Abstract
The actin family consists of conventional actin and actin-related proteins (ARPs), and the members show moderate similarity and share the same basal structure. Following the finding of various ARPs in the cytoplasm in the 1990s, multiple subfamilies that are localized predominantly in the nucleus were identified. Consistent with these cytological observations, subsequent biochemical analyses revealed the involvement of the nuclear ARPs in ATP-dependent chromatin-remodeling and histone acetyltransferase complexes. In addition to their contribution to chromatin remodeling, recent studies have shown that nuclear ARPs have roles in the organization of the nucleus that are independent of the activity of the above-mentioned complexes. Therefore, nuclear ARPs are recognized as novel key regulators of genome function, and affect not only the remodeling of chromatin but also the spatial arrangement and dynamics of chromatin within the nucleus.
Key words: actin-related protein, ARP, nucleus, chromatin, nuclear organization, chromatin remodeling
Introduction
Genome function is governed by both the organization of chromatin on the chromosomes and the spatial arrangement of chromatin within the cell nucleus. The nucleus is a dynamic organelle that is functionally compartmentalized, and chromatin is organized spatially into different nuclear compartments with the aid of the nuclear architecture. It has been found that both local changes in chromatin structure and alterations in the spatial organization of the nucleus occur during cell differentiation and development. Recent studies have indicated that the nuclear envelope, which includes the nuclear membrane, lamins and the nuclear pore complex (NPC), has important roles in the function of the genome. However, there is limited information about the architecture of the inner regions of the nucleus, and the molecular composition and dynamics of higher order nuclear structures remain unclear. The universal importance of the nuclear architecture in eukaryotic cells suggests that some evolutionarily conserved protein families will be involved in the organization and dynamics of the nucleus.
The actin family has been conserved as highly as histone molecules during the evolution of eukaryotes. In the cytoplasm, actin has central roles in the organization and dynamics of the cytoskeleton. Given the high degree of conservation and the structural roles of actin, researchers hypothesized that actin and/or related molecules are involved in the architecture of the nucleus. However, the presence of actin or related molecules in the nucleus had not been established. Therefore, this hypothesis was regarded with skepticism for a long time. However, recent studies have changed this situation. It was found that, although actin is actively exported from the nucleus,1–3 a certain amount of actin is constitutively localized in the nucleus.4–7 Importantly, in the middle of the 1990s, it was also found that multiple subfamilies of actin-related proteins (ARPs), which are members of the actin family, accumulate in the nucleus.8,9
In this review, we will focus on ARPs that accumulate in the nucleus, and will describe their important contributions to the local alteration of chromatin structure and also to the spatial organization of the nucleus. Nuclear ARPs have been identified in many organisms, including plants,10 insects11 and protists.12,13 However, here we will deal mainly with ARPs from budding yeast and vertebrates.
The Actin Family, Consisting of Conventional Actin and Actin-related Proteins (ARPs)
Within a few decades of the first isolation of actin from muscle in 1941, it was shown that the actin filament is the main architectural element of the cytoskeleton. The dynamics of the cytoskeleton are regulated by the assembly/disassembly of actin filaments, which depends on the exchange of adenosine diphosphate and adenosine triphosphate (ADP/ATP),14 and on association with various actin-binding proteins.
For a long time, it was thought that the actin family consisted only of isoforms of conventional actin, including α actin (muscle actin) and β actin (cellular actin). The amino acid sequences of these actin isoforms show significant evolutionary conservation, and the similarity among yeast actin, human α actin and human β actin is more than 95%. However, the discovery in 1992 of proteins that show moderate (30 to 70%) similarity to conventional actins revealed that the actin family consists of not only conventional actin but also related proteins.15–18 These molecules that are related to actins are now referred to as actin-related proteins (abbreviated as ARPs). By 1994, four subfamilies of ARPs had been reported in budding yeast,19–21 Drosophila11,22 and vertebrates.23 In budding yeast, conventional actin is named Act1 and the four ARPs were called Act2 to Act5 according to the order of their identification (refer to Table 1 for the correspondence of Act2 to Act5 to the current nomenclature).
Table 1.
Members of the actin family in budding yeast
| Name | Previous name | Similarity to actin (%) | Subcellular localization | |
| actin | Act1 | (100) | cytoplasm | |
| ARP | Arp1 | Act5 | 69 | cytoplasm |
| Arp2 | Act2 | 69 | cytoplasm | |
| Arp3 | Act4 | 60 | cytoplasm | |
| Arp4 | Act3 | 53 | nucleus | |
| Arp5 | 51 | nucleus | ||
| Arp6 | 46 | nucleus | ||
| Arp7 | 44 | nucleus | ||
| Arp8 | 44 | nucleus | ||
| Arp9 | 40 | nucleus | ||
| Arp10 | 38 | cytoplasm |
After the entire genome sequence of the budding yeast Saccharomyces cerevisiae had been determined in 1996,24 all the genes for ARPs in the yeast genome were identified.25 The actin family in budding yeast was found to comprise one conventional actin and ten ARPs. The ten ARPs were then classified according to their similarity to actin and designated Arp1 to Arp10 in place of the previous nomenclature, which included Act2 to Act5 (Table 1). In the new classification, Arp1 is the most similar to actin and Arp10 is the least similar to actin. Most Arps that have been identified in other organisms have been shown to be an ortholog of one of the 10 ARPs of budding yeast, and therefore, the 10 ARP subfamilies that correspond to the yeast ARPs are commonly used as the classification of ARPs in all eukaryotes.25,26 Genetic and biochemical results support the idea that each of the ARP subfamilies possesses individual properties and functions (see below).
Evolution and Structural Features of ARPs
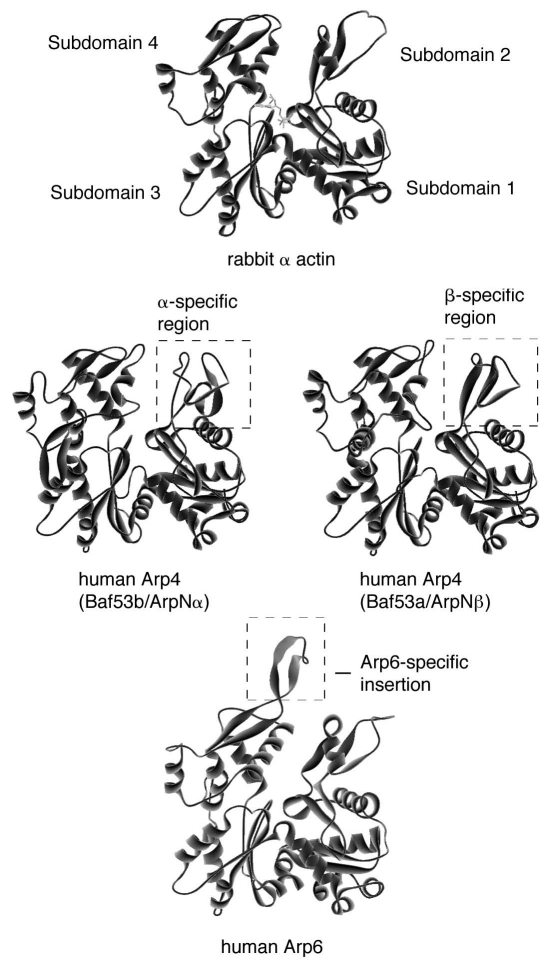
The actin molecule consists of two domains, which comprise the subdomains 1 and 3, and 2 and 4, respectively (Fig. 1), and the central ATP binding pocket is in the cleft between these two domains.27 Deduced 3-dimensional structures of ARPs show that they possess the conserved basal structure of actin, including the central ATP-binding pocket. In contrast, the surface features of ARPs are less conserved. For example, all nuclear ARPs contain a single or multiple insertion domain(s) as compared with actin.26,28 The predicted structure of human Arp6 is shown in Figure 1 as an example.
