituted mutant Gαq proteins
Human Gαq contains five endogenous Cys residues which are located at positions 9, 10, 144, 219, and 330 (highlighted in yellow in Fig. 1b). We generated a mutant version of Gαq by replacing C144, C219, and C330 with alanine. In the following, we refer to this construct as ‘Gαq-C-less’. This construct still contained two native Cys residues, C9 and C10, which proved to be essential for Gαq function (assessed in a second messenger assay; see below). In fact, it has been demonstrated that these two Cys residues are subject to palmitoylation which is required for proper Gq localization and function28,29.
By using the Gαq-C-less construct as a template, we introduced single Cys substitutions into Gαq domains that have been implicated in receptor/G protein interactions12–18. We first replaced the C-terminal five amino acids of Gαq-C-less individually with Cys (E355C, Y356C, N357C, L358C, and V359C; Fig. 1b). Guided by a model of the Gq heterotrimer (see Methods for details), we also introduced single Cys residues into surface-exposed sites of the N-terminal domain (R19C, R20C, R27C, R30C, and R31C) and the α4/β6 loop (P318C, S320C, D321C, and K322C; Fig. 1b).
All mutant Gαq subunits retain functional activity
To study whether the various mutant Gαq constructs retained functional activity, we co-transfected HEK 293 cells with the different mutant Gαq constructs and a plasmid coding for the porcine α2A-adrenergic receptor (α2A-AR). Importantly, in this system, the α2A-AR agonist, UK14304, triggers IP accumulation only in the presence of co-transfected Gαq (Supplementary Table 2)30. We found that all 14 Cys mutant Gαq constructs displayed robust functional activity in this assay system (Supplementary Table 2), indicating that the various Cys substitutions did not disrupt receptor/Gq and Gαq/effector (phospholipase Cβ) coupling.
General strategy used to identify M3R/Gαq contact sites
To detect specific M3R/Gαq contact sites, we co-expressed the 18 Cys mutant M3Rs with the 14 Cys mutant Gαq constructs, in all possible combinations (252 total). Subsequently, we exposed membranes prepared from co-transfected COS-7 cells to the oxidizing agent Cu(II)-(1,10-phenanthroline)3 (CuPhen; 100 μM) to promote the formation of disulfide bonds between vicinal Cys residues. We monitored the successful formation of disulfide bonds by visualizing cross-linked receptor/G protein complexes via Western blotting carried out under non-reducing conditions. Since the Gαq- and M3′(3C)-Xa-based constructs both have a molecular mass of ~40 kDa, the cross-linked receptor/Gαq complexes are predicted to have a size of ~80 kDa. All cross-linking reactions were carried out in the absence or presence of the muscarinic agonist, carbachol.
Cross-linking analysis of C-terminal mutant Gαq subunits
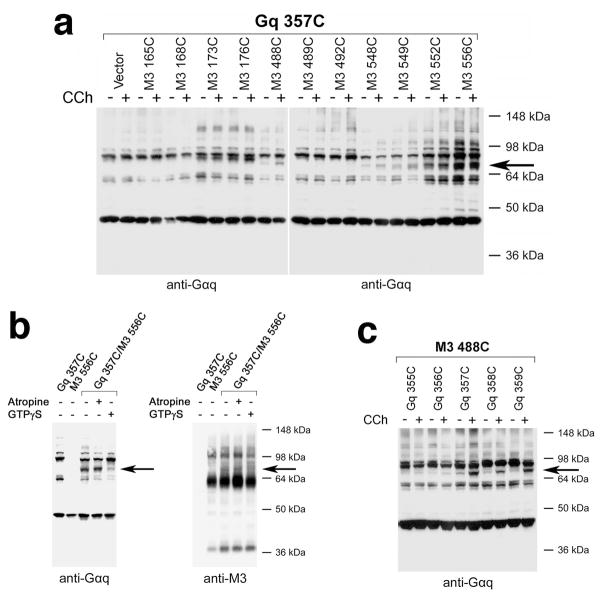
The five Gαq constructs containing C-terminal Cys substitutions (E355C, Y356C, N357C, L358C, and V359C) gave qualitatively similar cross-liking patterns (see next paragraph). The cross-linked receptor/Gαq complexes (~80 kDa in size) are indicated by arrows in Fig. 2a which shows cross-linking data obtained with a representative Gαq construct (Gαq-L357C; for additional cross-linking data see Supplementary Figs. 1, 2). As expected, these bands could be detected by both anti-Gαq and anti-M3R antibodies (shown for the representative Gαq-L357C/M3R-T556C combination in Fig. 2b). We did not observe any ~80 kDa bands when cells had been transfected with different mutant G protein or M3R constructs alone (Fig. 2b; also see Supplementary Fig. 3). These findings convincingly demonstrate that the ~80 kDa bands correspond to cross-linked receptor/Gαq complexes. Both the anti-Gαq and anti-M3R antibodies detected several additional immunoreactive bands which are likely to present non-crosslinked Gαq and M3R species as well as adducts with other proteins, consistent with the outcome of co-immunoprecipitation studies (see below).
Figure 2.
Cross-linking of Gαq subunits containing C-terminal Cys substitutions with Cyssubstituted mutant M3Rs. (a) Cross-link formation between the N357C Gαq construct and selected mutant M3Rs. (b) Effect of atropine and GTPγS treatment on cross-link formation between N357C-Gαq and the T556C mutant M3R. (c) Agonist-promoted cross-link formation between Gαq subunits containing C-terminal Cys substitutions and the A488C mutant M3R. To induce cross-link formation, membrane proteins prepared from transfected COS-7 cells were incubated with 100 μM Cu-Phen, either in the absence (−) or presence (+) of 1 mM carbachol (CCh) (a, c). The cross-linking data shown in panel (b) were obtained in the absence of carbachol. Note that GTPγS (100 μM) treatment greatly reduced the intensities of the observed cross-linking signals, while atropine (10 μM), an inverse muscarinic agonist, had no effect on the efficiency of cross-link formation (b). Western blotting studies were carried out under non-reducing conditions. The antibodies used are indicated underneath the blots. The blots shown are representative of two to four independent experiments. The bands corresponding to cross-linked receptor/Gαq complexes are indicated by arrows.
