Abstract
The nuclear pore complex (NPC) acts as a selective gate that mediates the bidirectional transport of macromolecules between the cytoplasm and the nucleus of eukaryotic cells. ‘Natively unfolded’ nucleoporins (Nups) with domains rich in phenylalanine-glycine (FG) repeats form the selective permeability barrier and provide binding sites for mobile transport receptors in the NPC. Understanding the structure and function of the FG-Nups barrier under real-time trafficking conditions is still a formidable challenge due to the dynamic nature of a channeled membranous environment. Recently, we have shown that three-dimensional (3D) density maps of transient interactions between the FG-Nups barrier and a cargo-free or a cargo-bound transport receptor in native NPCs can be obtained by an advanced single-molecule fluorescence microscopy approach. Moreover, we found that these interaction sites are spatially clustered into distinct groups in the periphery around a central axial channel with a diameter of approximately 10–20 nm in the NPC. The 3D distribution of interaction sites may indicate some native properties of the FG-Nups barrier. Here we speculate that the selective permeability barrier in the NPC could be formed by clustered FG-Nups.
Key words: nucleocytoplasmic transport, single-molecule tracking, three-dimensional distribution, real-time trafficking, super-resolution microscopy, single-molecule fluorescence
NPC Composition and Function
In eukaryotic cells, the exchange of genetic materials generated by transcription in the nucleus and proteins synthesized in the cytoplasm is mediated by nuclear pore complexes (NPCs). The NPC is a large assembly (∼60–100 MDa) that is embedded in the nuclear envelope (NE) and composed of approximately 30 different protein components, known as nucleoporins (Nups), each present in an integer multiple of eight copies.1–5 These Nups form a selective gate that allows passive diffusion of small molecules (<20–40 kDa) and transport-receptor-facilitated translocation of larger molecules (up to 25–50 MDa).6–8 Electron microscopy has revealed the general morphology of the NPC at nanometer-level resolution. The central nuclear pore is about 40–90 nm in length, with a minimum internal diameter of about 40–75 nm and an external diameter of about 120 nm. Flexible filaments extend about 50 nm into the cytoplasm, and a basket structure extends about 75 nm into the nucleus.8–10 Thus, a transiting substrate can potentially interact continuously with the Nups over a distance spanning about 200 nm.
The Structure and Function of FG-Nups in the NPC
In vertebrates, the approximately 30 different Nups can be classified as transmembrane Nups (∼10% of all Nups), structural Nups (∼50% of all Nups) and FG-Nups (∼40% of all Nups).1,8,11,12 The first two types typically contain either an alpha-solenoid or a beta-propeller fold, or in some cases both as separate structural domains. In contrast, the unique FG-Nups exhibit structural characteristics of ‘natively unfolded’ proteins, i.e., they are highly flexible proteins that lack ordered secondary structure. These disordered proteins include amino-acid sequences that contain many repeats of phenylalanine-glycine (FG) separated by spacer sequences. The core units of FG repeats can be further defined as predominately FG, GLFG (Gly-Leu-Phe-Gly) or FXFG (Phe-any-Phe-Gly).12
The FG-Nups are arranged in distinct sub-structural locations in the NPC, and the general morphology of the NPC is conserved from yeast to humans. In vertebrates, two Nups containing predominantly FXFG (Nup 153 and Nup 50) are found exclusively on the nuclear-basket side of the NPC. Four FG-repeat-containing Nups (POM 121, Nup 54, Nup 58 and Nup 45), one FXFG-Nup (Nup 62) and one GLFG-Nup (Nup 98) are distributed centrally or symmetrically around the scaffold of the NPC. On the cytoplasmic-fibrils side, there are two FG-Nups (Nup 214 and hCG1) and a single FXFG-Nup (Nup 358).5 In earlier studies, it was speculated that the asymmetric Nups serve as initial or terminal docking sites and likely as unidirectional indicators for transport substrates.13 However, later investigations on depletion of those asymmetric FG-Nups indicated there is almost no harm to transport viability.14
The FG-Nups have been found to be critical for nucleocytoplasmic transport: first, the FG-Nups form a highly selective barrier in the NPC to gate the essential bidirectional exchange of macromolecules between the cytoplasm and nucleus.6–8,11,12 Second, the ‘natively unfolded’ or ‘intrinsically disordered’ confirmation of FG-Nups allows the barrier to adapt multiple transient configurations to promote rapid association or disassociation rates for various transport cargos.11,12,15 Third, the FG-Nups barrier remains selective while simultaneously mediating multiple robust and efficient transport pathways. However, the functional analysis of nucleocytoplasmic transport remains a challenge in native NPCs due to the size of NPCs, the uncrystallizable FG-Nups and the dynamic massive transport. To advance our understanding of the transport mechanism, determining the native structure of the FG-Nups in the NPC and the transport receptor-FG Nups interactions is undoubtedly central to the process.
