Abstract
Cardiac hypertrophy and failure are accompanied by a reprogramming of gene expression that involves transcription factors and chromatin remodeling enzymes. Little is known about the roles of histone methylation and demethylation in this process. To understand the role of JMJD2A, a histone trimethyl demethylase, in cardiac hypertrophy, we generated mouse lines with heart-specific Jmjd2a deletion (hKO) and overexpression (Jmjd2a-Tg). Jmjd2a hKO and Jmjd2a-Tg mice had no overt baseline phenotype, but did demonstrate altered responses to cardiac stresses. While inactivation of Jmjd2a resulted in an attenuated hypertrophic response to transverse aortic constriction–induced (TAC-induced) pressure overload, Jmjd2a-Tg mice displayed exacerbated cardiac hypertrophy. We identified four-and-a-half LIM domains 1 (FHL1), a key component of the mechanotransducer machinery in the heart, as a direct target of JMJD2A. JMJD2A bound to the FHL1 promoter in response to TAC, upregulated FHL1 expression, and downregulated H3K9 trimethylation. Upregulation of FHL1 by JMJD2A was mediated through SRF and myocardin and required its demethylase activity. The expression of JMJD2A was upregulated in human hypertrophic cardiomyopathy patients. Our studies reveal that JMJD2A promotes cardiac hypertrophy under pathological conditions and suggest what we believe to be a novel mechanism for JMJD2A in reprogramming of gene expression involved in cardiac hypertrophy.
Introduction
Heart failure is a major public health problem and a leading cause of mortality in Western countries. It is the final common pathway for a wide spectrum of cardiovascular diseases including hypertension, coronary artery disease, myocardial ischemia, valve abnormalities, and inherited or acquired cardiomyopathies. There is no single unifying mechanism that explains the development and progression of heart failure. Nevertheless, heart failure is frequently preceded by left ventricular hypertrophy (LVH), and LVH is a major predictor for progressive heart disease and an adverse prognosis (1).
Myocardial hypertrophy is an adaptive response of cardiac muscle to altered conditions caused by a large number of physiological and pathological stimuli (2). Cardiac hypertrophy, even though it is physiological or compensatory in the beginning, can become pathological or maladaptive, leading to heart failure if left untreated. Pathological hypertrophy and heart failure are accompanied by a reprogramming of cardiac gene expression and activation of “fetal” genes that correlate with loss of cardiac functions (3–5). Therefore, elucidation of the mechanism or mechanisms involved in the reprogramming of cardiac gene expression in hypertrophic and failing hearts could provide us information for designing drugs to manipulate and normalize cardiac gene expression in a “transcriptional therapy” for cardiac hypertrophy and failure (6).
The transcriptional regulation of gene expression involves not only transcription factors but also posttranslational modifications of the histone tails. Histone modifications can alter chromatin conformations that allow the accessibility of transcription factors and the recruitment of the transcriptional complex on the promoter/enhancer and transcriptional regions of the genes. There are at least 7 distinct types of modifications found on histones, including methylation and demethylation (7–9). Different histone methylation patterns can provide specialized binding surfaces that recruit protein complexes containing chromatin remodeling and transcriptional activation/repression activity. Multiple histone methyltransferase and demethylases have been identified. Studies have suggested their roles in multiple aspects of development across various species and in diseases such as cancer and neurological disorders (10–18). However, the role of histone demethylases in the reprogramming of cardiac gene expression in hypertrophic and failing hearts remains elusive. A genome-wide histone methylation profile for heart failure showed a differential marking of trimethylation of H3K4 and H3K9 (H3K4me3 and H3K9me3) in cardiomyocytes during development of heart failure in both animal models and human (19), suggesting that the enzymes responsible for methylation and demethylation of H3K4me3 and H3K9me3 may play a role in cardiac hypertrophy and heart failure.
JMJD2A/KDM4A is a member of the JmjC domain–containing family JMJD2 of histone demethylases, including JMJD2B, JMJD2C, JMJD2D, and JMJD1B (12, 14, 18). JMJD2 proteins are lysine trimethyl–specific histone demethylases that catalyze the demethylation of trimethylated H3K9 (H3K9me3) and H3K36 (H3K36me3). Genome-wide studies show that H3K9me3 is enriched in heterochromatin (15), which predicts that the H3K9me3 demethylase activities of JMJD2 proteins may act as transcriptional activators. Consistent with this model, JMJD2A and JMJD2D were reported to be coactivators for the androgen receptor (20). On the other hand, JMJD2A has also been reported to repress N-CoR–regulated transcription in cell-culture studies (13, 21, 22). These results suggest that the function of JMJD2A may be context dependent and emphasize the need for direct analysis of JMJD2A function in specific physiological settings. Of relevance to the study presented here, it is notable that the roles of histone methylation/demethylation and the corresponding enzymes in the reprogramming of cardiac gene expression during cardiac hypertrophy and failure have not been elucidated.
To understand the role of JMJD2A in the reprogramming of gene expression in hypertrophic hearts, we generated conditional heart-specific Jmjd2a KO (Jmjd2a hKO) mice and mouse lines that overexpress JMJD2A in the postnatal heart (Jmjd2a-Tg). Our studies demonstrate a role for JMJD2A in promoting cardiac hypertrophy under pathologic conditions. We identified four-and-a-half LIM domains 1 (FHL1) as a target of JMJD2A. We have found that JMJD2A binds to the FHL1 promoter in response to hypertrophic stimuli and upregulates the transcription of FHL1. We have further observed that the transcriptional activity of JMJD2A is mediated through SRF/myocardin. JMJD2A promotes recruitment of SRF/myocardin to the SRF-targeted genes. JMJD2A is upregulated in human hypertrophic cardiomyopathy (HCM) patients. Taken together, our studies show that JMJD2A promotes cardiac hypertrophy and suggest what we believe is a novel mechanistic interplay among SRF, myocardin, and JMJD2A in the reprogramming of gene expression during hypertrophic remodeling.
Results
Generation and characterization of Jmjd2a hKO.
JMJD2A is expressed ubiquitously and higher in the heart, the skeletal muscle, and the liver in mice (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI46277DS1). To study the biological function of JMJD2A in the heart, we generated mice with conditional alleles for Jmjd2a and mice with deleted Jmjd2a in the heart using α-MHC–Cre (ref. 23 and Figure 1, A–D). The deletion of exon 3 in the Jmjd2afl/fl×α-MHC–Cre mouse (Jmjd2a hKO) resulted in a significant reduction of JMJD2A protein in the heart compared with control mice (Jmjd2afl/fl) (Figure 1D). The residual JMJD2A in hKO hearts may come from the contribution of noncardiomyocytes in the heart and/or incomplete deletion of Jmjd2a in the cardiomyocyte by the α-MHC–Cre transgene. Jmjd2a hKO mice are viable and no overt baseline cardiac phenotypes were observed for mice up to 6 months of age. The cardiac function of adult Jmjd2a hKO mice was indistinguishable from that of control mice as assayed by echocardiograph (data not shown).