All five Gαq subunits containing C-terminal Cys substitutions (E355C, Y356C, N357C, L358C, and V359C) could be cross-linked to multiple mutant M3Rs. These mutant M3Rs contained Cys substitutions in the i2 loop (L173C and R176C) and within the N-terminal portion of H8 (T549C, T552C, and T556C) (Fig. 2a; Supplementary Figs. 1, 3, and 4). All five mutant Gαq subunits cross-linked most efficiently with the T556C mutant M3R (Fig. 2a; Supplementary Fig. 1). All Cu-Phen-induced cross-links were confirmed by using the short, bi-functional, and irreversible chemical cross-linker, bis-maleimidoethane (BMOE; 0.5 mM), which can cross-link vicinal Cys residues (shown for a representative receptor/G protein combination, Gαq-L358C/M3R-T556C, in Supplementary Fig. 3b).
Interestingly, essentially all cross-links observed in the absence of the muscarinic agonist, carbachol, were also detectable in the presence of this ligand (Fig. 2a; Supplementary Fig. 1). One possible explanation for this finding is that the M3R is pre-coupled to Gq prior to receptor activation, and that M3R activation by carbachol does not lead to the dissociation of the receptor/Gq complex due to the lack of GTP (or other guanine nucleotides) in the incubation buffer. Consistent with this concept, pretreatment of membrane preparations with GTPγS (100 μM), a hydrolytically stable GTP analog predicted to promote the dissociation of G protein heterotrimers and receptor/G protein complexes by displacing GDP from the Gα subunit, reduced the intensities of the observed receptor/Gαq cross-linking signals by ~40–70% (n=4; shown for the Gαq-L357C/M3RT556C combination in Fig. 2b; also see Supplementary Fig. 3c).
To exclude the possibility that the cross-linking signals observed in the absence of carbachol were caused by agonist-independent receptor signaling (constitutive M3R activity), we carried out cross-linking studies in the presence of atropine (10 μM), an inverse muscarinic agonist31,32. We found that atropine treatment had no significant effect on the intensities of the observed disulfide cross-linking signals (shown for the Gαq-L357C/M3R-T556C combination in Fig. 2b; also see Supplementary Fig. 3c), suggesting that the cross-linking pattern obtained in the absence of carbachol reflects the structure of the inactive M3R/Gq complex.
Although most cross-links involving the C-terminus of Gαq occurred with similar efficiency in the absence or presence of carbachol, we noted that carbachol treatment greatly enhanced the efficiency of disulfide cross-linking in several cases. We found that carbachol strongly promoted cross-linking between the A488C mutant M3R and the last three amino acids of Gαq (N357C, L358C, and V359C; Fig. 2c). Agonist treatment also led to slightly stronger cross-linking signals between the T549C and T552C mutant M3Rs and several Gαq subunits containing C-terminal Cys substitutions (Supplementary Fig. 4).
Co-immunoprecipitation and mass spectrometry studies
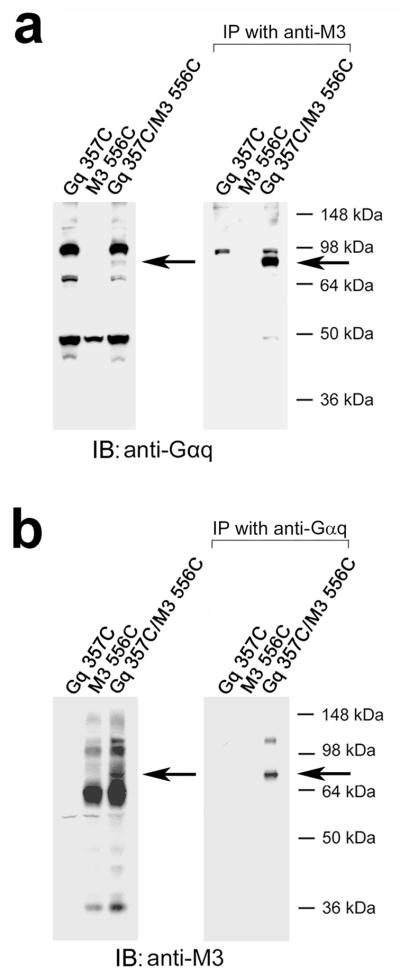
To further confirm that the ~80 kDa immunoreactive species observed in the cross-linking experiments do in fact represent Gαq/M3R complexes, we carried out a series of co-immunoprecipitation experiments. Specifically, we studied the representative N357C-Gαq/T556C-M3R pair and two other receptor/G protein combinations described in more detail below (R31C-Gαq/L173C-M3R, and D321C-Gαq/K548C-M3R. Initially, we incubated membrane proteins prepared from COS-7 cells that had been co-transfected with the three different receptor/G protein pairs with either 100 μM Cu-Phen (N357CG αq/T556C-M3R and R31C-Gαq/L173C-M3R) or 0.5 mM BMOE (D321C-Gαq/K548C-M3R) to induce cross-link formation. Following membrane lysis, we immunoprecipitated M3R-containing proteins with a polyclonal anti-M3 antibody, followed by Western blotting studies using an anti-Gαq monoclonal antibody. In a reciprocal fashion, we immunoprecipitated Gαq-containing proteins with the anti-Gαq antibody, followed by Western blotting studies using the anti-M3R antibody. In all co-immunoprecipitation experiments, the ~80 kDa band was the only immunoreactive species that was consistently detectable by both the anti-M3 and anti-Gαq antibodies (shown for the representative N357C-Gαq/T556C-M3R combination in Fig. 3; for additional co-immunoprecipitation data, see Supplementary Fig. 5). We did not observe this band using samples derived from cells that had been transfected with the individual receptor or G protein constructs alone (Fig. 3; Supplementary Fig. 5). These findings strongly support the notion that the ~80 kDa immunoreactive bands observed in the cross-linking experiments correspond to cross-linked receptor/Gαq complexes.
Figure 3.