Interactions between the FG-Nups and Transport Receptors
Facilitated translocation requires direct interactions between the transiting molecules and the NPC. Large nucleocytoplasmic transport cargos must contain specific targeting signals, generally referred to as nuclear localization signals (NLS) for nuclear import or nuclear export signals (NES) for nuclear export.16 The cargos can be specifically recognized by soluble accessory proteins that mediate transient interactions with FG-Nups—importins (for nuclear import) or exportins (for export). Importin β (Imp β)-type transport receptors account for the majority of the nuclear transport pathways. These receptors transport between the nucleus and cytoplasm, capture cargo molecules and mediate them from one side of the NE to the other. Association and dissociation of cargo-transporter complexes are guided by a concentration gradient of RanGTP across the NE, which is triggered through the RanGTP binding domains present in the transport receptors. A high nuclear-RanGTP concentration promotes cargos binding to exportins and the dislocation of substrates from importins, while cytoplasmic conditions with low levels of RanGTP enhance substrates released from exportins but allow importin-cargo complexes to form.17 Ran is constantly exported from the nucleus by importins and exportins. Continued transport cycles therefore require a renewal of the nuclear RanGTP pool, which is accomplished by nuclear transport factor 2 (NTF2)-mediated import of RanGDP, followed by RCC1 (Ran exchange factors)-mediated nucleotide exchange to RanGTP.18
Biochemical measurements and computational simulations have clearly indicated that there are multiple FG repeats on each FG-Nups and several FG-binding sites on transport receptors, for example, approximately ten FG-binding sites on Imp β1 and two binding sites on NTF2.38–45 Numerous pieces of evidence support the idea that facilitated translocation is accomplished through a series of low-affinity binding events between the transiting molecules and the FG-Nups along the NPC.19–28 Single-molecule imaging of nucleocytoplasmic transport in a permeabilized or live-cell system demonstrates that a transport receptor or a transport-receptor-cargo complex does not move through the NPC in a directional or linear fashion but instead proceeds through Brownian motion with transport times of ∼5–10 ms.29–33 Interactions between transport receptors and FG domains typically have nanomolar to micromolar binding affinities.34,35 Thus, the transport process suggests multiple transient and low-affinity NPC-receptor interactions. Indeed, transport-receptor mutants with increased affinity for binding FG repeats, such as Imp β1 (45–462) and NTF2 (N77Y) mutants, impair nucleocytoplasmic transport.35–37 Overall, rapid, low-affinity interactions between transport receptors and the FG-Nups are necessary for efficient facilitated translocations.
Proposed Transport Models
Because of the limited knowledge of the native configuration of disordered FG-Nups and the transient dynamic interactions in the NPC, it is still difficult to define the precise mechanism of nucleocytoplasmic transport. Numerous nuclear translocation models have been postulated, but none of them completely cover all currently known properties of nuclear transport. In addition, these models are not excluded from each other completely, and some of them are even ambiguous, which makes it a very difficult task to distinguish or validate the models by experimental design. Overall, there are at least three major disputes among the proposed models: (1) How do the FG-Nups form a selective permeability barrier in the native NPC? (2) How do transport receptors interact with the FG-Nups barrier to mediate translocation of larger cargo molecules through the NPC? (3) Where are the pathways for passive diffusion and facilitated translocations located in the NPC?