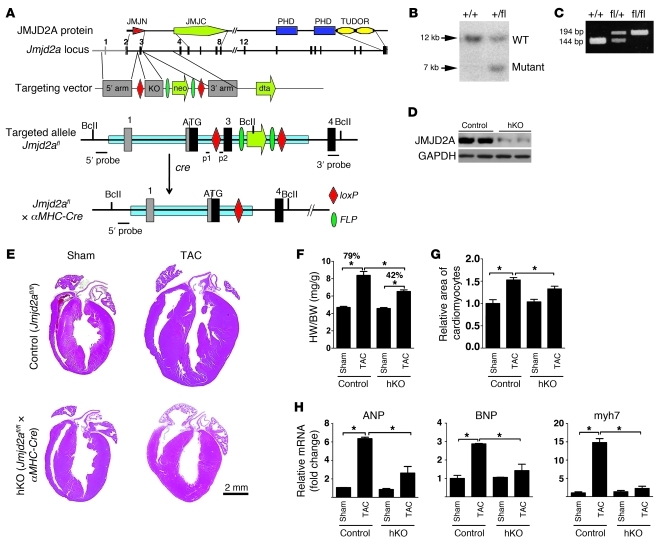
Figure 1. Generation and assessment of a cardiac-specific Jmjd2a KO mouse model.
(A) Strategy used to delete Jmjd2a in mouse cardiomyocytes and generate the Jmjd2a hKO model. Schematic protein structure of JMJD2A (14) and maps of the WT Jmjd2a locus, the targeting vector, the floxed allele, and the excised allele are shown. Exons are shown in bars. Exon 3 is flanked by loxP sites. (B) Southern blot of tail DNA from a WT (+/+) and a heterozygous (fl/+) mouse using 5′ probe shown in A after digestion with BclI. (C) PCR genotyping on genomic DNA isolated from a WT (+/+), a heterozygous (fl/+), and a homozygous floxed (fl/fl) mouse using primers p1/p2 indicated in A. (D) Western blot demonstrating significantly reduced JMJD2A protein in the heart homogenate of Jmjd2a hKO mice at 4 weeks. (E) H&E stains of representative paraffin sections of control and hKO hearts 3 weeks after sham and TAC operation. (F) HW/BW ratios of control and hKO mice 3 weeks after sham and TAC operation (n = 10–14 per group). The HW/BW ratio after TAC operation was increased 79% compared with sham operation in control mice, whereas it was only increased 42% in hKO mice. (G) Myocyte cross-sectional areas from control and hKO mice (>200 myocytes per heart in randomly selected filed) were assessed (n = 6–8 per group). (H) mRNA transcripts of ANP, BNP, and myh7 in control and hKO mouse hearts after 3 weeks sham and TAC operation. RNAs were normalized to internal GAPDH expression and presented as the fold change relative to control sham samples (n = 4–6 per group). Values are mean ± SEM. *P < 0.05.
Jmjd2a hKO mice have attenuated hypertrophic responses.
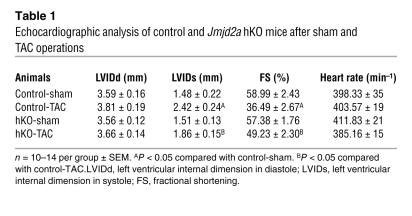
To determine whether Jmjd2a hKO mice have altered cardiac responses under pathological conditions, we subjected hKO and control (Jmjd2afl/fl) littermates to transverse aortic constriction (TAC), which causes pathological cardiac hypertrophy due to increased afterload (24). Three weeks after TAC, Jmjd2a hKO hearts were significantly smaller than those of control mice (Figure 1E). This is reflected in a significantly smaller increase in heart weight/body weight (HW/BW) ratio (Figure 1F) and cross-sectional cardiomyocyte areas (Figure 1G) in hKO mice compared with those of controls. hKO mice also showed better preserved cardiac function after TAC compared with TAC-operated control mice, as indicated by echocardiograph (Table 1). A blunted hypertrophic response in hKO mice was further indicated by significantly decreased expression of the “fetal gene” markers ANP, BNP, and myh7 in response to TAC-induced pressure overload compared with that of control mice (Figure 1H). To address the possible effect of Cre-transgene on the cardiac phenotype, we also subjected Jmjd2a+/+×α-MHC–Cre mice to sham and TAC operation and compared the phenotype with those of Jmjd2afl/fl littermates. No significant differences of hypertrophic responses were observed between the 2 genotypes (Supplemental Figure 2), suggesting that the hypertrophic response we observed in hKO mice is not due to the effect of the α-MHC–Cre transgene.
Table 1 .
Echocardiographic analysis of control and Jmjd2a hKO mice after sham and TAC operations
Overexpression of Jmjd2a in postnatal hearts exacerbates the hypertrophic response to TAC-induced hypertrophy.
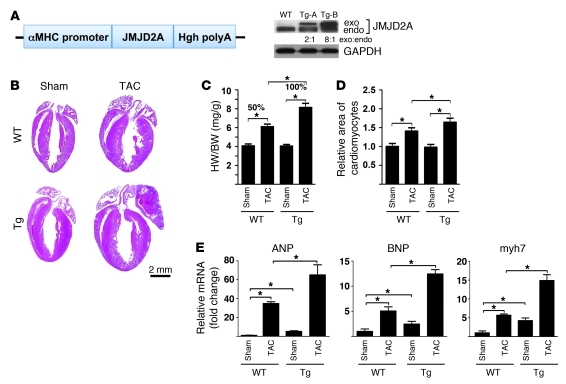
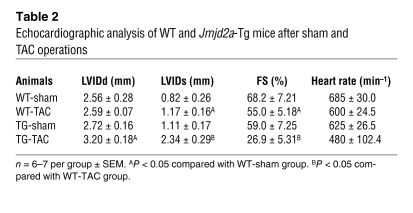
To determine whether gain of function of JMJD2A promotes cardiac hypertrophy, we generated transgenic mice in FVB background with JMJD2A expressed specifically in the postnatal heart (Figure 2A). Four founder mice were obtained. Two transgenic lines (Tg-A and Tg-B) with modest JMJD2A expression shown by Western blot analysis (Figure 2A) were established. The results presented here are those obtained with the transgenic line B. In the absence of stress, Jmjd2a-Tg mice displayed normal cardiac morphologies and functions similar to those of WT littermates (data not shown). Three weeks after TAC, Jmjd2a-Tg mice manifested an exacerbated hypertrophic response (Figure 2B) compared with that of WT littermates with significantly increased HW/BW ratio (Figure 2C). Analysis of cross-sectional cardiomyocyte areas also revealed a further increase in cardiomyocyte size after TAC in Tg mice compared with that of WT littermates (Figure 2D) and more fibrosis (data not shown). TAC Jmjd2a-Tg mice also showed significant loss of cardiac function compared with TAC WT mice (Table 2). An increase in hypertrophic response in TAC Jmjd2a-Tg mice is further indicated by the significantly larger increases of the expression of cardiac fetal gene markers such as ANP, BNP, and myh7 compared with those of WT littermates (Figure 2E). Similar exacerbated hypertrophic responses to TAC were also observed with the transgenic line A (Supplemental Figure 3), suggesting that the observed phenotypes are specifically due to JMJD2A. We also noticed a difference in the TAC-induced hypertrophic response between the WT mice in Figure 2C and control/WT mice in Figure 1F. This may be due to the difference of the genetic background of the KO (129/C57BL6) and transgenic (FVB) mice.