Co-immunoprecipitation experiments confirming the identity of a representative Gαq/M3R complex. (a, b) Co-immunoprecipitation of the N357C-Gαq/T556C-M3R complex. Co-immunoprecipitation studies were carried out with lysates prepared from COS-7 cells co-expressing the N357C-Gαq/T556C-M3R combination. For control purposes, COS-7 cells were also transfected with N357C Gαq or T556C M3R alone. To induce cross-link formation, membrane proteins were incubated with 100 μM Cu-Phen. Following membrane lysis, M3R-containing proteins were immunoprecipitated with a polyclonal anti-M3 antibody, followed by Western blotting studies using an anti-Gαq monoclonal antibody (a). In a reciprocal fashion, Gαq-containing proteins were immunoprecipitated with the anti-Gαq antibody, followed by immunoblotting studies using the anti-M3R antibody (b). Co-immunoprecipitation experiments were carried out as described under Supplementary Methods. The panels to the left show Western blots of samples prior to immunoprecipitation. In all co-immunoprecipitation experiments, the ~80 kDa band was the only immunoreactive species that was consistently detectable by both the anti-M3 and anti-Gαq antibodies. The blots shown are representative of two independent experiments. The bands corresponding to cross-linked receptor/Gαq complexes are indicated by arrows.
We also confirmed the structural identity of a putative Gαq/M3R complex (N357C-Gαq/T556C-M3R) via LC/MS/MS analysis (see Supplementary Results and Supplementary Fig. 6 for details).
A cross-link between αN of Gαq and the i2 loop of the M3R
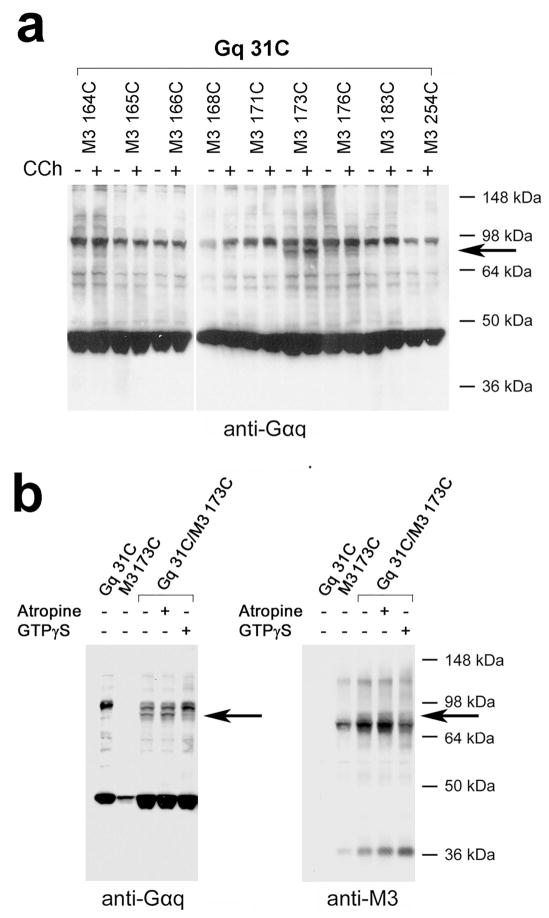
To identify potential contact sites between the αN helix of Gαq (in the Gq heterotrimer) and the intracellular surface of the M3R, we examined Cu-Phen-dependent cross-link formation between the R19C, R20C, R27C, R30C, and R31C mutant Gαq subunits (Fig. 1b) and the 18 Cys-substituted mutant M3Rs (Fig. 1a). Four of the five G protein constructs (R19C, R20C, R27C, and R30C) failed to display any significant cross-linking with any of the analyzed mutant M3Rs, independent of the absence or presence of carbachol (Supplementary Fig. 7). However, we observed a clear cross-linking signal between the R31C-Gαq subunit and the L173C receptor (Fig. 4). As expected, the band corresponding to the cross-linked R31C-Gαq/L173C-M3R complex could be detected by both anti-Gαq and anti-M3R antibodies (Fig. 4b; Supplementary Fig. 8). Moreover, this Gαq/M3R adduct was not observed when cells had been transfected with the R31C-Gαq or L173C-M3R constructs alone (Fig. 4b; Supplementary Fig. 8), providing further evidence for the specificity of the observed interaction (for detection of the R31C-Gαq/L173C-M3R complex in co-immunoprecipitation studies, see Supplementary Fig. 5a, b). The intensity of the R31C-Gαq/L173C-M3R cross-linking signal remained essentially unaffected by the addition of the agonist, carbachol (1 mM; Fig. 4a; Supplementary Fig. 8), or the inverse agonist, atropine (10 μM; Fig. 4b), but was significantly reduced by pretreatment of membranes with GTPγS (100 μM; Fig. 4b).
Figure 4.
Cross-link formation between a Cys residue introduced into the αN helix of Gαq (R31C) and a Cys residue substituted into the i2 loop of the M3R (L173C). (a) Cross-link formation between R31C Gαq and the L173C mutant M3R. (b) Effect of atropine and GTPγS treatment on the formation of the Gαq-R31C/M3R-L173C complex. While GTPγS (100 μM) treatment greatly reduced the intensity of the observed cross-linking signal, atropine (10 μM), an inverse muscarinic agonist, had no effect on the efficiency of Gαq-R31C/M3R-L173C complex formation. Membranes prepared from transfected COS-7 cells were incubated with 100 μM Cu-Phen in the absence (−) or presence (+) of 1 mM carbachol (CCh). The cross-linking data shown in panel (b) were obtained in the absence of carbachol. Western blotting studies were carried out under non-reducing conditions. The antibodies used are indicated underneath the blots. The blots shown are representative of three independent experiments. The bands corresponding to cross-linked receptor/Gαq complexes are indicated by arrows.