Figure 1.
The deduced three-dimensional structure of actin-related proteins. The three-dimensional structures of isoforms of human Arp4 (Baf53b/ArpNα and Baf53a/ArpNβ) and human Arp6 were deduced with the program SWISS-Model,121,122 visualized with the program WebLab Viewer, and compared with that of rabbit skeletal muscle actin. The α- and β-specific regions and an Arp6-specific insertion are shown.
ARPs appear to share a common ancestor with conventional actin, and they comprise a branch of the actin family. The latter forms part of the superfamily of proteins that possess an ATP binding pocket, which includes the 70 kDa heat shock cognate protein (Hsc70) and hexokinase,29 and even the prokaryotic proteins, FtsA, MreB and StbA.27 These comparative studies show that the actin superfamily, which is evolutionarily related to actin, is much more divergent than previously thought.
The Budding Yeast Arp4 as the First Identified Nuclear ARP
Before the entire genome sequence of budding yeast had been determined, we discovered an ARP that was the third member of the actin family and named it Act3 (Table 1).19 According to the subsequent classification described above, Act3 is now called Arp4.25 Act3/Arp4 is essential for cell viability and possesses two unique hydrophilic insertion segments, named insertion I and insertion II.19
Arp4 was assumed to be localized in the cytoplasm when it was first identified. However, insertion II of Arp4 exhibits weak similarity to the chromosomal protein HMG-14 and insertion I contains a putative nuclear localization signal.8 As a consequence, the cytological subcellular localization of Arp4 was examined by indirect immunofluorescence analysis and by using GFP-fusion proteins, and it was shown that Arp4 is localized predominantly in the nucleus.8 This report was the first example of the accumulation of a member of the actin family in the nucleus and indicated the possible involvement of ARPs in nuclear functions.
Genetic studies indicated independently that Arp4 is involved in chromatin function.30 Budding yeast contains a retrovirus-like transposable element, Ty1, which is flanked by two copies of a domain with a strong promoter, the δ element. Insertion of Ty1 or the δ element into the 5′ region of genes often causes inactivation of the adjacent gene as a result of interference or competition between transcription signals in the δ element and in the native promoter of the gene.30 Selection for extragenic suppression of Ty1 or δ elements in activated genes has identified numerous SPT (Suppressor of Ty) mutations, and most of the loci were shown to be involved in transcriptional regulation through the modification of chromatin structure.31–34 ARP4 was found to belong to the SPT genes because certain non-lethal mutations in ARP4 suppressed the transcriptional defect caused by the insertion of a δ element.35
Identification of Multiple ARPs that Accumulate in the Nucleus in Various Organisms
In light of the nuclear localization of Arp4, the subcellular localization of other ARPs of budding yeast was investigated, and unexpectedly more than half of them, including Arp5, Arp6, Arp7, Arp8 and Arp9, were localized predominantly in the nucleus9 (Table 1). In contrast, Arp1, Arp2, Arp3 and Arp10 were localized in the cytoplasm.9 This suggests that ARPs are as important in the nucleus as in the cytoplasm.
The presence of ARPs in the nucleus is not limited to yeast. In vertebrates, the first ARPs shown to accumulate in the nucleus were human orthologs of Arp4, named BAF53 or ArpN.36,37 Whereas yeast possesses a sole member (Arp4) of the Arp4 subfamily, humans possess two very similar isoforms, which are named BAF53a/ArpNβ and BAF53b/ArpNα (accession numbers AB015907 and AB015906, respectively).37,38 Although these two isoforms are encoded by individual genes, they are extremely similar (97%), with the exception of short stretches of amino acids, named the α- and β-specific regions (Fig. 1). Interestingly, whereas BAF53a/ArpNβ is expressed in all tissues, BAF53b/ArpNα is detected only in the brain,37,39 where it is expressed in neural cells, and during the neural differentiation of embryonic carcinoma (EC) cells.38,40 Indeed, mice lacking BAF53b/ArpNα have defects in neuronal development.41
Arp5, Arp6 and Arp8 have been shown to accumulate in the nucleus of vertebrate cells, in addition to the isoforms of Arp4.28,42,43 Among these vertebrate nuclear ARPs, at least Arp4 and Arp5 have been shown to shuttle between the cytoplasm and the nucleus.42,44 The distribution of the nuclear ARPs in mitotic cells was also tested, and it was shown that Arp4 and Arp8, but not Arp5 and Arp6, accumulated in mitotic chromosomes.42,43,45
ARPs Identified in Chromatin-Remodeling Complexes
Advances in chromatin research have revealed that two classes of protein complex in the nucleus play a major role in the remodeling of chromatin structure: (1) the ATP-dependent chromatin-remodeling (ADCR) complex and (2) histone modification complexes. The nuclear ARPs have been found to be essential subunits of these chromatin-remodeling complexes in various organisms; in many cases, monomeric actin is also found in the complexes that contain ARPs (Table 2). In budding yeast, actin and Arp4 are part of the NuA4 histone acetyltransferase (HAT) complex,46 and are also found in the INO80 and SWR1 ADCR complexes.47,48 In addition, the INO80 complex contains Arp5 and Arp8,47 and the SWR1 complex contains Arp6.48 Arp7 and Arp9 are found in the RSC and SWI/SNF ADCR complexes.49 Importantly, the compositions of these complexes with respect to ARPs are conserved evolutionarily. Actin, Arp4, Arp5 and Arp8 are found in the vertebrate INO80 complex50 as well as in the yeast INO80 complex. The Tip60 HAT complex, which is the counterpart of the yeast NuA4 complex, also contains actin and Arp4.51 The SRCAP52 and p400 ADCR complexes,53 which are counterparts of the yeast SWR1 complex, contain actin and Arp4, and in addition the former contains Arp6. Although Arp7 and Arp9 have not been identified in vertebrates,26 the vertebrate Brg/BRM ADCR complexes, which are counterparts of the yeast SWI/SNF and RSC complexes, contain actin and Arp4 instead (Table 2).36,39,54 In addition, a vertebrate complex involved in histone methylation contains Arp4.55
Table 2.