Figure 2. JMJD2A promotes cardiac hypertrophy in response to TAC-induced pressure overload.
(A) Schematic diagram of cDNA encoding a flag-tagged JMJD2A in an expression vector containing the α-MHC promoter (left panel). 4 founder mice were obtained. 2 transgenic lines (Tg-A and Tg-B) with modest JMJD2A expression shown by Western blot analysis with anti-JMJD2A antibody (right panel) were established. The exogenous JMJD2A protein level is about 2-fold higher in the Tg-A line and 8-fold higher in the Tg-B line compared with that of endogenous JMJD2A. (B) H&E stains of paraffin section of WT and Jmjd2a-Tg (Tg) mouse hearts 3 weeks after sham and TAC operations. (C) HW/BW ratio (n = 6–7 per group) and (D) relative areas of cardiomyocytes (n = 3–4 per group) in WT and Jmjd2a-Tg sham- and TAC-operated animals. (E) Transcript levels of fetal gene markers, including ANP, BNP, and myh7, were quantified by real-time qRT-PCR, normalized against levels of internal GAPDH, and expressed as the fold change relative to that of sham-operated WT animals (n = 3 per group). The HW/BW ratio after TAC operation was increased 50% compared with sham operation in WT mice, whereas it was increased 100% in Tg mice. Values are mean ± SEM. *P < 0.05.
Table 2 .
Echocardiographic analysis of WT and Jmjd2a-Tg mice after sham and TAC operations
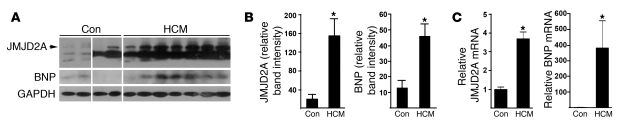
To determine whether JMJD2A is pathophysiologically relevant to human cardiac hypertrophy, we examined the protein levels of JMJD2A in the hearts of patients with HCM and non-HCM individuals by Western blot analysis. As shown in Figure 3, JMJD2A protein was significantly upregulated in HCM patients, and the upregulation was paralleled with that of BNP. An additional band was detected in human JMJD2A Western blot analysis. Whether this band is an alternative splice variant or degradation of JMJD2A remains to be determined. We also performed quantitative RT-PCR (qRT-PCR) analysis on the transcript of JMJD2A and BNP and observed similar upregulation of JMJD2A and BNP in HCM patients compared with controls. These data, together with those of loss-of- and gain-of-function studies of JMJD2A in mice, suggest that JMJD2A is a prohypertrophic factor under pathological conditions.
Figure 3. JMJD2A is upregulated in human hypertrophic hearts.
(A) Heart tissue samples of human HCM patients and unmatched non-HCM controls (con) were subjected to Western blot analysis using antibodies against JMJD2A, BNP, and GAPDH. Consistent with the HCM phenotype, BNP was upregulated in HCM samples. The lanes were run on the same gel but were noncontiguous. (B) More importantly, JMJD2A was significantly upregulated in HCM samples (n = 7) compared with samples from the non-HCM patients (n = 4). GAPDH was used as the loading control. (C) Relative mRNA levels of JMJD2A and BNP in control (n = 3) and HCM patients (n = 7) measured by qRT-PCR. Values are mean ± SEM. *P < 0.05.
Jmjd2a upregulates the expression of FHL1 and binds to the FHL1 promoter in vivo in response to TAC.
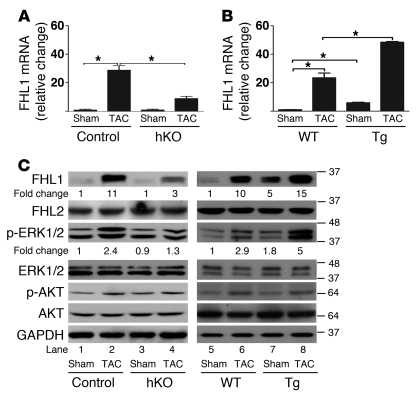
To understand the molecular mechanism or mechanisms that underlie the prohypertrophic function of JMJD2A, we performed gene profiling experiments with cDNAs from the hearts of WT, Jmjd2a hKO, and Jmjd2a-Tg mice. Of the genes whose expressions were altered in the hKO and transgenic mouse hearts in comparison with those of control/WT mice, we noticed that FHL1 was significantly changed (Figure 4). FHL1 was upregulated in response to TAC, which was significantly reduced in Jmjd2a hKO mice compared with controls (Figure 4, A and C) and exacerbated in transgenic mouse hearts (Figure 4, B and C). No change of expression for FHL2, a close member of the FHL protein family, was observed in either hKO or Tg mouse hearts compared with either control or WT mice, respectively (Figure 4C). We also noticed that, although both mRNA and protein levels of FHL1 were consistently altered in Jmjd2a hKO and/or in Jmjd2a-Tg mouse hearts when compared with their respective controls, the fold change in protein level was less than that of mRNA, suggesting an alternative posttranslational mechanism for FHL1 regulation independent of JMJD2A.
Figure 4. JMJD2A upregulates the expression of FHL1.
qRT-PCR analysis of transcript levels of FHL1 from (A) control (Jmjd2afl/fl) and Jmjd2a hKO mouse hearts (n = 4–6) and (B) WT and Jmjd2a-Tg mouse hearts (n = 3–4) 21 days after sham and TAC surgery. Values are mean ± SEM. *P < 0.05. (C) Western blot of heart lysates of sham- and TAC-operated control and Jmjd2a hKO mice (left panel) and sham- and TAC-operated WT and Jmjd2a-Tg mice (right panel) using antibodies against FHL1, FHL2, phospho-ERK1/2, ERK1/2, phospho-AKT, and AKT. Representative of 4–6 experiments was shown. The relative fold changes for FHL1 and p-ERK were quantified by ImageJ. GAPDH was used as the loading control.