An agonist-induced α4/β6 loop (Gαq)/H8 (M3R) cross-link
To elucidate potential points of interactions between the α4/β6 loop of Gαq and the cytoplasmic side of the M3R, we studied cross-link formation between the P318C, S320C, D321C, and K322C mutant Gαq subunits (Fig. 1b) and the 18 Cys-substituted mutant M3Rs (Fig. 1a). Since it proved difficult to detect K322C-Gαq via Western blotting, we excluded this construct from further studies. By using Cu-Phen as an oxidizing agent, we were unable to detect any significant, reproducible cross-links between the P318C, S320C, and D321C Gαq constructs and any of the 18 mutant M3Rs (Supplementary Fig. 9), independent of the absence or presence of carbachol (1 mM). We therefore carried out additional cross-linking studies with the bifunctional, irreversible chemical cross-linking reagent, BMOE. Independent of the absence or presence of carbachol, BMOE (0.5 mM) treatment failed to induce any significant, reproducible cross-links between most mutant Gαq (P318C, S320C, or D321C)/M3R combinations (Fig. 5a; Supplementary Fig. 10). However, in the absence of carbachol, incubation with BMOE (0.5 mM) led to faint cross-linking signals between the D321C-Gαq subunit and three mutant M3Rs containing Cys substitutions within the N-terminal segment of H8 (K548C, T549C, and T552C; Fig. 5a, b). Strikingly, in all three cases, carbachol treatment greatly enhanced the intensities of the cross-linked adducts (Fig. 5a, b), consistent with the outcome of co-immunoprecipitation studies (shown for the representative D321C-Gαq/K548C-M3R complex in Supplementary Fig. 5c, d). These agonist-induced increases in cross-linking efficiency could be abolished or greatly reduced by pretreatment of membranes with GTPγS (100 μM) or atropine (10 μM) (shown for the representative D321C-Gαq/K548C-M3R combination in Fig. 5b).
Figure 5.
Agonist-induced cross-link formation between a Cys residue introduced into the α4/β6 loop of Gαq (D321C) and Cys residues substituted into the N-terminal segment of H8 of the M3R. (a) Agonist-dependent cross-link formation between the D321C Gαq subunit and the K548C, T549C, and T552C mutant M3Rs. (b) Effect of atropine and GTPγS treatment on the formation of the Gαq-D321C/M3R-K548C complex. Note that treatment with either GTPγS (100 μM) or atropine (10 μM) prevented the increase in carbachol-induced cross-link formation. Membranes prepared from transfected COS-7 cells were incubated with 0.5 mM BMOE in the absence (−) or presence (+) of 1 mM carbachol (CCh). Western blots were probed with a monoclonal anti-Gαq antibody (a) or with both anti-Gαq and anti-M3R antibodies (b). The blots shown are representative of three independent experiments. The bands corresponding to cross-linked receptor/Gαq complexes are indicated by arrows.
Cell surface expression of cross-linked M3R/Gαq pairs
To confirm that the cross-linked Cys-substituted M3R/Gαq pairs were present on the cell surface, we incubated intact COS-7 cells expressing different M3R/Gαq combinations with the membrane-impermeable biotinylation reagent, sulfo-NHS-S-S-biotin (see Supplementary Methods for details). After treatment of cell membranes with BMOE (0.5 mM) and membrane lysis, we isolated biotinylated cell surface proteins by using agarose-conjugated streptavidin. We then analyzed biotinylated proteins via Western blotting. These studies confirmed the presence of biotinylated M3R/Gαq complexes, indicating that M3R/Gαq cross-linking occurred at the cell surface (shown for the representative N357C-Gαq/T556C-M3R and D321C-Gαq/K548C-M3R combinations in Supplementary Fig. 11).
Effect of receptor densities on Gαq/M3R cross-linking
In the present study, many of the modified M3Rs were expressed at rather high levels (>10 pmol/mg). To examine whether the observed cross-linking patterns could also be observed at lower (more physiological) receptor levels, we drastically reduced the expression levels 17 of three key mutant M3Rs (T556C, L173C, and D321C) to ~1 pmol/mg (by reducing the amount of transfected receptor DNA; see Methods for details). Following co-expression with N357C-Gαq (T556C-M3R), R31C-Gαq (L173C-M3R), or D321C-Gαq(K548C-M3R), respectively, the resulting cross-linking patterns were qualitatively similar to those obtained when the mutant receptors were expressed at considerably higher levels (Supplementary Fig. 12). The N357C-Gαq/T556C-M3R and R31C-Gαq/L173C-M3R combinations showed basal disulfide cross-linking that was not affected by the presence of carbachol, whereas the D321C-Gαq/K548C-M3R pair displayed pronounced agonist-inducible disulfide cross-linking (Supplementary Fig. 12).
To compare the expression levels of Gαq endogenously expressed in COS-7 cells with those of the transiently expressed modified Gαq subunits, we determined the intensities of immunoreactive Gαq bands via scanning densitometry. This analysis showed that endogenous Gαq and the mutationally modified Gαq subunits were expressed at approximately similar levels (arbitrary units: endogenous Gαq, 100; mutant Gαq subunits; 140 ± 18; n=18). Taken together, these findings indicate that it is highly likely that the observed disulfide cross-linking patterns are physiologically relevant.
Discussion
In the present study, we observed several M3R/Gαq cross-links even in the absence of activating ligands. These cross-links were not affected by treatment of samples with the inverse muscarinic agonist, atropine31,32, suggesting that they were not caused by agonist-independent M3R signaling but most likely reflect the existence of preformed M3R/Gαq complexes33–37. One of these agonist-independent cross-links occurred between a residue located on the surface-exposed side of the αN helix of Gαq (R31C) and a residue located in the i2 loop of the M3R (L173C). Interestingly, most class I GPCRs contain a leucine residue or another relatively bulky lipophilic amino acid at the position corresponding to L173 in the M3R38,39. Site-directed mutagenesis studies with different GPCRs have shown that the presence of this residue is critical for productive receptor/G proteins interactions (see, for example, refs. 38, 39; also see Supplementary Table 1). Our cross-linking data therefore support the novel concept that direct contacts between the i2 loop of the receptor and the αN helix of Gα play a central role in productive receptor/G protein coupling.