Chromatin-remodeling complexes including nuclear ARPs
| Organisms | Complex | Function of the complex | Genome function | Enzymatic component | Actin family-component |
| Budding yeast | INO80 | ATP-dependent chromatin remodeling | transcription DNA repair, DNA replication | Ino80 | actin, Arp4, Arp5, Arp8 |
| SWR1 | ATP-dependent chromatin remodeling (H2A.Z-deposition) | transcription DNA repair | Swr1 | actin, Arp4, Arp6 | |
| SWI/SNF | ATP-dependent chromatin remodeling | transcription DNA repair | Snf2 | Arp7, Arp9 | |
| RSC | ATP-dependent chromatin remodeling | transcription DNA repair | Sth1 | Arp7, Arp9 | |
| NuA4 | histone acetylation | transcription DNA repair | Esa1 | actin, Arp4 | |
| Human | INO80 | ATP-dependent chromatin remodeling | transcription DNA repair | INO80 | actin, Arp4, Arp5, Arp8 |
| SRCAP | ATP-dependent chromatin remodeling (H2A.Z-deposition) | transcription | SRCAP | actin, Arp4, Arp6 | |
| p400 | ATP-dependent chromatin remodeling (H2A.Z-deposition) | transcription | p400 | actin, Arp4 | |
| BAF/PBAF human SWI/SNF | ATP-dependent chromatin remodeling | transcription | BRM or Brg1 | actin, Arp4 | |
| Tip60 | histone acetylation | transcription DNA repair | Tip60 | actin, Arp4 |
The helicase-SANT-associated (HSA) domain has been identified as a motif that binds actin, Arp4, Arp7, Arp8 and Arp9 and recruits them to the chromatin-remodeling complexes.56 The HSA domain is found in the catalytic ATPase subunits of certain chromatin remodeling enzymes (Ino80, Swi2/Snf2, Sth1 and Swr1 of yeast, and Brg1, BRM and p400 of humans), and also in a component (Eaf1) of the NuA4 HAT complex. The stable association of the ARPs with these components could contribute to the regulation of the ATPase activity of chromatin-remodeling complexes. In contrast, motifs for the association of Arp5 and Arp6 have not been identified in enzymatic components of the complexes. Yeast Arp5 associates with Rvb proteins, which are highly conserved proteins related to the helicase subset of the AAA+ family of ATPases, and contributes to the assembly of the INO80 complex.57 Yeast Arp6 forms a subcomplex with Swc6 in the absence of Swr1, and is required for the association of the SWR1 complex with the histone variant H2A.Z, which is deposited in chromatin by the complex.58 Therefore, although Arp5 and Arp6 do not regulate the ATPase activity of the remodeling complexes directly through interaction with the catalytic subunit, they might modulate the function of the complexes through other mechanisms.
As components of these chromatin-remodeling complexes, nuclear ARPs are involved in multiple genome functions, including transcription,47,57,59–61 chromosome segregation,62,63 DNA repair61,64–68 and DNA replication.61,69–72 The findings of these studies indicate that nuclear ARPs are indispensable for a wide variety of genome functions, and further analyses of the nuclear ARPs should contribute to disclosing epigenetic mechanisms that govern these functions.
Nuclear ARP Molecules Outside the Chromatin-Remodeling Complexes
In addition to their important roles in genome function as components of the chromatin-remodeling complexes, recent analyses have indicated that nuclear ARPs also associate with other proteins that are not found in these complexes.61 Gel-filtration analysis showed that only a proportion of Arp4 and Arp6 is found in the chromatin-remodeling complexes.46,73,74 Consistently, yeast arp6Δ cells show a different pattern of gene expression to swr1Δ cells.74 In addition, by genetic screenings, Arp5 and Arp6, but not Ino80 and Swr1, were identified as proteins involved in vacuolar protein sorting75 and Arp5 was shown to be involved in nuclear architecture.76 Proteomics analysis indicated that Arp5, Arp6 and Arp8, but not other components of chromatin-remodeling complexes, are present in the fructose-1,6-bisphosphatase-associated fraction.77
It has been shown that vertebrate Arp8 and other components of the INO80 complex have different roles during mitosis than during interphase. Whereas Arp8 is co-immunoprecipitated with Ino80 in interphase cells as expected, it is not associated with Ino80 in mitotic cells.43 Consistent with this observation, Ino80 and Arp5 are dispersed throughout the cell during mitosis, but Arp8 is restricted to mitotic chromosomes. Knockdown of Ino80, Arp5 and Arp8 showed that only knockdown of Arp8 caused defects in chromosome alignment,43 which implied that Arp8 has a function that is independent of the INO80 complex.
It has also been shown that Arp2 and Arp3 are localized not only in the cytoplasm but also in the nucleus and are involved in transcription.78 Arp2 and Arp3 are not found in chromatin-remodeling complexes; however, they do form the Arp2/3 complex, which promotes actin polymerization. Taken together with observations that actin is required for the repositioning of genes within the nucleus,4,79–82 it is thought that Arp2 and Arp3 have a role in gene regulation through their contribution to local actin polymerization in the nucleus.
Contribution of Arp6 to the Spatial Arrangement of Chromatin
More recently, yeast Arp6 has been shown to have an ability to anchor chromatin to the nuclear periphery that is independent of chromatin remodeling activity.74 It is known that spatial positioning of chromatin in the interphase nucleus is important for transcriptional regulation.83 Cytological observations have revealed that transcriptionally inactive heterochromatin is localized preferentially at the nuclear periphery. In addition, it has been shown recently that some euchromatic genes are anchored to the nuclear periphery in budding yeast. For example, highly transcribed ribosomal protein (RP) genes associate with the NPC, and also inducible genes such as INO1, GAL1 and HXK1 are relocated to the nuclear envelope when they become activated.84–87 In addition, the spatial positioning of chromatin in the interphase nucleus contributes to genome stability. Recent studies have revealed that DNA double-strand breaks that cannot be repaired in situ and collapsed replication forks are relocated to the NPC for repair.88–91 However, little is known about the molecular mechanisms that are involved in the positioning of chromatin within the nucleus.
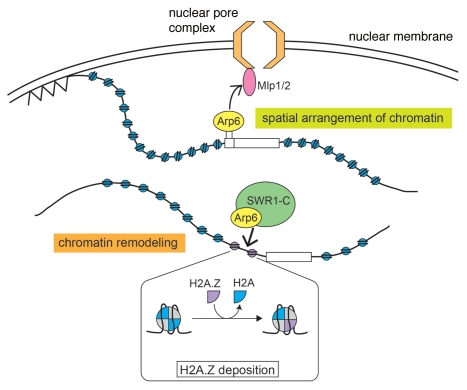
As mentioned above, Arp6 is a component of the yeast SWR1 and vertebrate SRCAP complexes, and these complexes function to exchange canonical H2A histone in nucleosomes for the histone variant H2A.Z.92 It has been shown that Arp6 is required for the deposition of H2A.Z by both SWR1 and SRCAP complexes.58,74,93 Indeed, a chromatin immunoprecipitation-DNA microarray (ChIP-chip) assay revealed that Arp6 binds to gene promoters together with Swr1, which demonstrates that Arp6 binds to these regions as a complex with Swr1. Furthermore, the binding of Swr1 to most promoters is impaired in a yeast arp6δ strain, and deletion of ARP6 and SWR1 caused similar effects on the transcription of some genes, including GAL1.74 These results indicate that Arp6 has an essential function in the organization of chromatin through the SWR1 complex.