Sheikh et al. have shown that FHL1 is a biomechanical stress sensor that mediates the MAPK-activated signaling pathway. FHL1 is required for pressure overload–induced cardiac hypertrophy, as deletion of FHL1 blunts the TAC-induced hypertrophic response (25). Upregulation of FHL1 leads to activation of the MAPK pathway (25). Consistent with this notion, we observed upregulation of p-ERK1/2 at baseline (Figure 4C) and a further increase in transgenic mouse hearts upon TAC (Figure 4C). Upregulation of p-ERK1/2 in response to TAC was attenuated in TAC-operated Jmjd2a hKO mice (Figure 4C). We also observed upregulation of fetal gene expression in FHL1-transduced neonatal cardiomyocytes (Supplemental Figure 4), suggesting that FHL1 can promote cardiac hypertrophy under cardiac stresses. In light of the functional importance of FHL1 in promoting cardiac hypertrophy, we next focused on investigating whether and how FHL1 is regulated by JMJD2A.
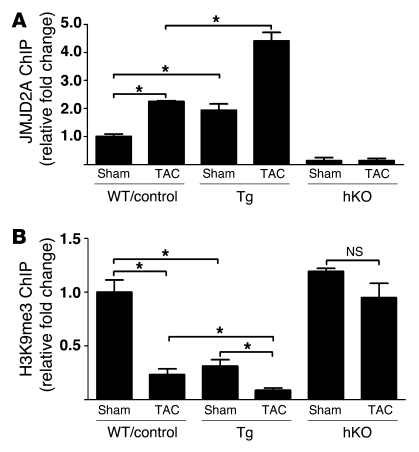
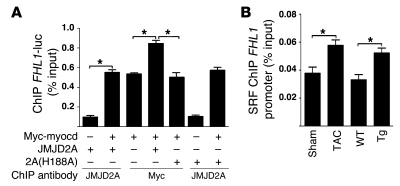
We first determined whether JMJD2A regulates FHL1 transcription by binding to the FHL1 promoter in vivo and in response to pressure overload using ChIP assays. As shown in Figure 5A, JMJD2A binds the FHL1 promoter in response to TAC in either WT or control (Jmjd2afl/fl) mice (Figure 5A). Binding of JMJD2A to the FHL1 promoter is specific, as a JMJD2A ChIP assay on the GAPDH promoter (a negative control) showed very little binding of JMJD2A to the GAPDH promoter (data not shown). Furthermore, ChIP assays with anti-JMJD2A antibodies in Jmjd2a hKO hearts showed very little binding (Figure 5A), which was similar to the results obtained using IgG control (data not shown). Significant amounts of JMJD2A were also found to bind the FHL1 promoter in Jmjd2a-Tg mouse hearts at baseline and were further augmented in TAC-operated Jmjd2a-Tg mouse hearts compared with those of WT counterparts (Figure 5A). ChIP assays with anti-H3K9me3 antibody indicated that binding of JMJD2A to the FHL1 promoter was associated with decreased levels of H3K9me3 (Figure 5B). The H3K9me3 level was also significantly downregulated in Jmjd2a-Tg mice and further decreased upon TAC (Figure 5B). Consistent with the role of JMJD2A in regulating the level of H3K9me3, no significant reduction of H3K9me3 was observed in association with TAC operation in Jmjd2a hKO mice (Figure 5B).
Figure 5. JMJD2A binds to the FHL1 promoter in vivo.
(A) JMJD2A-chromatin complexes were immunoprecipitated from lysates of sham- and TAC-operated WT, control (Jmjd2afl/fl), Jmjd2a-Tg, and Jmjd2a hKO mouse hearts, with anti-JMJD2A antibody and quantified by qPCR with primers in the FHL1 promoter. The amounts of immunoprecipitated complex were normalized against DNA purified from sonicated cell lysates (input) and expressed as relative to that of sham-operated WT/control. (B) ChIP was performed on aliquots of lysates from A using an anti-H3K9me3 antibody. The amounts of chromatin in the FHL1 promoter associated with H3K9me3 were measured by qPCR, normalized against input, and expressed relative to that of sham-operated WT/controls. JMJD2A binds the FHL1 promoter in response to TAC, and binding of JMJD2A is associated with decreased levels of H3K9me3. Values are expressed as mean ± SEM of 3 independent experiments. *P < 0.05.
Jmjd2a upregulates the transcription of FHL1 through SRF and myocardin.
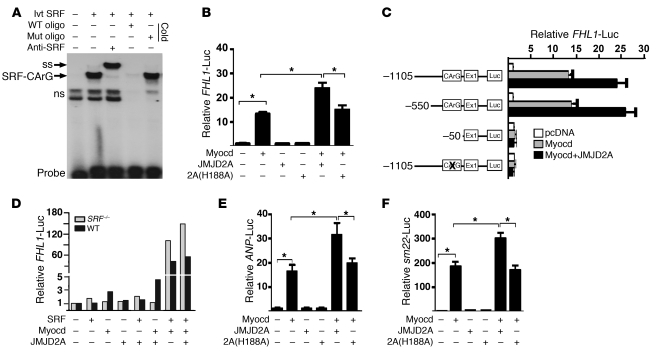
To understand how JMJD2A is recruited to the FHL1 promoter, we examined the 5′-upstream genomic sequences of the FHL1 promoter and identified a conserved serum responsive factor–binding (SRF-binding) site (Supplemental Figure 5), known as the CArG box (26). A gel mobility shift assay indicated that this CArG box is a functional SRF-binding site, as it formed a specific nucleotide-protein complex with SRF (Figure 6A). We next cloned the 1.9-kb promoter sequence containing this CArG box in front of a luciferase reporter plasmid and tested the ability of JMJD2A to transactivate the reporter. Myocardin (myocd) is a SRF cofactor and activates transcription of SRF-dependent genes in cardiac and smooth muscle cells (26–30). Myocardin activated the transcription of FHL1, which was further upregulated by JMJD2A (Figure 6B). The ability of JMJD2A to increase the transcription of FHL1 requires its demethylase activity, as a demethylase-inactive mutant of JMJD2A, 2A (H188A), failed to augment myocardin-activated FHL1 transcription (Figure 6B). Activation of FHL1 transcription by myocardin and JMJD2A was SRF-dependent, as mutation of the CArG box abolished the transactivation of FHL1 by myocardin or myocardin plus JMJD2A (Figure 6C). Furthermore, the ability of myocardin/JMJD2A to activate the transcription of FHL1 was abolished in SRF-null cells (Figure 6D). These results indicate that JMJD2A is a cofactor of SRF/myocardin and its catalytic activity is required for this coactivation. Since the cardiac “fetal” genes ANP and sm22 are known SRF/myocardin-targeted genes, we tested to determine whether JMJD2A can augment the transcription of these genes. As expected, JMJD2A upregulated myocardin-activated transcription of ANP and sm22 in a demethylase-dependent manner as well (Figure 6, E and F, respectively).
Figure 6. JMJD2A upregulates FHL1 transcription synergistically with SRF/myocardin.