The ‘virtual gating/polymer brush’ model.
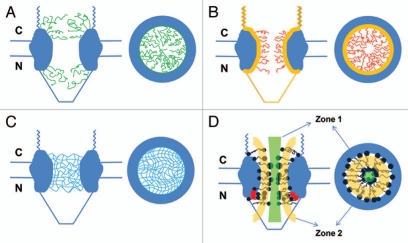
The ‘virtual gating’ model views the NPC as a catalyst that can lower the activation energy for the translocation process. It suggests that both sides of the NPC channel, crowded by FG-Nups, form a highly selective entropic barrier for large molecules (Fig. 1).3 Binding of transport receptors to FG repeats would overcome this entropic barrier and would provide a significant kinetic advantage for cargos that are bound to transport receptors (Fig. 1A).3 The ‘polymer brush’ model further suggests that the non-interacting hydrophobic FG-Nups can be pushed aside by receptor-cargo complexes (Fig. 1A). Recently, in vitro-purified FG filaments of Nup153 were experimentally observed to be collapsed by Imp β1 to their anchoring sites. The more Imp β1 was added, the more the FG filaments of Nup 153 were collapsed toward their anchoring sites.38
Figure 1.
Nucleocytoplasmic transport models. Structural models are presented in both a side view (perpendicular to the NE) and a top view (cross-section through the center of the NPC). (A) The ‘virtual gating/polymer brush’ model. Non-interacting FG-Nups (green curves) extend as ‘repulsive bristles’ or ‘polymer brushes’ to form an entropic barrier on both sides of the NPC. The entropic barrier is unfavorable for larger cargo molecules, but transport receptors binding to these FG domains can overcome the barrier and gain access into and through the NPC for themselves and their cargo complexes. (B) The ‘reduction of dimensionality’ model. FG domains (yellow layer) continuously coat the inner surface of the NPC to provide continuous binding sites for transport receptors and their cargo complexes. Transport complexes move along the coated surface with a random two-dimensional walk. The spacer sequence (red curves) between FG repeats may form a physical barrier for larger cargo molecules alone and constitute a narrow axial channel for passive diffusion of smaller molecules. (C) The ‘selective phase/hydrogel’ model. Hydrophobic interactions among FG repeats generate a sieve-like hydrogel meshwork (blue mesh). Transport receptors and the transport complexes bind to and dissolve into the meshwork to complete the translocation. Smaller molecules diffuse through multiple gaps in the meshwork. (D) The ‘forest’ model. FG-Nups contain FG domains with two conformations: the globular-collapsed conformation (dark blue dots for low charge and cohesive, red dots for high charge and non-cohesive) and the extended-coil conformation (dark blue lines). The globular-collapsed domains form a transport zone 1 in the middle of the NPC, and the extended-coil domains form a peripheral zone 2.
The ‘reduction of dimensionality’ model.
The ‘reduction of dimensionality’ model proposes that the FG repeats of FG-Nups coat the inner surface of the NPC, and that transport receptors or cargo complexes conduct two-dimensional movement along the inner wall by continuous interactions with the FG repeats. The spacer sequences between the FG repeats would form a physical barrier for large molecules and leave a narrow central axial channel for passive diffusion of small molecules in the NPC (Fig. 1B).46
The ‘selective phase/hydrogel’ model.
In contrast to the above models, the ‘selective phase’ model proposes that a sieve-like meshwork is formed within the NPC through weak hydrophobic interactions between FG repeats.47,48 Here, the size of the FG mesh determines the upper size limits of the diffusion gates for small molecules and mechanically restricts the access of large molecules. Therefore, there will be multiple narrow channels for passive diffusion of small molecules in the NPCs. However, the binding of transport receptors to FG repeats can dissolve the FG-FG meshwork, and this binding therefore facilitates exclusive access to the NPC for large receptor-cargo complexes (Fig. 1C). Interestingly, it was recently shown that the FG-repeat domain of a specific Nup can form a hydrogel-like structure in vitro by hydrophobic interactions between aromatic rings.48 This hydrogel-like structure can also display selective properties that are reminiscent of the gating behavior of NPCs.