In contrast, in some other regions of the genome, including RP genes and telomeres, Arp6 binds to chromatin independently of Swr1. Interestingly, these regions have been shown to associate with the nuclear envelope.87,94 Deletion of ARP6 impaired both the association of RP genes with the NPC and the localization of an RP gene to the nuclear periphery. In addition, the artificial binding of Arp6-LexA to a chromatin region caused the region to relocate to the nuclear periphery in both wild-type and swr1Δ cells. These results suggest that Arp6 has a role in the spatial arrangement of chromatin and that this function of Arp6 is independent of its role in the SWR1 complex (Fig. 2).
Figure 2.
Roles of Arp6 in the local alteration of chromatin structure and the spatial organization of the nucleus. Arp6 has an essential function in chromatin remodeling through the deposition of H2A.Z by the SWR1 complex (lower chromatin). In addition, Arp6 has a role in the spatial arrangement of chromatin through its interaction with the myosin-like proteins Mlp1 and Mlp2 (upper chromatin). This function of Arp6 is independent of the SWR1 complex.
The myosin-like proteins Mlp1 and Mlp2 are components of the NPC and are located on the nucleoplasmic surface.95 To test the possibility that Arp6 has a functional relationship with the myosin-like proteins, mlp1Δ mlp2Δ strains were utilized in the chromatin relocation assay with Arp6-LexA. It was found that the perinuclear relocation activity of Arp6 was impaired in these strains, which suggested that a direct or indirect interaction between Arp6 and Mlp1/2 is involved in the Arp6-mediated anchoring of chromatin to the nuclear envelope. Yra1, which interacts physically with Mlp1/Mlp2, has been identified as a binding partner of Arp6.96 Therefore, it seems likely that the Mlp-Yra1-Arp6 interaction contributes to the perinuclear localization of RP genes bound by Arp6 (Fig. 2).
Possible Molecular Features Shared between Actin and Nuclear ARPs
The molecular mechanisms by which ARPs contribute to the organization of chromatin and the nucleus have not been fully elucidated yet. However, ARPs are assumed to share molecular features with actin, and it is likely that these features are involved in their function. In the following sections, molecular features that are possibly conserved between actin and nuclear ARPs are discussed. Analysis of such features will be useful for a better understanding of the molecular function of ARPs in the nucleus.
ATP-Binding Pocket of Actin and ARPs
Actin binds ATP and this ability is necessary for its function. Although the ATP-binding ability of the nuclear ARPs has not been tested thoroughly, yeast Arp4 at least possesses ATP-binding ability.73 Interestingly, binding of ATP to Arp4 causes Arp4 to dissociate from the other components of nuclear complexes (assumed to be the NuA4, INO80 and SWR1 chromatin-remodeling complexes), whereas inhibition of ATP binding to Arp4 increases the amount of Arp4 that is incorporated into the complexes.73 Given that the presence of Arp4 in the chromatin-remodeling complexes is essential for their function, it is expected that the association/dissociation of Arp4 caused by ATP binding is involved in the functional regulation of the complexes. This feature of Arp4 seems to resemble the assembly/disassembly of actin filaments, which is also regulated by the exchange of ATP/ADP.
Serine14 and Glycine156 of actin, which are located in different loops close to the bound ATP, are crucial for the structure of the ATP-binding pocket.73,97 Consistent with the analysis of ATP binding mentioned above, mutation of the amino acids in Arp4 that correspond to S14 and G156 affects the dissociation of Arp4 from nuclear complexes. This result suggests that Arp4 shares the structure of the ATP-binding pocket with actin. Interestingly, features of the amino acid sequences around S14 and G156 are conserved not only in the nuclear ARPs but also in members of the actin superfamily, including Hsc70, hexokinase and prokaryotic MreB and FtsA. Given that the actin fold and ATP-binding ability are conserved among the members of the actin superfamily, it is most likely that the nuclear ARPs share the structure of the ATP-binding pocket with the other members.
In contrast, the mutations in the ATP-binding pocket of Arp4 did not decrease the acetylation of histones.73 Similarly, mutations in the predicted ATP-binding sites of Arp5 and Arp8 did not decrease expression of the INO1 gene, which is a target of the INO80 complex.98 Taken together with our observation that the ATP-binding arp4 mutants show defects in the dissociation, but not association, of Arp4-containing complexes, these findings suggest that the ATP-binding pocket of nuclear ARPs is essential for the clearance of the complexes, but dispensable for their activity.
Various Intermolecular-Interaction Targets of Actin and ARPs
Actin is known to interact with a variety of proteins, including DNaseI, myosins and various actin-binding proteins. In addition to components of chromatin-remodeling complexes, various other proteins have been determined to associate with ARPs. It is plausible that not only the ATP-binding pocket but also the participation in multiple intermolecular interactions are common features among members of the actin family.
The association of yeast Arp4 with core histones was the first reported intermolecular interaction of the nuclear ARPs.99 The ability of Arp4 to interact with histones is assumed to contribute to the recruitment of the chromatin-remodeling complexes that contain Arp4 (NuA4, INO80 and SWR1) onto chromatin. It has also been shown that Arp4 preferentially associates with histone H2A that has been phosphorylated at serine 129 (γ-H2AX), and it is assumed that this property is involved in the recruitment of these remodeling complexes to sites of DNA damage.100 Arp8 also interacts with core histones H3 and H4,43,98 in both yeast and vertebrates. Interestingly, human Arp8 recruits the INO80 complex to DNA damage sites in a γ-H2AX-independent manner.101 High throughput analyses with budding yeast have indicated that Arp5 and Arp7 interact with core histones.96,102 These observations seem to imply that interaction with histones might be a common characteristic of the nuclear ARPs. The isoelectric point of nuclear ARPs is lower than that of cytoplasmic ARPs (a mean of 5.1 vs. 6.1 in budding yeast), and this feature might contribute to their association with core histones, which possess high isoelectric points. In addition to core histones, various chromatin proteins have roles in the organization of chromatin. Heterochromatin protein 1 (HP1) is known to play a crucial role in the organization of heterochromatin. Vertebrate Arp6 associates with HP1 and is co-localized in regions of pericentric heterochromatin.28
The domains of the nuclear ARPs that are responsible for intermolecular interactions have not been elucidated in detail yet. However, some observations have suggested the involvement of the insertion domains that are specific for each nuclear ARP. Insertion II of yeast Arp4 is responsible for the interaction with core histones mentioned above.99 In addition, the brain-specific isoform of human Arp4, BAF53b/ArpNα, interacts with the transcriptional corepressor CtBP through the α-specific region (Fig. 1).103 BAF53b/ArpNα is structurally distinct from the other isoform BAF53a/ArpNβ in this region,103 and the corresponding β-specific region of BAF53a/ArpNβ (Fig. 1) does not associate with CtBP,103 but is co-precipitated with a HAT enzyme.104 Therefore, it is expected that the distinct intermolecular interactions in which the insertion domains participate are responsible for the individual functions of specific ARPs in the nucleus.