(A) A 32P-labeled oligonucleotide probe containing the CArG box from the FHL1 promoter was incubated with the in vitro–translated (ivt) SRF in the presence or absence of anti-SRF antibody or a 100-fold molar excess of unlabeled, WT, or mutant (mut) oligonucleotides. SRF forms a complex with the WT oligonucleotide probe (SRF-CArG) that can be super-shifted (ss) by an anti-SRF antibody. ns, nonspecific nucleoprotein complex. (B) A 1.9-kb FHL1 promoter–driven luciferase construct was transfected along with the plasmids indicated into QBI293A cells. FHL1-luciferase activities were measured 24 hours after transfection and normalized against cotransfected β-galactosidase. (C) Responsiveness of the deletion and site-specific mutants of the FHL1-luc reporter to myocardin and JMJD2A in QBI293A cells. Deletion or site-specific mutation of the SRF-binding site CArG box impairs the responsiveness of the promoter to myocardin and JMJD2A. (D) Relative activity of the FHL1-luc reporter in SRF-null and WT ES cells in the presence and absence of cotransfected expression plasmids as indicated. Representative of 3 independent experiments is shown. (E) Relative luciferase activity of the ANP-luc reporter and (F) the sm22-luc reporter in QBI293A cells transfected with the plasmids indicated. Myocardin-activated ANP- and sm22-luc reporters that can be further upregulated by WT but not mutant JMJD2A. Data shown are mean ± SEM of 3 independent experiments. *P < 0.05.
Hebbar and Archer have shown that all 4 core histones (H2A, H2B, H3, and H4) and the linker histone H1 are associated with transiently transfected DNA despite altered histone H1 stoichiometry and an absence of nucleosome positioning on transfected DNA (31). As demethylated H3K9me3 could provide binding sites to recruit transcription factors, the inability of the demethylase-inactive mutant JMJD2A (H188A) to augment myocardin-activated FHL1-luc reporter suggests that there may be less myocardin binding on the FHL1-luc promoter. We performed ChIP assays to test this hypothesis. As shown in Figure 7A, although the expression levels of myocardin in transfected cells were similar (data not shown), there was more myocardin bound on the FHL1 promoter in cells transfected with both JMJD2A and myocardin than in cells transfected with myocardin alone or myocardin with the catalytically inactive mutant JMJD2A (Figure 7A). An increased amount of SRF was also found to be associated with the endogenous FHL1 promoter chromatin in vivo in either Jmjd2a-Tg mouse heats or hypertrophic mouse hearts after TAC surgery (Figure 7B).
Figure 7. JMJD2A enhances the binding of SRF/myocardin to the FHL1 promoter.
(A) QBI293A cells were transfected with the plasmids indicated. ChIP assays were performed 24 hours later using anti-myc and anti-JMJD2A antibodies as indicated. (B) ChIP assays were performed with lysates of sham- and TAC-operated WT mouse hearts and with WT and Jmjd2a-Tg mouse hearts using anti-SRF antibody. Chromatins associated with SRF on the FHL1 promoter were quantified using qPCR. Data are expressed as mean ± SEM of 3 independent experiments. *P < 0.05.
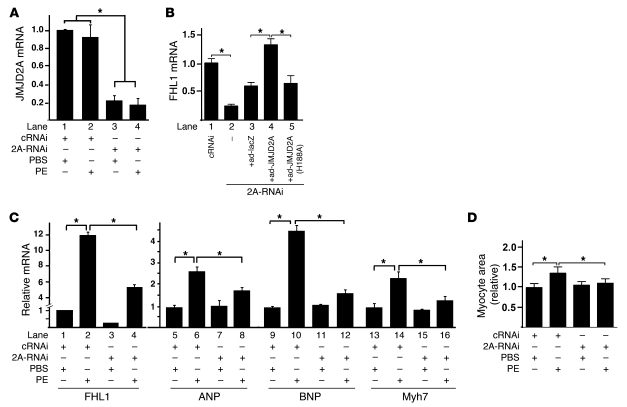
To test whether the demethylase activity of JMJD2A is required for activation of FHL1 transcription in vivo, we performed gene knockdown (KD) experiments in rat neonatal cardiomyocytes using JMJD2A-specific siRNA duplexes. The mRNA level of JMJD2A was significantly KD by JMJD2A-specific siRNAs compared with control nonspecific siRNA (80% reduction; Figure 8A). Consistent with what we observed in Jmjd2a hKO mice, downregulation of JMJD2A in cardiomyocytes resulted in a decrease in the FHL1 transcription at baseline (Figure 8B). Reexpression of JMJD2A in the JMJD2A KD cardiomyocytes using adenoviral transduction restored FHL1 expression (Figure 8B), whereas the mutant JMJD2A (H188A) did not (Figure 8B). We also determined whether JMJD2A is involved in phenylephrine-stimulated (PE-stimulated) myocyte hypertrophy. PE had no effects on the transcript level of JMJD2A (Figure 8A) and significantly upregulated FHL1 transcription (Figure 8C). Upregulation of FHL1 by PE was significantly attenuated in JMJD2A KD cells (Figure 8C). Upregulation of fetal genes by PE, including ANF, BNP, and myh7 (Figure 8C), and of the cardiomyocyte size (Figure 8D) in JMJD2A KD cells were significantly attenuated, suggesting that JMJD2A mediates the hypertrophic effect of PE on cardiomyocytes.
Figure 8. Downregulation of JMJD2A attenuates the expression of FHL1 and PE-activated fetal gene program in cardiomyocytes.
Neonatal rat ventricular myocytes were transfected with nonspecific control siRNA (cRNAi) or JMJD2A-specific siRNA (2A-RNAi). Forty-eight hours after transfection, cells were treated with PBS (vehicle) or with PE (10 μM) (A, C, and D) or with various adenoviruses as indicated (B). Relative transcript levels of JMJD2A (A), FHL1 (B, lanes 1–5, and C, lanes 1–4), and fetal genes (C, lanes 5–16) were determined 24 hours after the treatment using qRT-PCR, normalized against internal GAPDH, and expressed relative to those of the vehicle-treated cRNAi transfected cells. (D) Cardiomyocytes were photographed, and areas were quantified using ImageJ and expressed relative to those of the vehicle-treated cRNAi transfected cells. Data are expressed as mean ± SEM of 3 independent experiments. *P < 0.05.
Discussion
A key finding in the current study is that JMJD2A promotes cardiac hypertrophy under pathological conditions. This is supported by both gain-of- and loss-of-function studies presented here. While overexpression of JMJD2A in mice exacerbates the hypertrophic response to pressure overload, inactivation of Jmjd2a blunts it (Figures 2 and 1, respectively). Since JMJD2A is upregulated in human HCM patients (Figure 3), we speculate that this upregulation of JMJD2A may play an active role in promoting cardiac hypertrophy in humans under cardiac stress conditions. It is noted that the mechanism by which the JMJD2A activity is regulated in mice may be different from that in humans, since we did not observe upregulation of JMJD2A expression in TAC-induced hypertrophy in our mouse model (data not shown). Rather, we found enhanced binding of JMJD2A to the promoter of prohypertrophic genes such as FHL1 in response to TAC.