The ‘forest’ model.
Based on the measurements of low-affinity interactions between the FG Nups, it has been found that the FG Nups anchored around the central plane of the NPC may form a meshwork of filaments, as predicted by the ‘selective-phase’ model, while few FG Nups anchored at the nuclear basket behave as repulsive filaments, as would be suggested by the ‘virtual-gate’ model.14 In a recent further study, analysis of the hydrodynamic properties of the FG-domains of FG-Nups revealed that there are five distinct domains: (a) collapsed-coil, low charge-content, cohesive domain, (b) collapsed-coil, high charge-content, non-cohesive domain, (c) extended-coil, non-cohesive domain, (d) folded domain and (e) NPC anchor domain.49 The clusters of collapsed-coil domains form the so-called ‘shrubs’, and the groups of extended-coil domains or the mixtures of extended-coil and collapsed-coil domains form ‘trees’. Thus, a forest-like structure of FG-Nups is formed in the NPC. In the ‘forest’ model, the collapsed-coil domains form a transport zone 1 in the middle of the NPC, and the extended-coil domains form a peripheral zone 2 (Fig. 1D). Small molecules, transport receptors and complexes pass through zone 1 or 2 depending on their size, surface charge and hydrophobicity.
High-resolution Snapshots of Transient Interactions between the FG-Nups Barrier and Transiting Molecules in Native NPCs
Examinations of the proposed models give rise to challenging questions such as how to non-invasively detect the structure of FG-barrier in native NPCs. Furthermore, it is not clear how to capture transient passive diffusion or transient transporter-FG-repeat interactions under real-time trafficking conditions. Electron microscopy (EM) and cryo-EM have been employed to reveal the architecture of NPCs. Moreover, the localizations of individual structured Nups have been determined by immuno-EM.5–9,12 However, the unstructured dynamic selective permeability FG-barrier and the transient dynamic diffusion or interactions cannot be measured directly by the above methods in either stained or freeze-isolated NPCs.
Because of its opportunities for non-invasive and sub-diffraction-limit imaging and tracking of bio-molecules in live cells and tissues, single-molecule fluorescence microscopy has been widely applied to elucidate many problems unresolved by conventional ensemble methods in biological and biomedical research. In fact, wide-field and narrow-field epi-fluorescence microscopy have been successfully used to track transport receptors and cargo complexes through the NPCs at the single-molecule level.29–33 A series of transport kinetics in the NPC have been determined in these studies, such as transport time, transport efficiency and the one-dimensional (1D) spatial locations of single transiting molecules. Recently, single-point edge-excitation sub-diffraction (SPEED) microscopy, a newly developed form of single-molecule fluorescence microscopy, was used to further advance the imaging and tracking of single transporting molecules through the NPC from 1D to 3D.52 SPEED microscopy is capable of tracking the locations of Alexa Fluor 647-labeled transiting molecules in a single native GFP-conjugated NPC with a spatiotemporal resolution of 9 nm at 400 µs. By SPEED microscopy, a 2D density map of transient interactions between transiting molecules and the FG-Nups can be plotted. A subsequent deconvolution algorithm applied to the 2D density map can further recover a 3D spatial-density distribution of those transient interactions in native NPCs. Thus, 3D transport passageways for transiting molecules through the NPC can be obtained under real-time trafficking conditions. We have applied the technique to determine the spatial locations of 3D transport pathways for Imp β1 through the NPC with and without a model import cargo of NLS-2xGFP (a nuclear localization sequence of PPKKKRKV and two copies of green fluorescence protein).52