Actin-binding proteins and myosin are known to be major binding partners of cytoplasmic actin. Although the interaction of ARPs with these molecules has not been analyzed in depth, some observations suggest that ARPs also interact with actin-binding proteins and members of the myosin family. Mouse Arp4 interacts with gelsolin,105 and the interaction is thought to contribute to the recruitment of the Brg/BRM complex, which includes Arp4, to gene promoters through the association of gelsolin with a nuclear receptor.106 Importantly, the residues in actin that bind gelsolin are highly conserved in Arp4 from yeasts to humans, which suggests that the interaction between Arp4 and gelsolin is evolutionarily conserved.107 Recently, it has been shown that the actin-bundling protein actinin-4 interacts with the INO80 complex, which contains actin, Arp4, Arp5 and Arp8, and participates in transcriptional regulation.108 Given that actinin in the cytoplasm associates with filamentous actin, it is supposed that a subcomplex that contains monomeric actin and some ARP molecules interacts with actinin-4 in the nucleus. Another actin-binding protein, profilin, also associates with ARPs in the nucleus.109,110 Indeed, a number of actin-binding proteins are found in the nucleus, and at least some of them would be expected to function in association with ARPs.
With respect to myosin, an isoform of myosin I has been shown to accumulate in the nucleus and has been named nuclear myosin I (NM1).111,112 NM1 is required for transcription by RNA polymerases I and II both in vitro and in vivo.111–117 It has also been shown that NM1 is required for the directional movement of active genes into the center of the nucleus and to the subcompartmental interchromatin granules, which harbor key factors for transcriptional elongation and pre-mRNA splicing.79,81 In addition, NM1 is required for the repositioning of entire chromosome territories after the removal of serum from the culture.82 Importantly, actin and Arp4 are required for this activity of NM1.79,81 These observations suggest that members of the actin family are involved in functional associations with members of the myosin family in the nucleus. It seems plausible that functional interactions between NM1 and nuclear ARPs have a major role in regulating the spatial positioning and the dynamics of chromatin within the nucleus.
Perspectives
Two hierarchical genome structures, the organization of chromatin on the chromosomes and the spatial arrangement of chromatin in the nucleus, have central roles in the regulation of genome functions, including the processes of transcription, DNA replication and repair. It can be supposed that nuclear ARPs are involved in various biological phenomena through their contribution to these hierarchical organizational processes.
Recently, it has been reported that Arp6 is involved in thermosensing in plants.118 Interestingly, Arabidopsis mutants in ARP6 were found to exhibit a global high-temperature response even at lower temperatures, which indicated that Arp6 is involved in the temperature-sensing machinery. Given that mutants of genes for the histone variant H2A.Z show similar phenotypes, the findings suggest that the abnormal temperature response in arp6 mutants results from a defect in the deposition of H2A.Z by the SWR1/SRCAP ADCR complex, which contains Arp6. Nucleosomes that contain H2A.Z have been shown to be involved in the formation of “nucleosome-free regions” in gene promoters in mammalian cells,119 and indeed, in Arabidopsis, nucleosomes that contain H2A.Z are evicted from chromatin at high temperature.118 Arp6 is evolutionarily conserved, and it is plausible that Arp6 also has this function in response to environmental changes in other eukaryotes. Although the major contribution of Arp6 during temperature sensing appears to be the deposition of H2A.Z,118,120 Arp6 also has an H2A.Z-independent function in the nucleus, which is described above.74 It would be interesting to determine the involvement of the H2A.Z-independent function of Arp6 in responses to environmental changes in plants and other organisms.
The organization of the nucleus changes during differentiation and development, and the disruption of this organization has been observed in multiple types of cancer cell. However, in contrast to the structure of chromatin, little is known about the factors that contribute to the architecture of the nucleus. Nuclear ARPs are novel molecules that have important functions at both hierarchical levels of organization. Given the evolutionary conservation and multiple important functions of actin and ARPs in the organization and dynamics of the cytoskeleton, further analyses of the nuclear ARPs will certainly contribute to understanding of the molecular basis and dynamics of the nuclear architecture.
References
- 1.Percipalle P, Zhao J, Pope B, Weeds A, Lindberg U, Daneholt B. Actin bound to the heterogeneous nuclear ribonucleoprotein hrp36 is associated with Balbiani ring mRNA from the gene to polysomes. J Cell Biol. 2001;153:229–236. doi: 10.1083/jcb.153.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stuven T, Hartmann E, Gorlich D. Exportin 6: A novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J. 2003;22:5928–5940. doi: 10.1093/emboj/cdg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wada A, Fukuda M, Mishima M, Nishida E. Nuclear export of actin: A novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J. 1998;17:1635–1641. doi: 10.1093/emboj/17.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gieni RS, Hendzel MJ. Actin dynamics and functions in the interphase nucleus: Moving toward an understanding of nuclear polymeric actin. Biochem Cell Biol. 2009;87:283–306. doi: 10.1139/O08-133. [DOI] [PubMed] [Google Scholar]
- 5.Grummt I. Actin and myosin as transcription factors. Curr Opin Genet Dev. 2006;16:191–196. doi: 10.1016/j.gde.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Pederson T, Aebi U. Actin in the nucleus: what form and what for? J Struct Biol. 2002;140:3–9. doi: 10.1016/s1047-8477(02)00528-2. [DOI] [PubMed] [Google Scholar]
- 7.Percipalle P, Visa N. Molecular functions of nuclear actin in transcription. J Cell Biol. 2006;172:967–971. doi: 10.1083/jcb.200512083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber V, Harata M, Hauser H, Wintersberger U. The actin-related protein Act3p of Saccharomyces cerevisiae is located in the nucleus. Mol Biol Cell. 1995;6:1263–1270. doi: 10.1091/mbc.6.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harata M, Oma Y, Tabuchi T, Zhang Y, Stillman DJ, Mizuno S. Multiple actin-related proteins of Saccharomyces cerevisiae are present in the nucleus. J Biochem. 2000;128:665–671. doi: 10.1093/oxfordjournals.jbchem.a022799. [DOI] [PubMed] [Google Scholar]
- 10.Kandasamy MK, Deal RB, McKinney EC, Meagher RB. Plant actin-related proteins. Trends Plant Sci. 2004;9:196–202. doi: 10.1016/j.tplants.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Fyrberg C, Ryan L, Kenton M, Fyrberg E. Genes encoding actin-related proteins of Drosophila melanogaster. J Mol Biol. 1994;241:498–503. doi: 10.1006/jmbi.1994.1526. [DOI] [PubMed] [Google Scholar]