Our studies suggest that JMJD2A may promote cardiac hypertrophy through FHL1. This is supported by the following evidence: FHL1 is known to mediate pressure overload–induced cardiac hypertrophy (25), and overexpression of FHL1 in cardiomyocyte upregulates fetal gene expression (Supplemental Figure 4). JMJD2A activates the transcription of FHL1 both in vitro and in vivo and in response to hypertrophic stimuli (Figures 4–6). The phenotype of cardiac inactivation of Jmjd2a is consistent with that of FHL1 KO mice (Figure 1). The MAPK-signaling pathway regulated by FHL1 is also affected by JMJD2A (Figure 4). It remains to be determined whether FHL1 is the major downstream effector of JMJD2A. This could be determined by examining the phenotype of Jmjd2a-Tg and FHL1-null compound mice and testing to determine whether inactivation of FHL1 blunts the TAC-induced hypertrophy in Jmjd2a-Tg mice. It is likely that JMJD2A may have other transcriptional targets during hypertrophic remodeling. We have shown that JMJD2A can synergistically activate ANP and sm22 transcription with SRF/myocardin in vitro (Figure 6). Myocardin/SRF has been shown to be involved in cardiac development and hypertrophic remodeling (32–33). Whether these and/or other SRF/myocardin-targeted genes mediate the prohypertrophic function of JMJD2A in vivo remains to be determined in the future. A genome-wide ChIP followed by deep sequencing (ChIP-seq) with chromatins from Jmjd2a-Tg and Jmjd2a hKO hearts would provide means to identify further potential JMJD2A targets involved in cardiac hypertrophy and heart failure.
It is well established that histone modifications play important roles in gene transcription. Over the past decades, a great deal has been learned regarding histone acetylation and its role in cardiac remodeling (6). By comparison, little is known about the function of histone methylation even though it is the most abundant form of histone modifications. Unlike histone acetylation, which is usually associated with gene activation, histone methylation can lead to either activation or repression of gene transcription, depending on the lysine residues, the degree of methylation status (mono-, di-, or trimethylation), and chromatin location. It was shown previously that JMJD1A, a di/monomethyl demethylase, can activate the expression of contractile genes in smooth muscle cells (34). Our studies indicate that trimethyl demethylation of H3K9me3 at the FHL1 promoter by JMJD2A in cardiomyocytes promotes gene transcription. As JMJD2A upregulates other SRF-regulated genes such as ANP and sm22, it is tempting to speculate that demethylation of H3K9 on promoters of SRF-targeted genes activates gene transcription.
JMJD2A could activate the transcription of SRF-targeted genes by either providing binding sites that stabilize/increase the affinity for transcription factors or preventing binding of H3K9me3 to heterochromatin binding protein HP-1. Binding of HP-1 to H3K9me3 was known to mediate conversion of euchromatin to heterochromatin, resulting in silencing of gene transcription. Our studies favor the former possibility as KD of JMJD2A did not abolish the ability of SRF/myocardin to activate FHL1 transcription (Supplemental Figure 6), suggesting that FHL1 was not inactivated in the absence of JMJD2A. Furthermore, we observed an increased amount of SRF/myocardin binding to the FHL1 promoter that is associated with upregulation of JMJD2A and decreased levels of H3K9me3. Taken together, our data suggest a feed-forward mechanism for the synergy between SRF/myocardin and JMDJ2A; SRF/myocardin recruits JMJD2A to the FHL1 promoter. In turn, JMJD2A demethylates H3K9me3, generating a surface/binding area for further recruitment of SRF/myocardin.
JMJD2A is a global regulator of chromatin remodeling and gene expression, and yet deletion of JMJD2A affects the transcription of only a handful of genes. While this finding may seem paradoxical, it is not unexpected. Gene expression is regulated through the action of transcription factors and histone-modifying enzymes. Many different histone-modifying enzymes, including HDACs, HATs, HMTs, and HDMs, contribute to the dynamic regulation of chromatin structure and function, with concomitant impacts on gene transcription. Unlike transcription factors that often have on-off effects on gene transcription, the effects of histone-modifying enzymes on gene transcription are often modulatory. This modulatory effect can be context- and gene-dependent such that only those genes exceeding the threshold will yield a phenotype and be identified. Therefore, it is not surprising that deletion of JMJD2A in the heart resulted in changes of the transcription levels of only a few target genes. However, it is worth noting that even though the number of genes whose expressions are affected by deletion of Jmjd2a is small, the genome-wide H3K9me3 marks affected by Jmjd2a deficiency may still be large. It will be interesting to identify these marks using ChIP-seq and to further investigate the relationship between JMJD2A-regulated H3K9me3 marks and other chromatin marks, i.e., histone code, for such a relationship may ultimately determine the transcriptional state of the gene as either active, repressed, or poised for activation.
In summary, our studies indicate that JMJD2A promotes cardiac hypertrophy in response to hypertrophic stimuli. JMJD2A demethylates H3K9me3 and activates transcription of prohypertrophic genes synergistically with SRF/myocardin. It is noted that the effect of JMJD2A on the expression of SRF/myocardin-targeted genes is modulatory, as loss of function of Jmjd2a does not abolish the ability of SRF/myocardin to activate gene transcription. JMJD2A could be a potential drug target for transcriptional therapy against cardiac hypertrophy and heart failure. It will be worth determining in the future whether small molecules designed to target the demethylase activity of JMJD2A could reduce and normalize the expression of SRF/myocardin-targeted genes that are upregulated during cardiac remodeling without compromising the physiological cardiac growth and functions of these genes that are part of the adaptive response to altered conditions.
Methods
Gene targeting.
A Jmjd2a-targeting vector was generated using the pGKNEO-F2L2-DTA vector that contains a neomycin resistance cassette flanked by frt and loxP sites and a diphtheria toxin gene cassette. The arms for homologous recombination were generated by high-fidelity PCR amplification of 129SvEv genomic DNA. The resulting vector was verified by restriction mapping and DNA sequencing. The targeting vector was linearized with PvuI and electroporated into 129SvEv-derived ES cells. ES cell clones were screened for homologous recombination by Southern blot analysis. Genomic DNA was digested with BclI, and successful loxP site incorporation was confirmed with a 5′ probe and PCR strategy. Targeted ES cells were injected into the blastocysts of C57BL/6 females to generate chimeric mice. Chimeras were bred to C57BL/6 females to achieve germ-line transmission. The Jmjd2afl/fl mice were intercrossed to α-MHC–Cre transgenic mice to obtain mice with Jmjd2a-null allele specifically in the heart. All experiments on control and JMJD2A hKO mice were conducted on a 129SvEv/C57BL6 mixed background.