The 3D spatial-density map of Imp β1-FG-repeat interactions obtained by the new single-molecule method revealed the spatial distribution of FG repeats available for Imp β1 molecules in the native NPCs. We have found that the spatial density of interaction sites gradually increases from both sides of the NPC and is highest in the central pore region. Moreover, cargo-free or cargo-bound Imp β1 rarely occupies an axial channel with a diameter of approximately 10–20 nm at its narrowest point through the NPC, and the interaction regions project extensively into the cytoplasm and the nucleoplasm (Figs. 2 and 3).52 The density maps of Imp β1-FG-repeat interactions detected under real-time trafficking conditions not only verified the estimated spatial distribution of the unstructured FG domains50,51 (Fig. 2), but also enabled us to further examine the previously proposed models. First, the symmetrical gradient distribution of interaction densities between Imp β1 and the FG-Nups may suggest a higher entropic barrier in the central pore and several lower barriers on either side, which is complementary to the proposal by the ‘virtual gating’ model that a single entropic barrier exists in the NPC.3,52 Such a gradient entropic barrier allows much easier diffusion of facilitated translocations through the NPC.52 Second, the axial channel seldom occupied by cargo-free and cargo-bound Imp β1 was further verified recently by SPEED microscopy as indeed being the major pathway for passive diffusion (Ma and Yang, unpublished data). This finding supports the prediction of a central primary pathway for small molecules by the ‘reduction of dimensionality’ model,46 but not the hypothesis of multiple small channels proposed by the ‘selective phase/hydrogel’ model.47,48 Third, the sparse and wide distribution of interaction sites in the cytoplasm and the nucleoplasm may suggest that the FG-Nups preferably adapt extended conformation on both sides of the central pore as speculated in the ‘virtual gating/polymer brush’ model.38 Finally, the spatial locations we observed of pathways for cargo-free and cargo-bound Imp β1 strongly support the predictions by the ‘forest’ model of distinct transport zones for transport receptors and receptor-cargo complexes.49
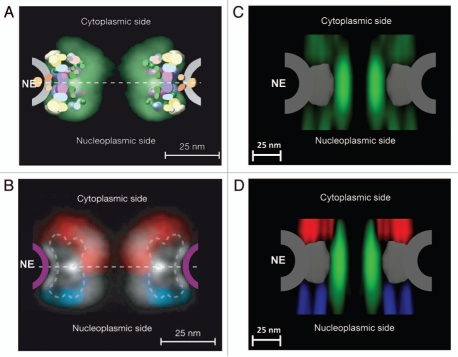
Figure 2.
Spatial-density distribution of the FG-repeat regions in the NPC. (A) Map density of Nups derived from the combined localization volumes of all structured domains (five different colors) and the normalized localization probability of all unstructured regions (green cloud). The estimated unstructured regions are based on integrated data from cryo-EM, immuno-electron microscopy, ultracentrifugation, immunoblotting, protein interaction assays, affinity purification and proteomics.50,51 NE indicates the location of the nuclear envelope. (B) Projection of the localization probabilities of the FG-repeat regions from all the FG-Nups is shown by a density plot. Red for the FG-Nups that are cytoplasmically disposed; blue for those nucleoplasmically disposed; white for those found equally on both sides. (A and B) are reprinted from reference 51 with permission from the publisher. (C) Projection of the normalized spatial-density map of interactions between Imp β1 and the FG repeats (green cloud) at the central pore region, detected by SPEED microscopy. (D) The same density map as in (C) is shown in different colored regions (red for the cytoplasm, blue for the nucleoplasm and green for the central pore).
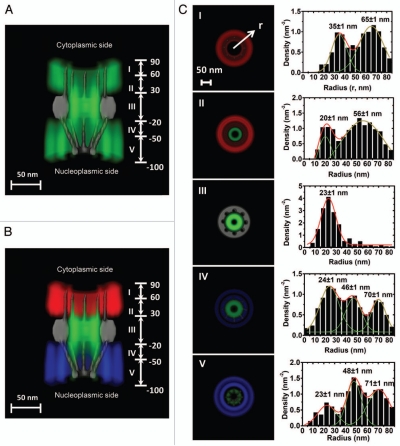
Figure 3.
A 3D spatial-density map of interaction sites between Imp β1 and the FG-Nups. (A) A complete 3D spatial-density map of interaction sites between Imp β1 and the FG-Nups. Cut-away view of the 3D spatial density map of Imp β1 (green cloud and brighter color indicates higher density) superimposed on the NPC architecture (grey). Five regions with distinct spatial location clusters for Imp β1 are marked from I to V. Numbers denote the distance from the central plane of the NPC in nanometers. (B) The same density map as in (A) is shown in different colored regions (red for the cytoplasm, blue for the nucleoplasm and green for the central pore). (C) Histograms of averaged spatial densities along the radii (r) at the cross-section of NPC in the range I to V. Major peaks were obtained by Gaussian fittings (green and red lines). Bin size: 5 nm.