Most M3R/Gαq contact sites identified in the inactive state of the M3R were also observed following activation of the M3R by carbachol. The most likely explanation for this finding is that the M3R is precoupled to Gq prior to receptor activation, and that carbachol-mediated M3R activation does not result in the dissociation of the receptor/Gq complex due to the lack of GTP (or other guanine nucleotides) in the incubation buffer. The M3R/Gαq cross-linking signals that we detected in the presence of carbachol therefore most likely reflect the structural organization of the M3R/Gq complex in which the guanine nucleotide binding pocket of the Gq heterotrimer is ‘empty’. It is well known that 19 heterotrimeric G proteins with an empty nucleotide binding pocket interact with GPCRs with very high affinity40.
We also detected several cross-links that showed very strong agonist dependence. Most strikingly, we identified a carbachol-induced cross-link between a residue located in the α4/β6 loop of Gαq (D321C) and the N-terminal segment of H8 of the M3R (K548C, T549C, and T552C). This finding suggests that receptor activation leads to a structural change at the receptor/Gq interface that increases the proximity between the N-terminal portion of H8 of the M3R and the α4/β6 loop of Gαq. The agonist carbachol also promoted significant cross-linking between a residue located at the cytoplasmic end of TM6 of the M3R (A488C) and the last three residues of Gαq (N357C, L358C, and V359C). Since M3R residue A488 (residue 6.33 according to the Ballesteros-Weinstein GPCR numbering system)41 is predicted to be buried in the TM core in the inactive conformation of the receptor5–8, this observation supports previous findings that GPCR activation involves an opening of the intracellular receptor surface, allowing the C-terminus of Gα to make productive interactions with previously buried TM5 and TM6 residues of the receptor11,42,43. Agonist treatment also led to more intense cross-linking signals between the N-terminal portion of H8 of the M3R (T549C and T552C) and the C-terminus of Gαq, indicating that these regions move closer to each other following M3R activation. However, we cannot completely exclude the possibility that the promiscuous cross-linking pattern observed with the mutant Gαq subunits containing C-terminal Cys substitutions is caused by the existence of multiple conformational states of the receptor/G protein complex.
It is likely that these agonist-promoted conformational changes at the M3R/Gαq interface play a key role in receptor-catalyzed GDP release from Gq. The C-terminus of Gα is linked to the β6/α5 loop of Gα via the α5 helix (note that the β6/α5 loop contains residues that are critical for guanine nucleotide binding). Interestingly, a recent biophysical study suggests that a rotational/translation movement of the α5 helix is critical for receptor (rhodopsin)-mediated GDP release from the G protein44. Analogously, the agonist-induced contact between H8 of the M3R and the α4/β6 loop of Gαq observed in the present study may alter the orientation of the β6/α5 loop by conformational changes that are propagated from the α4/β6 loop to the adjacent β6 strand (also see refs. 21 and 45).
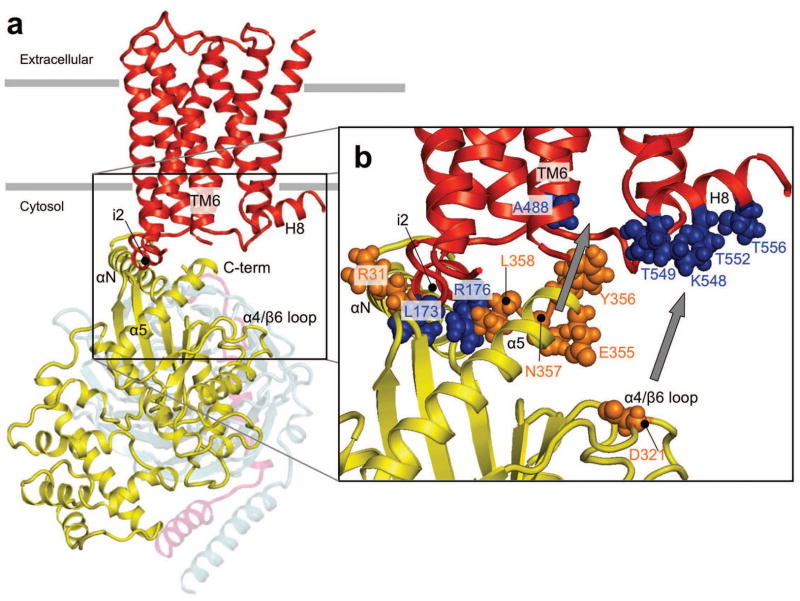
The observed M3R/Gαq cross-linking patterns provided many structural constraints that allowed us to generate a model of the M3R/Gαqβγ complex in its inactive state (Fig. 6) (see Supplementary Methods and Supplementary Table 3 for details). A key feature of this model is that the αN helix of Gαq is located adjacent to the i2 loop of the M3R. The highly flexible C-terminal portion of Gαq points diagonally towards the intracellular opening of the M3R, where it can make contacts with residues located on the i2 loop and H8. Moreover, the N-terminus of Gαq and the C-terminus of Gγ, both of which carry lipid modifications, are in proximity of the plasma membrane. The agonist-promoted cross-links between the extreme C-terminus of Gαq and position A488 of the M3R (bottom of TM6) and between the α4/β6 loop of Gαq and H8 support a model in which agonist-induced conformational changes on the intracellular receptor surface allow Gα (including the α4/β6 loop) to move closer towards and interact more extensively with the receptor (indicated by arrows in Fig. 6).
Figure 6.
Molecular model of the M3R/Gq complex. (a) Side view of the complex between the inactive state of the M3R and the Gq heterotrimer. The M3R is shown in red, Gαq in yellow, Gβ in grey, and Gγ in pink. (b) Enlargement of the M3R/Gαq interface. M3R residues are shown in blue, Gαq residues in orange. The structure shown represents the complex between the resting state of the M3R in contact with the Gq heterotrimer (for details, see Supplementary Methods). The interaction between R31C in the αN helix of Gαq and L173C in the i2 loop of the M3R provided a key contact site for the docking process. The C-terminus of Gαq points toward the intracellular core of the receptor, between the i2 loop and H8. The finding that the C-terminal residues of Gαq can interact with multiple M3R residues in the i2 loop (L173 and R176) and the N-terminal portion of H8 (T549, T552 and T556) suggests that the extreme C-terminus of Gαq is conformationally highly flexible. Agonist-induced M3R activation promoted the formation of cross-links between the extreme C-terminus of Gαq and the cytoplasmic end of TM6 (A488C) (grey arrow) and the N-terminal segment of H8 (T549C and T552C) of the M3R. Moreover, in the activated state of the receptor, a residue in the α4/β6 loop of Gαq (D321C) could be cross-linked to the N-terminal portion of H8 (K548C, T549C, and T552C) (grey arrow). The potential functional implications of theses structural changes are discussed in the text.
