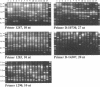
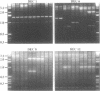
Abstract
The RAPD (random amplified polymorphic DNA) fingerprinting method, which utilizes low stringency PCR amplification with single primers of arbitrary sequence to generate strain-specific arrays of anonymous DNA fragments, was calibrated relative to the widely used, protein-based multilocus enzyme electrophoretic (MLEE) typing method. RAPD fingerprinting was carried out on five isolates from each of 15 major groups of Escherichia coli strains that cause diarrheal disease worldwide (75 isolates in all). Each group consisted of isolates that were not distinguishable from one another by MLEE typing using 20 diagnostic enzyme markers. In our RAPD tests, three or more distinct subgroups in each MLEE group were distinguished with each of five primers, and 74 of the 75 isolates were distinguished when data obtained with five primers were combined. Thus, RAPD typing is far more sensitive than MLEE typing for discriminating among related strains of a species. Despite their different sensitivities, the same general relationships among strains were inferred from MLEE and RAPD data. Thus, our results recommend use of the RAPD method for studies of bacterial population genetic structure and evolution, as well as for epidemiology.
Full text
PDF
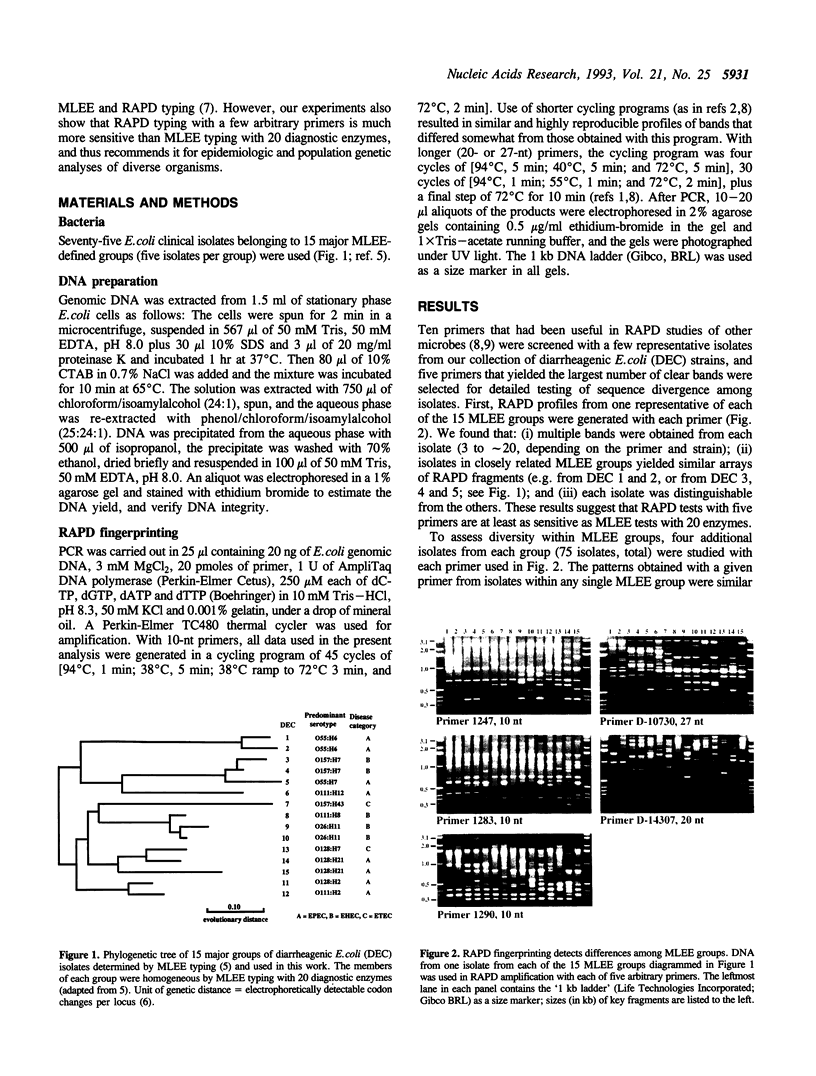
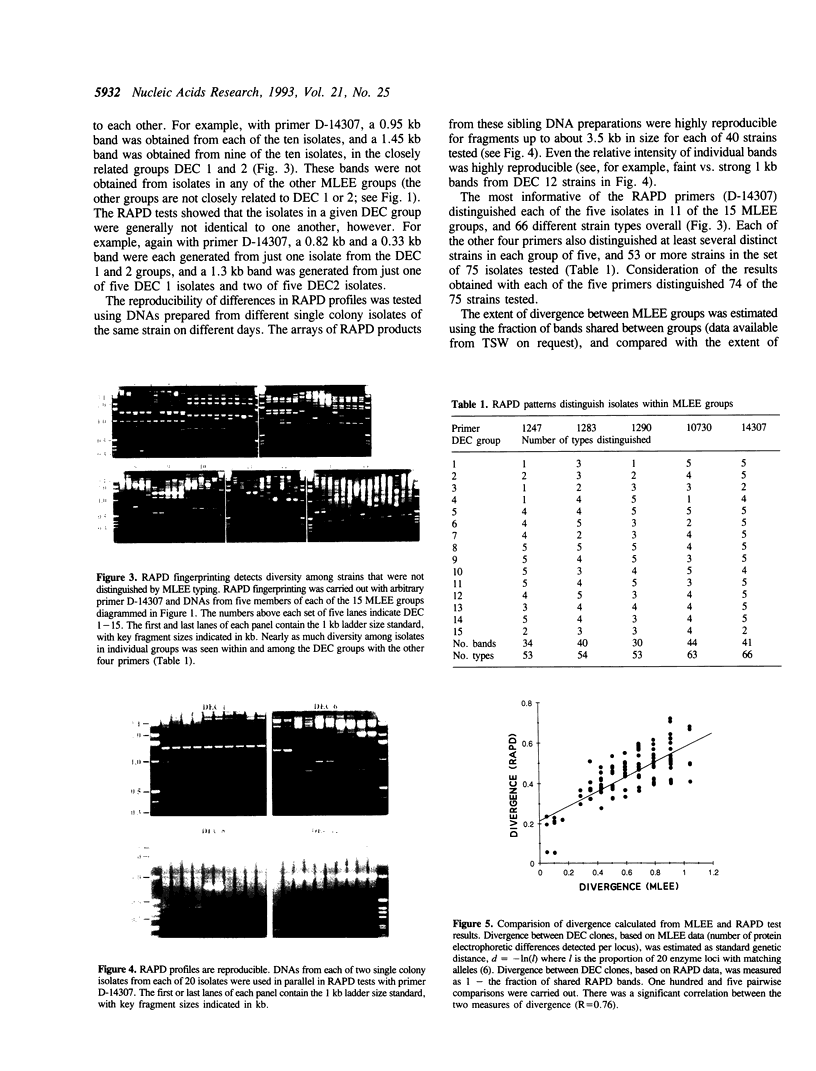

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akopyanz N., Bukanov N. O., Westblom T. U., Kresovich S., Berg D. E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992 Oct 11;20(19):5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeit R. D., Arthur M., Dunn R., Kim C., Selander R. K., Goldstein R. Resolution of recent evolutionary divergence among Escherichia coli from related lineages: the application of pulsed field electrophoresis to molecular epidemiology. J Infect Dis. 1990 Feb;161(2):230–235. doi: 10.1093/infdis/161.2.230. [DOI] [PubMed] [Google Scholar]