Generation of Jmjd2a-Tg mice.
A fragment containing cDNA for human JMJD2A with an N-terminal Flag tag was subcloned into the SalI/HindIII sites between the mouse cardiac α-MHC promoter and the human growth hormone gene polyadenylation sequence (35). The plasmid was digested free of vector sequence with BamHI, purified, and microinjected into fertilized eggs of FVB mice to generate transgenic mice according to standard procedures. Founder and offspring mice were genotyped by PCR (Supplemental Table 1).
Surgical procedures and echocardiography.
TAC and sham surgery were performed on control, Jmjd2a hKO, WT, and heterozygote transgenic mice at 8 to 10 weeks of age as described previously (24). Echocardiography was performed on mice before sacrifice. All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Texas Southwestern Medical Center.
Measurement of cardiomyocyte area.
Cardiomyocyte size was assessed by measuring the mean cardiomyocyte cross-sectional area for each sham-operated and TAC-operated heart. More than 200 randomly selected cardiomyocytes cut in cross-section and with a central nucleus on H&E-stained heart sections were measured with ImageJ.
RNA and cDNA preparation, real-time qRT-PCR, and microarray analysis.
Total RNA was isolated with TRIzol reagent (Invitrogen) following manufacturer’s protocol. First-strand cDNA was made using Superscript III Reverse Transcriptase (Invitrogen). SYBR-based qRT-PCR was used to examine relative levels of selected mRNAs. All data were normalized to an internal standard (GAPDH; ΔCT method). Sequences for gene-specific primer pairs are listed in Supplemental Table 1. Microarray analyses were performed using RNAs of mouse hearts from WT sham, WT TAC, Jmjd2a-Tg sham, Jmjd2a-Tg TAC, control, and Jmjd2a hKO mice with Illumina bead array platform. All microarray data were deposited into the NCBI GEO repository (GSE27689).
ChIP.
ChIP was performed as described with minor modifications (36). Protein-chromatin complexes were immunoprecipitated with antibodies against JMJD2A (Bethyl), trimethylated H3K9 (Abcam), and SRF (Santa Cruz Biotechnology Inc.). Immunoprecipitated chromatin fragments were quantified by SYBR-based qPCR, normalized using the percent input method (Invitrogen).
Protein and Western blot.
Tissue protein was extracted with T-PER buffer (Pierce) according to the manufacture’s protocol. After SDS-PAGE separation, the protein was probed on nylon membrane (Bio-Rad). The following antibodies were used: JMJD2A (Bethyl), FHL1 (Abcam), FHL2 (Abcam), p-ERK1/2, ERK1/2, p-AKT, AKT (Cell Signaling), BNP (Santa Cruz Biotechnology Inc.), and GAPDH (Santa Cruz Biotechnology Inc.).
Gel shifts.
Gel shifts were performed as described (37). Briefly, in vitro–translated SRF (Promega) was incubated with 32P-labeled oligonucleotide probe in binding buffer at room temperature for 20 minutes. DNA and protein complexes were separated on 8% nondenaturing polyacrylamide gels. Competition experiments were performed by addition of a 100-fold molar excess of non–radio-labeled WT or mutant oligonucleotide probes. Supershift assay was performed with addition of anti-SRF antibody for 15 minutes following incubation of SRF and radioactive probe in binding buffer.
Plasmids, cell culture, transfection, siRNA, and luciferase reporter assays.
The expression vector of human JMJD2A was made by subcloning PCR-amplified inserts into Flag-tagged pcDNA3.1 (Invitrogen). FHL1-luciferase reporter was constructed by subcloning PCR-amplified inserts corresponding to the promoter sequence of FHL1 from mouse genomic DNA (–1105 to +832) into the pGL3-basic vector (Promega). Point mutation of the FHL1 promoter was performed using the QuikChange kit (Stratagene). Primers used in the cloning are listed in Supplemental Table 1. ANP- and sm22-luciferase reporter constructs, SRF and myocardin expression plasmids, and SRF-null ES cells were gifts from E. Olson (University of Texas Southwestern). QBI-293A cells (ATCC) were maintained in complete medium (DMEM supplemented with 10% fetal bovine serum, 2 mM glutamine, and penicillin/streptomycin). Rat neonatal ventricular cardiomyocytes were prepared as described (38). Cells were transfected with a combination of plasmids indicated for each experiment using Fugene 6 (Roche). siRNA duplex (smart pool) was purchased from Dharmacon and transfected using Lipofectamine 2000 (Invitrogen). Cell lysates were assayed for luciferase expression using a luciferase assay kit (Promega). Relative promoter activities were expressed as relative luminescence units normalized for cotransfected β-galactosidase activities in the cell.
Adenoviruses expressing JMJ2A and FHL1 and neonatal cardiomyocyte culture.
Adenoviruses harboring FHL1, WT, and mutant forms of JMJD2A were constructed and packaged with AdEasy XL Adenoviral system (Stratagene) according to the manufacturer’s protocol. The recombined constructs were used to package and amplify adenovirus in QBI-293A cells.
Patient samples and controls.
Human hypertrophic heart tissue samples were obtained from patients undergoing surgical myectomy who were previously diagnosed with HCM. Control non-HCM adult human ventricular tissue samples were obtained from unmatched victims of motor vehicle accidents (2 cases) and patients previously diagnosed with atrial fibrillation (2 cases). Tissue procurement was based on the receipt of written patient-informed consent and approved by the institutional review boards (Peking Union Medical College). Western blot analysis of the tissue lysates was performed using standard protocols with antibody against JMJD2A (Bethyl), BNP (Santa Cruz Biotechnology Inc.), and GAPDH (Santa Cruz Biotechnology Inc.).
Statistics.
All data are shown as mean ± SEM. When comparing multiple groups, 1-way ANOVA was used. Student’s t test (2-tailed) was used to compare the difference between 2 groups. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This study was supported by a Scientist Development grant from the American Heart Association, Texas Advanced Research Program 010019-0070-2007, and RO1 HL085749 from the National Heart, Lung, and Blood Institute (to Z.P. Liu). Work in J.A. Hill’s lab is supported by HL-075173, HL-080144, HL-090842, UO1-RFA-HL09-010, and T32 HL007360-31 from the NIH, and 0640084N and DeHaan Cardiac Myogenesis Research Center grants from the American Heart Association.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(6):2447–2456. doi:10.1172/JCI46277.