In the present study, we focused on identifying receptor contact sites for three key regions of Gαq known to be critically involved in receptor/G protein interactions. However, it should be noted that several other Gα regions as well as sites on the G protein βγ complex have also been implicated in receptor/G protein coupling12–14.
The M3R/Gαq cross-linking patterns observed in the present study can be explained by the interaction of one M3R monomer with one Gαq subunit. However, our data do not rule out the existence of M3R dimers or oligomers46,47.
In summary, the present study represents the first analysis examining receptor/Gα interaction sites in a systematic and comprehensive fashion. Importantly, all experimental data were obtained with functional receptor and Gα proteins present in a native membrane environment (in situ). Since heterotrimeric G proteins as well as most GPCRs share a high degree of structural homology, our findings should be of broad general relevance.
METHODS
Materials
Carbamylcholine chloride (carbachol), atropine sulfate, cupric sulfate (CuSO4), UK14304, 1,10-phenanthroline, and N-ethylmaleimide were purchased from Sigma. BMOE and sulfo-NHS-S-S-biotin were from Thermo Scientific. [3H]NMS (70.0 Ci/mmol) and myo-[3H]inositol (20 Ci/mmol) were obtained from PerkinElmer Life Sciences. CuSO4 was mixed with 1,10-phenanthroline at a molar ratio of 1:3 (ref. 32). The concentrations indicated in the text for the Cu(II)-(1,10-phenanthroline)3 complex (Cu-Phen) refer to molar copper concentrations.
Construction of Cys-substituted mutant M3R and Gαq constructs
All receptor Cys substitutions were introduced into a pCD-based expression plasmid coding for a modified version of the rat M3R referred to as M3′(3C)-Xa (Fig. 1a)23. This receptor construct lacks most endogenous Cys residues, except for C140, C220, and C532 which proved to be essential for M3R function, all five N-glycosylation sites contained within the extracellular N-terminal domain, as well as the central portion of the i3 loop (A274–K469; this region was replaced with two adjacent factor Xa cleavage sites)23. Cys residues were substituted into the M3′ (3C)-Xa construct by using the QuikChange™ site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. Using a similar strategy, we generated a mutant version of human Gαq (in the pcDNA3.1 vector), termed Gαq-C-less, in which three of the five native Cys residues were replaced with alanine (C144, C219, and C330; Fig. 1b). All Cys substitutions were introduced into the Gαq-C-less construct. The identity of all mutant M3R and Gαq constructs was confirmed by sequencing the entire coding sequences.
Transient expression of Cys-substituted mutant M3R and Gαq constructs in cultured cells
For radioligand binding and functional studies, all receptor constructs were transiently expressed in COS-7 cells, as described previously32. To increase receptor expression levels, transfected cells were incubated with 1 μM atropine for the last 24 h of culture. For disulfide cross-linking experiments, COS-7 cells were co-transfected with receptor and Gαq plasmid DNAs (2.5 μg each per 100 mm dish), using conditions similar to those described previously32. To lower the expression levels for selected mutant M3Rs, COS-7 cells were co-transfected with 0.05 μg of receptor DNA, 2.45 μg vector DNA, and 2.5 μg Gαq DNA (per 100 mm dish). To examine whether the various mutant Gαq constructs retained functional activity, we transiently co-expressed the different mutant Gαq constructs with a pCMV4-based plasmid coding for the porcine α2A-adrenergic receptor (α2A-AR)30. Specifically, 3 × 105 HEK 293 cells were seeded into 25 cm2 flasks 24 h prior to transfections. Cells were co-transfected with 1 μg of the α2A-AR construct and 3 μg of mutant Gαq DNA. About 24 h later, transfected cells were split into 12-well plates for PI hydrolysis assays.
Preparation of membranes from transfected COS-7 cells
COS-7 cells were harvested ~48 h after transfections, and membranes were prepared for radioligand binding and disulfide cross-linking studies as described32.
Radioligand binding studies
The various mutant M3Rs were characterized in [3H]NMS saturation and in [3H]NMS/carbachol competition binding assays. Binding studies were carried out using membranes prepared from transfected COS-7 cells, as described in detail previously32.
Measurement of receptor-mediated PI hydrolysis
To examine whether the various mutant M3Rs retained the ability to activate G proteins, we determined carbachol-mediated increases in intracellular inositol monophosphate (IP) levels. PI assays were carried out essentially as described32. To verify that the different mutant Gαq subunits retained functional activity, we also performed PI assays with HEK 293 cells co-transfected with the porcine α2A-AR and the different Gαq mutant constructs. Transfected HEK-293 cells grown in 12-well plates were incubated with 3 μCi/ml myo-[3H]inositol for the last 24 h of culture. We then determined the ability of the α2A-AR agonist, UK14304, to stimulate increases in intracellular IP levels32. PI data were analyzed using the nonlinear curve-fitting program Prism 4.0 (GraphPad).
Disulfide cross-linking and solubilization of Cys-substituted Gαq and M3R proteins
Disulfide cross-linking studies were carried out as described in detail previously32. In brief, receptor/Gαq-containing membranes prepared from co-transfected COS-7 cells were incubated either with the oxidizing agent, Cu-Phen (100 μM), or with the irreversible chemical cross-linker, BMOE (0.5 mM), in the absence or presence of 1 mM carbachol. Reactions were carried out for 10 min at 22 °C and then terminated by the addition of either EDTA and N-ethylmaleimide (10 mM each; Cu-Phen-induced cross-linking) or 10 mM DTT (BMOE-mediated cross-linking), followed by a 10-min incubation on ice. Subsequently, membrane proteins were solubilized by incubating samples with 1.2% digitonin (Roche Applied Science)32. Samples were then stored at −70 °C or used directly for SDS-PAGE.