- 12.Raza S, Sahasrabuddhe AA, Gupta CM. Nuclear localization of an actin-related protein (ORF LmjF21.0230) in Leishmania+ Mol Biochem Parasitol. 2007;153:216–219. doi: 10.1016/j.molbiopara.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Gordon JL, Beatty WL, Sibley LD. A novel actin-related protein is associated with daughter cell formation in Toxoplasma gondii. Eukaryot Cell. 2008;7:1500–1512. doi: 10.1128/EC.00064-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheterline P, Clayton J, Sparrow J. Protein profile: Actin. In: Sheterline P, editor. Protein Profile. 1994. p. 1. [Google Scholar]
- 15.Clark SW, Meyer DI. Centractin is an actin homologue associated with the centrosome. Nature. 1992;359:246–250. doi: 10.1038/359246a0. [DOI] [PubMed] [Google Scholar]
- 16.Lees-Miller JP, Helfman DM, Schroer TA. A vertebrate actin-related protein is a component of a multisubunit complex involved in microtubule-based vesicle motility. Nature. 1992;359:244–246. doi: 10.1038/359244a0. [DOI] [PubMed] [Google Scholar]
- 17.Lees-Miller JP, Henry G, Helfman DM. Identification of act2, an essential gene in the fission yeast Schizosaccharomyces pombe that encodes a protein related to actin. Proc Natl Acad Sci USA. 1992;89:80–83. doi: 10.1073/pnas.89.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwob E, Martin RP. New yeast actin-like gene required late in the cell cycle. Nature. 1992;355:179–182. doi: 10.1038/355179a0. [DOI] [PubMed] [Google Scholar]
- 19.Harata M, Karwan A, Wintersberger U. An essential gene of Saccharomyces cerevisiae coding for an actin-related protein. Proc Natl Acad Sci USA. 1994;91:8258–8262. doi: 10.1073/pnas.91.17.8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muhua L, Karpova TS, Cooper JA. A yeast actin-related protein homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell. 1994;78:669–679. doi: 10.1016/0092-8674(94)90531-2. [DOI] [PubMed] [Google Scholar]
- 21.Clark SW, Meyer DI. ACT3: A putative centractin homologue in S. cerevisiae is required for proper orientation of the mitotic spindle. J Cell Biol. 1994;127:129–138. doi: 10.1083/jcb.127.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frankel S, Heintzelman MB, Artavanis-Tsakonas S, Mooseker MS. Identification of a divergent actin-related protein in drosophila. J Mol Biol. 1994;235:1351–1356. doi: 10.1006/jmbi.1994.1090. [DOI] [PubMed] [Google Scholar]
- 23.Clark S, Staub O, Clark I, Holzbaur E, Paschal B, Vallee R, et al. Beta-centractin: characterization and distribution of a new member of the centractin family of actin-related proteins. Mol Biol Cell. 1994;5:1301–1310. doi: 10.1091/mbc.5.12.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, et al. Life with 6,000 Genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 25.Poch O, Winsor B. Who's who among the Saccharomyces cerevisiae actin-related proteins? A classification and nomenclature proposal for a large family. Yeast. 1997;13:1053–1058. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1053::AID-YEA164>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 26.Muller J, Oma Y, Vallar L, Friederich E, Poch O, Winsor B. Sequence and comparative genomic analysis of actin-related proteins. Mol Biol Cell. 2005;16:5736–5748. doi: 10.1091/mbc.E05-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabsch W, Holmes K. The actin fold. FASEB J. 1995;9:167–174. doi: 10.1096/fasebj.9.2.7781919. [DOI] [PubMed] [Google Scholar]
- 28.Ohfuchi E, Kato M, Sasaki M, Sugimoto K, Oma Y, Harata M. Vertebrate Arp6, a novel nuclear actin-related protein, interacts with heterochromatin protein 1. Eur J Cell Biol. 2006;85:411–421. doi: 10.1016/j.ejcb.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Holmes KC, Sander C, Valencia A. A new ATP-binding fold in actin, hexokinase and Hsc70. Trends Cell Biol. 1993;3:53–59. doi: 10.1016/0962-8924(93)90161-s. [DOI] [PubMed] [Google Scholar]
- 30.Jiang YW, Stillman DJ. Epigenetic effects on yeast transcription caused by mutations in an actin-related protein present in the nucleus. Genes Dev. 1996;10:604–619. doi: 10.1101/gad.10.5.604. [DOI] [PubMed] [Google Scholar]
- 31.Clark-Adams CD, Norris D, Osley MA, Fassler JS, Winston F. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 1988;2:150–159. doi: 10.1101/gad.2.2.150. [DOI] [PubMed] [Google Scholar]
- 32.Eisenmann DM, Dollard C, Winston F. SPT15, the gene encoding the yeast TATA binding factor TFIID, is required for normal transcription initiation in vivo. Cell. 1989;58:1183–1191. doi: 10.1016/0092-8674(89)90516-3. [DOI] [PubMed] [Google Scholar]
- 33.Swanson MS, Winston F. SPT4, SPT5 and SPT6 Interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics. 1992;132:325–336. doi: 10.1093/genetics/132.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roeder GS, Beard C, Smith M, Keranen S. Isolation and characterization of the SPT2 gene, a negative regulator of Ty-controlled yeast gene expression. Mol Cell Biol. 1985;5:1543–1553. doi: 10.1128/mcb.5.7.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harata M, Zhang Y, Stillman DJ, Matsui D, Oma Y, Nishimori K, et al. Correlation between chromatin association and transcriptional regulation for the Act3p/Arp4 nuclear actin-related protein of Saccharomyces cerevisiae. Nucleic Acids Res. 2002;30:1743–1750. doi: 10.1093/nar/30.8.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 37.Harata M, Mochizuki R, Mizuno S. Two isoforms of a human actin-related protein show nuclear localization and mutually selective expression between brain and other tissues. Biosci Biotechnol Biochem. 1999;63:917–923. doi: 10.1271/bbb.63.917. [DOI] [PubMed] [Google Scholar]
- 38.Olave I, Wang W, Xue Y, Kuo A, Crabtree GR. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 2002;16:2509–2517. doi: 10.1101/gad.992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuroda Y, Oma Y, Nishimori K, Ohta T, Harata M. Brain-specific expression of the nuclear actin-related protein ArpNa and its involvement in mammalian SWI/SNF chromatin remodeling complex. Biochem Biophys Res Commun. 2002;299:328–334. doi: 10.1016/s0006-291x(02)02637-2. [DOI] [PubMed] [Google Scholar]
- 41.Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, et al. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 42.Kitayama K, Kamo M, Oma Y, Matsuda R, Uchida T, Ikura T, et al. The human actin-related protein hArp5: nucleo-cytoplasmic shuttling and involvement in DNA repair. Exp Cell Res. 2009;315:206–217. doi: 10.1016/j.yexcr.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 43.Aoyama N, Oka A, Kitayama K, Kurumizaka H, Harata M. The actin-related protein hArp8 accumulates on the mitotic chromosomes and functions in chromosome alignment. Exp Cell Res. 2008;314:859–868. doi: 10.1016/j.yexcr.2007.11.020. [DOI] [PubMed] [Google Scholar]