Furthermore, the single-molecule studies of the Imp β1-FG-repeat interactions have provided more new insights into the structure of ‘native unfolded’ FG-Nups in the native NPC. As shown in Figure 3, the interaction sites between Imp β1 and the FG repeats clearly form several different regions along the NPC axis and are further divided into distinct interaction groups in each region. Apparently, the interaction sites are not evenly distributed in the NPC; instead, they form spatial clusters inside and outside the central nuclear pore. Several unique features for these clustered interaction sites were observed: (1) these interaction clusters cannot be formed without the presence of both Imp β1 and the FG domains in the same regions; (2) the clustered interaction sites between Imp β1 and the FG repeats statistically remained at almost the same spatial locations when the same transport conditions were set up in the permeabilized cell system; and (3) more distinct interaction groups with lower spatial densities project into the cytoplasm and the nucleoplasm than in the central pore region. The interaction regions with higher spatial densities indicate that there are more or longer effective interactions between Imp β1 and the FG-Nups. However, in the rarely Imp β1-occupied axial channel serving as the major pathway for passive diffusion of small molecules, there could be no FG repeats or few FG repeats with very low binding affinities for Imp β1. Therefore, the heterogeneously distributed clusters are driven by the interactions between Imp β1 and the FG-Nups. Finally, the observed configurations of interaction sites do not support that the FG repeats are evenly distributed in the NPC, and clearly agree with the descriptions that FG-Nups may form ‘bundles’ or ‘shrubs’ and ‘trees’ as suggested by computational simulations and the ‘forest’ model.43–45,49
Outlook
Ultimate verification of nucleocytoplasmic transport mechanism requires us to close more fundamental gaps in our understanding of nucleocytoplasmic transport: (1) How do the simultaneous import and export coordinate to enhance robust and effective trafficking through the native NPC, specifically when a very large cargo complex is passing through the NPC, such as during export of a ribosomal subunit? (2) How does the conformation of the FG-Nups barrier change to handle massive trafficking conditions? (3) How can we unify the proposed models to cover all current knowledge of nuclear transport? Undoubtedly, new experimental designs and further investigations are needed to seek answers to the above questions. A combination of multiple methodologies and a systematic analysis of the obtained data will continually advance our understanding of the nucleocytoplasmic transport mechanism.
Acknowledgements
We thank the foundation support from National Institute of Health (GM094041-01) and the Research Capacity Enhancement Grant (Bowling Green State University).
Abbreviations
- NPC
nuclear pore complex
- Imp β1
importin β1
- Nups
nucleoporins
- FG
phenylalanine-glycine
- SPEED
single-point edge-excitation sub-diffraction
References
- 1.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rout MP, Aitchison JD. The nuclear pore complex as a transport machine. J Biol Chem. 2001;276:16593–16596. doi: 10.1074/jbc.R100015200. [DOI] [PubMed] [Google Scholar]
- 3.Rout MP, Aitchison JD, Magnasco MO, Chait BT. Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 2003;13:622–628. doi: 10.1016/j.tcb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture and transport mechanism. J Cell Biol. 2000;148:635–641. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suntharalingam M, Wente SR. Peering through the pore: nuclear pore complex structure, assembly and function. Dev Cell. 2003;4:775–789. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- 6.Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 7.Fried H, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci. 2003;60:1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fahrenkrog B, Aebi U. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol. 2003;4:757–766. doi: 10.1038/nrm1230. [DOI] [PubMed] [Google Scholar]