Western blot analysis
SDS-PAGE and Western blotting studies were carried out as described previously32. In brief, samples containing 20 μg of solubilized membrane protein were incubated with Laemmli loading buffer for 30 min at 37 °C, either under nonreducing (Cu-Phen-induced cross-linking) or under reducing conditions (BMOE-mediated cross-linking), in the absence or presence of 10% β-mercaptoethanol, respectively. Blots were first probed with an anti-Gαq monoclonal antibody directed against residues 22–31 of human Gαq (BD Biosciences), followed by incubation with secondary goat anti-mouse HRP conjugate (Calbiochem). This antibody failed to detect the R27C and R30C mutant Gαq subunits with high efficiency (note that R27 and R30 are contained within the antibody recognition sequence). However, we were able to detect these two mutant G protein constructs using a rabbit polyclonal anti-Gαq antibody directed against the C-terminal domain (residues 350–359) of mammalian Gαq/11 (Calbiochem). All Gαq blots were routinely stripped by using OneMinute Western Blot Stripping Buffer (GM Biosciences) and reprobed with a rabbit anti-M3R polyclonal antibody directed against the C-terminal 18 amino acids of the rat M3R32. Immunoreactive proteins were visualized by using SuperSignal West Pico Chemiluminescent Substrate (Pierce) and autoradiography32.
Construction of homology models and docking of Gq to the M3R
The procedures used for constructing a molecular model of the M3R/Gq complex are described in detail under Supplementary Methods. The model that was most compatible with the observed cross-linking data is shown in Fig. 6 (see text for details). The coordinates on which this model is based are provided as a pdb file (M3R_Gq.pdb) under Supplementary Information.
Data analysis
Data were presented as mean ± s. e. m. for the indicated number of experiments. Sigmoidal concentration-response data were analyzed using the nonlinear curve-fitting program Prism 4.0 (GraphPad). Statistical significance was determined using the Student’s t-test.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIDDK, NIH, and utilized the high-performance computational capabilities of the Biowulf Linux cluster at the NIH, Bethesda, MD (http://biowulf.nih.gov).
Footnotes
Authors’ Contributions
J.H. designed and conducted most of the biochemical experiments and wrote the paper. Y.W., X.Z., and J.L. were involved in the pharmacological characterization of the mutant proteins. J.R.L. carried out the mass spectrometry studies. S.C. carried out the molecular modeling studies and wrote the paper. J.K. performed modular modeling studies. J.W. designed the experiments and wrote the paper.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
References
- 1.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Lagerström MC, Schiöth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 3.Regard JB, Sato IT, Coughlin SR. Anatomical profiling of G protein-coupled receptor expression. Cell. 2008;135:561–571. doi: 10.1016/j.cell.2008.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabrera-Vera TM, et al. Insights into G protein structure, function, and regulation. Endocr Rev. 2003;24:765–781. doi: 10.1210/er.2000-0026. [DOI] [PubMed] [Google Scholar]
- 5.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 6.Warne T, et al. Structure of a β1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherezov V, et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasmussen SG, et al. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 9.Jaakola VP, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 11.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 12.Oldham WM, Hamm HE. How do receptors activate G proteins? Adv Protein Chem. 2007;74:67–93. doi: 10.1016/S0065-3233(07)74002-0. [DOI] [PubMed] [Google Scholar]
- 13.Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- 14.Johnston CA, Siderovski DP. Receptor-mediated activation of heterotrimeric G-proteins: current structural insights. Mol Pharmacol. 2007;72:219–230. doi: 10.1124/mol.107.034348. [DOI] [PubMed] [Google Scholar]
- 15.Wess J. Molecular basis of receptor/G-protein-coupling selectivity. Pharmacol Ther. 1998;80:231–264. doi: 10.1016/s0163-7258(98)00030-8. [DOI] [PubMed] [Google Scholar]
- 16.Hamm HE, et al. Site of G protein binding to rhodopsin mapped with synthetic peptides from the α subunit. Science. 1988;241:832–835. doi: 10.1126/science.3136547. [DOI] [PubMed] [Google Scholar]
- 17.Onrust R, et al. Receptor and βγ binding sites in the α subunit of the retinal G protein transducin. Science. 1997;275:381–384. doi: 10.1126/science.275.5298.381. [DOI] [PubMed] [Google Scholar]
- 18.Conklin BR, Farfel Z, Lustig KD, Julius D, Bourne HR. Substitution of three amino acids switches receptor specificity of Gqα to that of Giα. Nature. 1993;363:274–276. doi: 10.1038/363274a0. [DOI] [PubMed] [Google Scholar]
- 19.Cai K, Itoh Y, Khorana HG. Mapping of contact sites in complex formation between transducin and light-activated rhodopsin by covalent crosslinking: use of a photoactivatable reagent. Proc Natl Acad Sci USA. 2001;98:4877–4882. doi: 10.1073/pnas.051632898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itoh Y, Cai K, Khorana HG. Mapping of contact sites in complex formation between light-activated rhodopsin and transducin by covalent crosslinking: use of a chemically preactivated reagent. Proc Natl Acad Sci USA. 2001;98:4883–4887. doi: 10.1073/pnas.051632998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnston CA, Siderovski DP. Structural basis for nucleotide exchange on Gαi subunits and receptor coupling specificity. Proc Natl Acad Sci USA. 2007;104:2001–2006. doi: 10.1073/pnas.0608599104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Wess J. Molecular biology of muscarinic acetylcholine receptors. Crit Rev Neurobiol. 1996;10:69–99. doi: 10.1615/critrevneurobiol.v10.i1.40. [DOI] [PubMed] [Google Scholar]