References
- 1.Levy D, Garrison RJ, Savage DD, Kannel WB, Castelli WP. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. N Engl J Med. 1990;322(22):1561–1566. doi: 10.1056/NEJM199005313222203. [DOI] [PubMed] [Google Scholar]
- 2.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358(13):1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 3.Abraham WT, et al. Coordinate changes in Myosin heavy chain isoform gene expression are selectively associated with alterations in dilated cardiomyopathy phenotype. Mol Med. 2002;8(11):750–760. [PMC free article] [PubMed] [Google Scholar]
- 4. Blaxall BC, Tschannen-Moran BM, Milano CA, Koch WJ. Differential gene expression and genomic patient stratification following left ventricular assist device support. Am J Coll Cardiol. 2003. 41 7 1096 1106 . 10.1016/S0735-1097(03)00043-3 [DOI] [PubMed] [Google Scholar]
- 5.Meyer K, Zhang L. Fetal programming of cardiac function and disease. Reprod Sci. 2007;14(3):209–216. doi: 10.1177/1933719107302324. [DOI] [PubMed] [Google Scholar]
- 6.McKinsey TA, Olson EN. Toward transcriptional therapies for the failing heart: chemical screens to modulate genes. J Clin Invest. 2005;115(3):538–546. doi: 10.1172/JCI24144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couture JF, Trievel RC. Histone-modifying enzymes: encrypting an enigmatic epigenetic code. Curr Opin Struct Biol. 2006;16(6):753–760. doi: 10.1016/j.sbi.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Bártová E, Krejcí J, Harnicarová A, Galiová G, Kozubek S. Histone modifications and nuclear architecture: a review. J Histochem Cytochem. 2008;56(8):711–721. doi: 10.1369/jhc.2008.951251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 10.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406(6796):593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 11.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Trojer P, Reinberg D. Histone lysine demethylases and their impact on epigenetics. Cell. 2006;125(2):213–217. doi: 10.1016/j.cell.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Klose RJ, et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine36. Nature. 2006;442(7100):312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 14.Whetstine JR, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125(3):467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 15.Barski A, et al. High resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 16.Lorbeck MT, et al. The histoe demethylase Dmel\Kdm4A controls genes required for life span and male–specific sex determination in Drosophila. Gene. 2010;450(1–2):8–17. doi: 10.1016/j.gene.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agger K, Christensen J, Cloos PA, Helin K. The emerging functions of histone demethylases. Curr Opin Genet Dev. 2008;18(2):159–168. doi: 10.1016/j.gde.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Nottke A, Colaiácovo MP, Shi Y. Developmental roles of the histone lysine demethylases. Development. 2009;136(6):879–889. doi: 10.1242/dev.020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneda R, et al. Genome-wide histone methylation profile for heart failure. Genes Cells. 2009;14(1):69–77. doi: 10.1111/j.1365-2443.2008.01252.x. [DOI] [PubMed] [Google Scholar]
- 20.Shin S, Janknecht R. Activation of androgen receptor by histone demethylases JMJD2A and JMJD2D. Biochem Biophys Res Commun. 2007;359(3):742–746. doi: 10.1016/j.bbrc.2007.05.179. [DOI] [PubMed] [Google Scholar]
- 21.Zhang D, Yoon HG, Wong J. JMJD2A is a novel N-CoR-interacting protein and is involved in repression of the human transcription factor achaete scute-like homologue 2 (ASCL2/Hash2). Mol Cell Biol. 2005;25(15):6404–6414. doi: 10.1128/MCB.25.15.6404-6414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray SG, et al. Functional characterization of JMJD2A, a histone deacetylase- and retinoblastoma-binding protein. J Biol Chem. 2005;280(31):28507–28518. doi: 10.1074/jbc.M413687200. [DOI] [PubMed] [Google Scholar]
- 23.Oka T, et al. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ Res. 2006;98(6):837–845. doi: 10.1161/01.RES.0000215985.18538.c4. [DOI] [PubMed] [Google Scholar]
- 24. Berry JM, Naseem RH, Rothermel BA, Hill JA. Models of cardiac hypertrophy and transition to heart failure. Drug Discovery Today: Disease Models. 2007. 4 4 197 206 . 10.1016/j.ddmod.2007.06.003 [DOI] [Google Scholar]
- 25.Sheikh F, et al. An FHL1-containing complex within the cardiomyocyte sarcomere mediates hypertrophic biomechanical stress responses in mice. J Clin Invest. 2008;118(12):3870–3880. doi: 10.1172/JCI34472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miano JM. Serum response factor: toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35(6):577–593. doi: 10.1016/S0022-2828(03)00110-X. [DOI] [PubMed] [Google Scholar]
- 27.McDonald OG, Owens GK. Programming smooth muscle plasticity with chromatin dynamics. Circ Res. 2007;100(10):1428–1441. doi: 10.1161/01.RES.0000266448.30370.a0. [DOI] [PubMed] [Google Scholar]
- 28.Wang D, et al. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105(7):851–862. doi: 10.1016/S0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 29.Wang DZ, Olson EN. Control of smooth muscle development by the myocardin family of transcriptional coactivators. Curr Opin Genet Dev. 2004;14(5):558–566. doi: 10.1016/j.gde.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol. 2007;292(1):C70–C81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- 31.Hebbar PB, Archer TK. Altered histone H1 stoichiometry and an absence of nucleosome positioning on transfected DNA. J Biol Chem. 2008;283(8):4595–4601. doi: 10.1074/jbc.M709121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xing W, et al. Myocardin induces cardiomyocyte hypertrophy. Circ Res. 2006;98(8):1089–1097. doi: 10.1161/01.RES.0000218781.23144.3e. [DOI] [PubMed] [Google Scholar]
- 33.Kontaraki JE, Parthenakis FI, Patrianakos AP, Karalis IK, Vardas PE. Myocardin gene regulatory variants as surrogate markers of cardiac hypertrophy – study in a genetically homogeneous population. Clin Genet. 2008;73(1):71–78. doi: 10.1111/j.1399-0004.2007.00932.x. [DOI] [PubMed] [Google Scholar]
- 34.Lockman K, Taylor JM, Mack CP. The histone demethylase, Jmjd1a, interacts with the myocardin factors to regulate SMC differentiation marker gene expression. Circ Res. 2007;101(12):e115–e123. doi: 10.1161/CIRCRESAHA.107.164178. [DOI] [PubMed] [Google Scholar]
- 35.Subramaniam A, Jones WK, Gulick J, Wert S, Neumann J, Robbins J. Tissue-specific regulation of the alpha-myosin heavy chain gene promoter in transgenic mice. J Biol Chem. 1991;266(36):24613–24620. [PubMed] [Google Scholar]
- 36.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1(1):179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 37.Zhou W, et al. FoxO4 inhibits NF-kappaB and protects mice against colonic injury and inflammation. Gastroenterology. 2009;137(4):1403–1414. doi: 10.1053/j.gastro.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu ZP, Olson EN. Suppression of proliferation and cardiomyocyte hypertrophy by CHAMP, a cardiac-specific RNA helicase. Proc Natl Acad Sci U S A. 2002;99(4):2043–2048. doi: 10.1073/pnas.261708699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.