- 9.Beck M, Forster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, et al. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- 10.Rout MP, Blobel G. Isolation of the yeast nuclear pore complex. J Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denning DP, Patel SS, Uversky V, Fink AL, Rexach M. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci USA. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rout MP, Wente SR. Pores for thought: nuclear pore complex proteins. Trends Cell Biol. 1994;4:357–365. doi: 10.1016/0962-8924(94)90085-x. [DOI] [PubMed] [Google Scholar]
- 13.Strawn LA, Shen T, Shulga N, Goldfarb DS, Wente SR. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat Cell Biol. 2004;6:197–206. doi: 10.1038/ncb1097. [DOI] [PubMed] [Google Scholar]
- 14.Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- 15.Pontius BW. Close encounters: why unstructured, polymeric domains can increase rates of specific macromolecular association. Trends Biochem Sci. 1993;18:181–186. doi: 10.1016/0968-0004(93)90111-y. [DOI] [PubMed] [Google Scholar]
- 16.Görlich D, Panté N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 17.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 18.Ribbeck K, Lipowsky G, Ken HM, Stewart M, Görlich D. NTF2 mediates nuclear import of Ran. EMBO J. 1998;17:6587–6598. doi: 10.1093/emboj/17.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 20.Allen NP, Huang L, Burlingame A, Rexach M. Proteomic analysis of nucleoporin interacting proteins. J Biol Chem. 2001;276:29268–29274. doi: 10.1074/jbc.M102629200. [DOI] [PubMed] [Google Scholar]
- 21.Damelin M, Silver PA. Mapping interactions between nuclear transport factors in living cells reveals pathways through the nuclear pore complex. Mol Cell. 2000;5:133–140. doi: 10.1016/s1097-2765(00)80409-8. [DOI] [PubMed] [Google Scholar]
- 22.Katahira J, Straesser K, Saiwaki T, Yoneda Y, Hurt E. Complex formation between Tap and p15 affects binding to FG-repeat nucleoporins and nucleocytoplasmic shuttling. J Biol Chem. 2002;277:9242–9246. doi: 10.1074/jbc.M110007200. [DOI] [PubMed] [Google Scholar]
- 23.Marelli M, Aitchison JD, Wozniak RW. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p and Nup170p. J Cell Biol. 1998;143:1813–1830. doi: 10.1083/jcb.143.7.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rout MP, Blobel G, Aitchison JD. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 25.Strasser K, Bassler J, Hurt E. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG and FG repeat nucleoporins is essential for nuclear mRNA export. J Cell Biol. 2000;150:695–706. doi: 10.1083/jcb.150.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strawn LA, Shen T, Shulga N, Goldfarb DS, Wente SR. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat Cell Biol. 2004;6:197–206. doi: 10.1038/ncb1097. [DOI] [PubMed] [Google Scholar]
- 27.Strawn LA, Shen T, Wente SR. The GLFG regions of Nup116p and Nup100p serve as binding sites for both Kap95p and Mex67p at the nuclear pore complex. J Biol Chem. 2001;276:6445–6452. doi: 10.1074/jbc.M008311200. [DOI] [PubMed] [Google Scholar]
- 28.Terry LJ, Wente SR. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J Cell Biol. 2007;178:1121–1132. doi: 10.1083/jcb.200704174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang W, Gelles J, Musser SM. Imaging of single-molecule translocation through nuclear pore complexes. Proc Natl Acad Sci USA. 2004;101:12887–12892. doi: 10.1073/pnas.0403675101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang W, Musser SM. Visualizing single molecules interacting with nuclear pore complexes by narrow-field epifluorescence microscopy. Methods. 2006;39:316–328. doi: 10.1016/j.ymeth.2006.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang W, Musser SM. Nuclear import time and transport efficiency depend on importin beta concentration. J Cell Biol. 2006;174:951–961. doi: 10.1083/jcb.200605053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kubitscheck U, Grunwald D, Hoekstra A, Rohleder D, Kues T, Siebrasse JP, et al. Nuclear transport of single molecules: dwell times at the nuclear pore complex. J Cell Biol. 2005;168:233–243. doi: 10.1083/jcb.200411005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dange T, Grünwald D, Grünwald A, Peters R, Kubitscheck U. Autonomy and robustness of translocation through the nuclear pore complex: a single-molecule study. J Cell Biol. 2008;183:77–86. doi: 10.1083/jcb.200806173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pyhtila B, Rexach M. A gradient of affinity for the karyopherin Kap95p along the yeast nuclear pore complex. J Biol Chem. 2003;278:42699–42709. doi: 10.1074/jbc.M307135200. [DOI] [PubMed] [Google Scholar]