- 23.Zeng FY, Hopp A, Soldner A, Wess J. Use of a disulfide cross-linking strategy to study muscarinic receptor structure and mechanisms of activation. J Biol Chem. 1999;274:16629–16640. doi: 10.1074/jbc.274.23.16629. [DOI] [PubMed] [Google Scholar]
- 24.Cai K, et al. Single-cysteine substitution mutants at amino acid positions 306–321 in rhodopsin, the sequence between the cytoplasmic end of helix VII and the palmitoylation sites: sulfhydryl reactivity and transducin activation reveal a tertiary structure. Biochemistry. 1999;38:7925–7930. doi: 10.1021/bi9900119. [DOI] [PubMed] [Google Scholar]
- 25.Ernst OP, et al. Mutation of the fourth cytoplasmic loop of rhodopsin affects binding of transducin and peptides derived from the carboxyl-terminal sequences of transducin α and γ subunits. J Biol Chem. 2000;275:1937–1943. doi: 10.1074/jbc.275.3.1937. [DOI] [PubMed] [Google Scholar]
- 26.Swift S, et al. Role of the PAR1 receptor 8th helix in signaling: the 7-8-1 receptor activation mechanism. J Biol Chem. 2006;281:4109–4116. doi: 10.1074/jbc.M509525200. [DOI] [PubMed] [Google Scholar]
- 27.Delos Santos NM, Gardner LA, White SW, Bahouth SW. Characterization of the residues in helix 8 of the human β1-adrenergic receptor that are involved in coupling the receptor to G proteins. J Biol Chem. 2006;281:12896–12907. doi: 10.1074/jbc.M508500200. [DOI] [PubMed] [Google Scholar]
- 28.Wedegaertner PB, Chu DH, Wilson PT, Levis MJ, Bourne HR. Palmitoylation is required for signaling functions and membrane attachment of Gqα and Gsα. J Biol Chem. 1993;268:25001–25008. [PubMed] [Google Scholar]
- 29.Edgerton MD, Chabert C, Chollet A, Arkinstall S. Palmitoylation but not the extreme amino-terminus of Gqα is required for coupling to the NK2 receptor. FEBS Lett. 1994;354:195–199. doi: 10.1016/0014-5793(94)01101-x. [DOI] [PubMed] [Google Scholar]
- 30.Chabre O, Conklin BR, Brandon S, Bourne HR, Limbird LE. Coupling of the α2A-adrenergic receptor to multiple G-proteins. A simple approach for estimating receptor-G-protein coupling efficiency in a transient expression system. J Biol Chem. 1994;269:5730–5734. [PubMed] [Google Scholar]
- 31.Spalding TA, Burstein ES, Brauner-Osborne H, Hill-Eubanks D, Brann MR. Pharmacology of a constitutively active muscarinic receptor generated by random mutagenesis. J Pharmacol Exp Ther. 1995;275:1274–1279. [PubMed] [Google Scholar]
- 32.Li JH, et al. Distinct structural changes in a G protein-coupled receptor caused by different classes of agonist ligands. J Biol Chem. 2007;282:26284–93. doi: 10.1074/jbc.M704875200. [DOI] [PubMed] [Google Scholar]
- 33.Rebois RV, Hébert TE. Protein complexes involved in heptahelical receptor-mediated signal transduction. Receptors Channels. 2003;9:169–194. [PubMed] [Google Scholar]
- 34.Nobles M, Benians A, Tinker A. Heterotrimeric G proteins precouple with G protein-coupled receptors in living cells. Proc Natl Acad Sci USA. 2005;102:18706–18711. doi: 10.1073/pnas.0504778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galés C, et al. Real-time monitoring of receptor and G-protein interactions in living cells. Nat Methods. 2005;2:177–184. doi: 10.1038/nmeth743. [DOI] [PubMed] [Google Scholar]
- 36.Clark MA, Sethi PR, Lambert NA. Active Gαq subunits and M3 acetylcholine receptors promote distinct modes of association of RGS2 with the plasma membrane. FEBS Lett. 2007;581:764–770. doi: 10.1016/j.febslet.2007.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lohse MJ, et al. Optical techniques to analyze real-time activation and signaling of G-protein-coupled receptors. Trends Pharmacol Sci. 2008;29:159–165. doi: 10.1016/j.tips.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Moro O, Lameh J, Högger P, Sadée W. Hydrophobic amino acid in the i2 loop plays a key role in receptor-G protein coupling. J Biol Chem. 1993;268:22273–22276. [PubMed] [Google Scholar]
- 39.Wacker JL, et al. Disease-causing mutation in GPR54 reveals the importance of the second intracellular loop for class A G-protein-coupled receptor function. J Biol Chem. 2008;283:31068–310678. doi: 10.1074/jbc.M805251200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bornancin F, Pfister C, Chabre M. The transitory complex between photoexcited rhodopsin and transducin. Eur J Biochem. 1989;184:687–98. doi: 10.1111/j.1432-1033.1989.tb15068.x. [DOI] [PubMed] [Google Scholar]
- 41.Ballesteros JA, Weinstein H. Integrated methods for modeling G-protein coupled receptors. Meth Neurosci. 1995;25:366–428. [Google Scholar]
- 42.Hubbell WL, Altenbach C, Hubbell CM, Khorana HG. Rhodopsin structure, dynamics, and activation: a perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Adv Protein Chem. 2003;63:243–290. doi: 10.1016/s0065-3233(03)63010-x. [DOI] [PubMed] [Google Scholar]
- 43.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci USA. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE. Mechanism of the receptor-catalyzed activation of heterotrimeric G proteins. Nat Struct Mol Biol. 2006;13:772–777. doi: 10.1038/nsmb1129. [DOI] [PubMed] [Google Scholar]
- 45.Kapoor N, Menon ST, Chauhan R, Sachdev P, Sakmar TP. Structural evidence for a sequential release mechanism for activation of heterotrimeric G proteins. J Mol Biol. 2009;393:882–897. doi: 10.1016/j.jmb.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 46.Javitch JA. The ants go marching two by two: oligomeric structure of G-protein-coupled receptors. Mol Pharmacol. 2004;66:1077–1082. doi: 10.1124/mol.104.006320. [DOI] [PubMed] [Google Scholar]
- 47.Milligan G, Bouvier M. Methods to monitor the quaternary structure of G protein-coupled receptors. FEBS J. 2005;272:2914–2925. doi: 10.1111/j.1742-4658.2005.04731.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.