- 44.Lee JH, Chang SH, Shim JH, Lee JY, Yoshida M, Kwon H. Cytoplasmic localization and nucleo-cytoplasmic shuttling of BAF53, a component of chromatin-modifying complexes. Mol Cells. 2003;16:78–83. [PubMed] [Google Scholar]
- 45.Lee K, Shim J, Kang M, Kim J, Ahn J, Yoo S, et al. Association of BAF53 with mitotic chromosomes. Mol Cells. 2007;24:288–293. [PubMed] [Google Scholar]
- 46.Galarneau L, Nourani A, Boudreault AA, Zhang Y, Héliot L, Allard S, et al. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol Cell. 2000;5:927–937. doi: 10.1016/s1097-2765(00)80258-0. [DOI] [PubMed] [Google Scholar]
- 47.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 48.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 49.Cairns BR, Erdjument-Bromage H, Tempst P, Winston F, Kornberg RD. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol Cell. 1998;2:639–651. doi: 10.1016/s1097-2765(00)80162-8. [DOI] [PubMed] [Google Scholar]
- 50.Jin J, Cai Y, Yao T, Gottschalk AJ, Florens L, Swanson SK, et al. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J Biol Chem. 2005;280:41207–41212. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- 51.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 52.Cai Y, Jin J, Florens L, Swanson SK, Kusch T, Li B, et al. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J Biol Chem. 2005;280:13665–13670. doi: 10.1074/jbc.M500001200. [DOI] [PubMed] [Google Scholar]
- 53.Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, et al. The p400 complex is an essential E1A transformation target. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 54.Kitagawa H, Fujiki R, Yoshimura K, Mezaki Y, Uematsu Y, Matsui D, et al. The chromatin-remodeling complex WINAC targets a nuclear receptor to promoters and is impaired in Williams syndrome. Cell. 2003;113:905–917. doi: 10.1016/s0092-8674(03)00436-7. [DOI] [PubMed] [Google Scholar]
- 55.Katoh Y, Ikura T, Hoshikawa Y, Tashiro S, Ohta M, Kera Y, et al. Methionine adenosyltransferase II serves as a transcriptional corepressor of Maf oncoprotein. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.02.018. in press. [DOI] [PubMed] [Google Scholar]
- 56.Szerlong H, Hinata K, Viswanathan R, Erdjument-Bromage H, Tempst P, Cairns BR. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat Struct Mol Biol. 2008;15:469–476. doi: 10.1038/nsmb.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jonsson ZO, Jha S, Wohlschlegel JA, Dutta A. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol Cell. 2004;16:465–477. doi: 10.1016/j.molcel.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 58.Wu WH, Alami S, Luk E, Wu CH, Sen S, Mizuguchi G, et al. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat Struct Mol Biol. 2005;12:1064–1071. doi: 10.1038/nsmb1023. [DOI] [PubMed] [Google Scholar]
- 59.Meagher RB, Kandasamy MK, McKinney EC, Roy E. Chapter 5. Nuclear actin-related proteins in epigenetic control. Int Rev Cell Mol Biol. 2009;277:157–215. doi: 10.1016/S1937-6448(09)77005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meagher RB, Kandasamy MK, Deal RB, McKinney EC. Actin-related proteins in chromatin-level control of the cell cycle and developmental transitions. Trends Cell Biol. 2007;17:325–332. doi: 10.1016/j.tcb.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 61.Dion V, Shimada K, Gasser SM. Actin-related proteins in the nucleus: life beyond chromatin remodelers. Curr Opin Cell Biol. 2010;22:383–391. doi: 10.1016/j.ceb.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 62.Ogiwara H, Enomoto T, Seki M. The INO80 chromatin remodeling complex functions in sister chromatid cohesion. Cell Cycle. 2007;6:1090–1095. doi: 10.4161/cc.6.9.4130. [DOI] [PubMed] [Google Scholar]
- 63.Minoda A, Saitoh S, Takahashi K, Toda T. BAF53/Arp4 homolog Alp5 in fission yeast is required for histone H4 acetylation, kinetochore-spindle attachment and gene silencing at centromere. Mol Biol Cell. 2005;16:316–327. doi: 10.1091/mbc.E04-06-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 65.Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, et al. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 66.van Attikum H, Gasser SM. ATP-dependent chromatin remodeling and DNA double-strand break repair. Cell Cycle. 2005;4:1011–1014. doi: 10.4161/cc.4.8.1887. [DOI] [PubMed] [Google Scholar]
- 67.Morrison AJ, Shen X. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol. 2009;10:373–384. doi: 10.1038/nrm2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 69.Vincent JA, Kwong TJ, Tsukiyama T. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat Struct Mol Biol. 2008;15:477–484. doi: 10.1038/nsmb.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, et al. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr Biol. 2008;18:566–575. doi: 10.1016/j.cub.2008.03.049. [DOI] [PubMed] [Google Scholar]
- 71.Papamichos-Chronakis M, Peterson CL. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol. 2008;15:338–345. doi: 10.1038/nsmb.1413. [DOI] [PubMed] [Google Scholar]
- 72.Falbo KB, Alabert C, Katou Y, Wu S, Han J, Wehr T, et al. Involvement of a chromatin remodeling complex in damage tolerance during DNA replication. Nat Struct Mol Biol. 2009;16:1167–1172. doi: 10.1038/nsmb.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sunada R, Görzer I, Oma Y, Yoshida T, Suka N, Wintersberger U, et al. The nuclear actin-related protein Act3p/Arp4p is involved in the dynamics of chromatin-modulating complexes. Yeast. 2005;22:753–768. doi: 10.1002/yea.1239. [DOI] [PubMed] [Google Scholar]
- 74.Yoshida T, Shimada K, Oma Y, Kalck V, Akimura K, Taddei A, et al. Actin-related protein Arp6 influences H2A.Z-dependent and -independent gene expression and links ribosomal protein genes to nuclear pores. PLoS Genet. 2010;6:1000910. doi: 10.1371/journal.pgen.1000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bonangelino CJ, Chavez EM, Bonifacino JS. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2486–2501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Teixeira MT, Dujon B, Fabre E. Genome-wide nuclear morphology screen identifies novel genes involved in nuclear architecture and gene-silencing in Saccharomyces cerevisiae. J Mol Biol. 2002;321:551–561. doi: 10.1016/s0022-2836(02)00652-6. [DOI] [PubMed] [Google Scholar]
- 77.Brown CR, Hung GC, Dunton D, Chiang HL. The TOR complex 1 is distributed in endosomes and in retrograde vesicles that form from the vacuole membrane and plays an important role in the vacuole import and degradation pathway. J Biol Chem. 2010;285:23359–23370. doi: 10.1074/jbc.M109.075143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoo Y, Wu X, Guan JL. A novel role of the actin-nucleating Arp2/3 complex in the regulation of RNA polymerase II-dependent transcription. J Biol Chem. 2007;282:7616–7623. doi: 10.1074/jbc.M607596200. [DOI] [PubMed] [Google Scholar]