- 35.Quimby BB, Leung SW, Bayliss R, Harreman MT, Thirumala G, Stewart M, et al. Functional analysis of the hydrophobic patch on nuclear transport factor 2 involved in interactions with the nuclear pore in vivo. J Biol Chem. 2001;276:38820–38829. doi: 10.1074/jbc.M105054200. [DOI] [PubMed] [Google Scholar]
- 36.Jäggi RD, Franco-Obregón A, Mühlhäusser P, Thomas F, Kutay U, Ensslin K. Modulation of nuclear pore topology by transport modifiers. Biophys J. 2003;84:665–670. doi: 10.1016/S0006-3495(03)74886-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lane CM, Cushman I, Moore MS. Selective disruption of nuclear import by a functional mutant nuclear transport carrier. J Cell Biol. 2000;151:321–332. doi: 10.1083/jcb.151.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim RY, Fahrenkrog B, Koser J, Schwarz-Herion K, Deng J, Aebi U. Nanomechanical basis of selective gating by the nuclear pore complex. Science. 2007;318:640–643. doi: 10.1126/science.1145980. [DOI] [PubMed] [Google Scholar]
- 39.Naim B, Brumfeld V, Kapon R, Kiss V, Nevo R, Reich Z. Passive and facilitated transport in nuclear pore complexes is largely uncoupled. J Biol Chem. 2007;282:3881–3888. doi: 10.1074/jbc.M608329200. [DOI] [PubMed] [Google Scholar]
- 40.Kamier O, Peters R. Permeability of single nuclear pores. Biophysical J. 1999;77:217–228. doi: 10.1016/S0006-3495(99)76883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otsuka S, Iwasaka S, Yoneda Y, Takeyasu K, Yoshimura SH. Individual binding pockets of importinbeta for FG-nucleoporins have different binding properties and different sensitivities to RanGTP. Proc Natl Acad Sci USA. 2008;105:16101–16106. doi: 10.1073/pnas.0802647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senay C, Ferrari P, Rocher C, Rieger KJ, Winter J, Platel D, et al. The Mtr2-Mex67 NTF2-like domain complex: structural insights into a dual role of Mtr2 for yeast nuclear export. J Biol Chem. 2003;278:48395–48403. doi: 10.1074/jbc.M308275200. [DOI] [PubMed] [Google Scholar]
- 43.Isgro TA, Schulten K. Binding dynamics of isolated nucleoporin repeat regions to importinbeta. Structure. 2005;13:1869–1879. doi: 10.1016/j.str.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 44.Isgro TA, Schulten K. Association of nuclear pore FG-repeat domains to NTF2 import and export complexes. J Mol Biol. 2007;366:330–345. doi: 10.1016/j.jmb.2006.11.048. [DOI] [PubMed] [Google Scholar]
- 45.Isgro TA, Schulten K. Cse1p-binding dynamics reveal a binding pattern for FG-repeat nucleoporins on transport receptors. Structure. 2007;15:977–991. doi: 10.1016/j.str.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 46.Peters R. Translocation through the nuclear pore complex: selectivity and speed by reduction-of-dimensionality. Traffic. 2005;6:421–427. doi: 10.1111/j.1600-0854.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- 47.Ribbeck K, Gorlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frey S, Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 49.Yamada J, Phillips JL, Patel S, Goldfien G, Calestagne-Morelli A, Huang H, et al. A bimodal distribution of two distinct categories of intrinsically-disordered structures with separate functions in FG nucleoporins. Mol Cell Proteomics. 2010;9:2205–2224. doi: 10.1074/mcp.M000035-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 51.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 52.Ma J, Yang W. Three-dimensional distribution of transient interactions in the nuclear pore complex obtained from single-molecule snapshots. Proc Natl Acad Sci USA. 2010;107:7305–7310. doi: 10.1073/pnas.0908269107. [DOI] [PMC free article] [PubMed] [Google Scholar]