- Clark A. G., Lanigan C. M. Prospects for estimating nucleotide divergence with RAPDs. Mol Biol Evol. 1993 Sep;10(5):1096–1111. doi: 10.1093/oxfordjournals.molbev.a040057. [DOI] [PubMed] [Google Scholar]
- Frost J. A., Cheasty T., Thomas A., Rowe B. Phage typing of Vero cytotoxin-producing Escherichia coli O 157 isolated in the United Kingdom: 1989-91. Epidemiol Infect. 1993 Jun;110(3):469–475. doi: 10.1017/s0950268800050895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersulyte D., Woods J. P., Keath E. J., Goldman W. E., Berg D. E. Diversity among clinical isolates of Histoplasma capsulatum detected by polymerase chain reaction with arbitrary primers. J Bacteriol. 1992 Nov;174(22):7075–7079. doi: 10.1128/jb.174.22.7075-7079.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J. G., Dykhuizen D. E., DuBose R. F., Hartl D. L. Phylogenetic analysis using insertion sequence fingerprinting in Escherichia coli. Mol Biol Evol. 1989 Jan;6(1):1–14. doi: 10.1093/oxfordjournals.molbev.a040531. [DOI] [PubMed] [Google Scholar]
- Smith J. M., Smith N. H., O'Rourke M., Spratt B. G. How clonal are bacteria? Proc Natl Acad Sci U S A. 1993 May 15;90(10):4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M., Neubauer K., Barnabé C., Guerrini F., Skarecky D., Ayala F. J. Genetic characterization of six parasitic protozoa: parity between random-primer DNA typing and multilocus enzyme electrophoresis. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1335–1339. doi: 10.1073/pnas.90.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990 Dec 25;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam T. S., Wolfe M. L., Wachsmuth I. K., Orskov F., Orskov I., Wilson R. A. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect Immun. 1993 May;61(5):1619–1629. doi: 10.1128/iai.61.5.1619-1629.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]