- 79.Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 80.Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, et al. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci USA. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mehta IS, Amira M, Harvey AJ, Bridger JM. Rapid chromosome territory relocation by nuclear motor activity in response to serum removal in primary human fibroblasts. Genome Biol. 2010;11:5. doi: 10.1186/gb-2010-11-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 84.Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: The nucleopore-promoter interaction of genes in yeast. Mol Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 85.Brickner JH, Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, et al. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 87.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 88.Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Abdallah P, Luciano P, Runge KW, Lisby M, Geli V, Gilson E, et al. A two-step model for senescence triggered by a single critically short telomere. Nat Cell Biol. 2009;11:988–993. doi: 10.1038/ncb1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 91.Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009;23:912–927. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 93.Matsuda R, Hori T, Kitamura H, Takeuchi K, Fukagawa T, Harata M. Identification and characterization of the two isoforms of the vertebrate H2A.Z histone variant. Nucleic Acids Res. 2010;38:4263–4273. doi: 10.1093/nar/gkq171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Taddei A, Hediger F, Neumann FR, Gasser SM. The function of nuclear architecture: a genetic approach. Annu Rev Genet. 2004;38:305–345. doi: 10.1146/annurev.genet.37.110801.142705. [DOI] [PubMed] [Google Scholar]
- 95.Strambio-de-Castillia C, Blobel G, Rout MP. Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol. 1999;144:839–855. doi: 10.1083/jcb.144.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 97.Görzer I, Schüller C, Heidenreich E, Krupanska L, Kuchler K, Wintersberger U. The nuclear actin-related protein Act3p/Arp4p of Saccharomyces cerevisiae is involved in transcription regulation of stress genes. Mol Microbiol. 2003;50:1155–1171. doi: 10.1046/j.1365-2958.2003.03759.x. [DOI] [PubMed] [Google Scholar]
- 98.Shen X, Ranallo R, Choi E, Wu C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell. 2003;12:147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- 99.Harata M, Oma Y, Mizuno S, Jiang YW, Stillman DJ, Wintersberger U. The nuclear actin-related protein of Saccharomyces cerevisiae, Act3p/Arp4, interacts with core histones. Mol Biol Cell. 1999;10:2595–2605. doi: 10.1091/mbc.10.8.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 101.Kashiwaba S, Kitahashi K, Watanabe T, Onoda F, Ohtsu M, Murakami Y. The mammalian INO80 complex is recruited to DNA damage sites in an ARP8 dependent manner. Biochem Biophys Res Commun. 2010;402:619–625. doi: 10.1016/j.bbrc.2010.10.066. [DOI] [PubMed] [Google Scholar]
- 102.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 103.Oma Y, Nishimori K, Harata M. The brain-specific actin-related protein ArpNa interacts with the transcriptional co-repressor CtBP. Biochem Biophys Res Commun. 2003;301:521–528. doi: 10.1016/s0006-291x(02)03073-5. [DOI] [PubMed] [Google Scholar]
- 104.Park J, Wood MA, Cole MD. BAF53 forms distinct nuclear complexes and functions as a critical c-Myc-interacting nuclear cofactor for oncogenic transformation. Mol Cell Biol. 2002;22:1307–1316. doi: 10.1128/mcb.22.5.1307-1316.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee YH, Campbell HD, Stallcup MR. Developmentally essential protein flightless I is a nuclear receptor coactivator with actin binding activity. Mol Cell Biol. 2004;24:2103–2117. doi: 10.1128/MCB.24.5.2103-2117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Archer SK, Behm CA, Claudianos C, Campbell HD. The flightless I protein and the gelsolin family in nuclear hormone receptor-mediated signalling. Biochem Soc Trans. 2004;32:940–942. doi: 10.1042/BST0320940. [DOI] [PubMed] [Google Scholar]
- 107.Archer SK, Claudianos C, Campbell HD. Evolution of the gelsolin family of actin-binding proteins as novel transcriptional coactivators. Bioessays. 2005;27:388–396. doi: 10.1002/bies.20200. [DOI] [PubMed] [Google Scholar]
- 108.Kumeta M, Yoshimura SH, Harata M, Takeyasu K. Molecular mechanisms underlying nucleocytoplasmic shuttling of actinin-4. J Cell Sci. 2010;123:1020–1030. doi: 10.1242/jcs.059568. [DOI] [PubMed] [Google Scholar]
- 109.Rawe VY, Payne C, Schatten G. Profilin and actin-related proteins regulate microfilament dynamics during early mammalian embryogenesis. Hum Reprod. 2006;21:1143–1153. doi: 10.1093/humrep/dei480. [DOI] [PubMed] [Google Scholar]
- 110.Hara Y, Yamagata K, Oguchi K, Baba T. Nuclear localization of profilin III-ArpM1 complex in mouse spermiogenesis. FEBS Lett. 2008;582:2998–3004. doi: 10.1016/j.febslet.2008.07.058. [DOI] [PubMed] [Google Scholar]
- 111.Pestic-Dragovich L, Stojiljkovic L, Philimonenko AA, Nowak G, Ke Y, Settlage RE, et al. A myosin I isoform in the nucleus. Science. 2000;290:337–341. doi: 10.1126/science.290.5490.337. [DOI] [PubMed] [Google Scholar]
- 112.Philimonenko VV, Zhao J, Iben S, Dingova H, Kysela K, Kahle M, et al. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol. 2004;6:1165–1172. doi: 10.1038/ncb1190. [DOI] [PubMed] [Google Scholar]
- 113.Nowak G, Pestic-Dragovich L, Hozαk P, Philimonenko A, Simerly C, Schatten G, et al. Evidence for the presence of myosin I in the nucleus. J Biol Chem. 1997;272:17176–17181. doi: 10.1074/jbc.272.27.17176. [DOI] [PubMed] [Google Scholar]
- 114.Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol. 2004;6:1094–1101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- 115.Kysela K, Philimonenko AA, Philimonenko VV, Janacek J, Kahle M, Hozak P. Nuclear distribution of actin and myosin I depends on transcriptional activity of the cell. Histochem Cell Biol. 2005;124:347–358. doi: 10.1007/s00418-005-0042-8. [DOI] [PubMed] [Google Scholar]
- 116.Fomproix N, Percipalle P. An actin-myosin complex on actively transcribing genes. Exp Cell Res. 2004;294:140–148. doi: 10.1016/j.yexcr.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 117.Percipalle P, Fomproix N, Cavellan E, Voit R, Reimer G, Kruger T, et al. The chromatin remodelling complex WSTF-SNF2 h interacts with nuclear myosin 1 and has a role in RNA polymerase I transcription. EMBO Rep. 2006;7:525–530. doi: 10.1038/sj.embor.7400657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 119.Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, et al. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Smith AP, Jain A, Deal RB, Nagarajan VK, Poling MD, Raghothama KG, et al. Histone H2A.Z regulates the expression of several classes of phosphate starvation response genes but not as a transcriptional activator. Plant Physiol. 2010;152:217–225. doi: 10.1104/pp.109.145532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Peitsch M. ProMod and Swiss-model: internet-based tools for automated comparative protein modelling. Biochem Soc Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- 122.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]