Abstract
Interphase nuclear architecture is disrupted and rapidly reformed with each cell division cycle. Successive cell generations exhibit a “memory” of this nuclear architecture, as well as for gene expression. Furthermore, many features of nuclear and mitotic chromosome structure are recognizably species and tissue specific. We wish to know what properties of the underlying chromatin structure may determine these conserved features of nuclear architecture. Employing a particular mouse autoimmune anti-nucleosome monoclonal antibody (PL2-6), combined with deconvolution immunofluorescence microscopy, we present evidence for a unique epitope (involving a ternary complex of histones H2A and H2B and DNA) which is localized only at the exterior chromatin surface of interphase nuclei and mitotic chromosomes in mammalian, invertebrate and plant systems. As only the surface chromatin region is identified with antibody PL2-6, we have assigned it the name “epichromatin”. We describe an “epichromatin hypothesis”, suggesting that epichromatin may have a unique evolutionary conserved conformation which facilitates interaction with the reforming post-mitotic nuclear envelope and a rapid return of interphase nuclear architecture.
Key words: interphase nuclear architecture, mitotic chromosomes, chromatin, epichromatin, evolution
Introduction
The appearance of histones and nucleosomes in the earliest eukaryotes laid the groundwork for an entirely new set of architectural principles for packaging of the DNA, not previously seen in the evolution of life. Describing these principles is a major goal of cell biology and encompasses understanding the interphase nucleus, the mitotic chromosome and other chromatin structural states.1–5 A major advance in our understanding of the organization of chromatin came with the description of chromosome territories (CTs),6,7 which demonstrated that the individuality of mitotic chromosomes is preserved, albeit in a swollen form, within the interphase nucleus. Evidence has also been presented that within these territories chromatin exists in “knot-free” globules, analogous to the nucleosomal “beads-on-a-string”; but at a larger scale, encompassing megabase lengths of chromosomal DNA.8 Compartmentalization of silenced heterochromatin to the periphery of the interphase nucleus is a common architectural theme. Current evidence supports that there are numerous lamina-associated peripheral chromatin domains, characterized by low gene-expression, which may facilitate global nuclear organization.9,10 To the extent that these architectural principles may be universal among the eukaryotes, one must ask how they are maintained through successive cell generations and during the extensive evolution of new species.
Anti-nuclear antibodies are a frequent occurrence in the sera of autoimmune humans and animals, being especially prevalent in systemic lupus erythematosus (SLE).11,12 These autoantibodies can be directed against a variety of nuclear antigens, including DNA, histones, nucleosomes and subnucleosomal particles. As part of a program to analyze the mechanisms underlying autoimmune disorders, several mouse strains have been identified that spontaneously develop SLE and are the source of a number of monoclonal anti-nucleosomal antibodies.13,14 We have employed some of these mouse monoclonal antibodies (mAbs) to probe nuclear structural domains. Historically, this approach was useful in identifying the histone H3 variants of centromeric regions employing CREST autoantibodies from scleroderma patients.15,16 We tested a panel of these mouse mAbs on rapidly growing U2OS and HL-60/S4 cells by deconvolution immunofluorescence microscopy with the aim of searching for chromatin substructures. One anti-nucleosome antibody (PL2-6) specifically stained a chromatin compartment at the periphery of interphase nuclei, as well as staining the surface of mitotic chromosomes. We suggest that this compartment represents a unique surface chromatin conformation, to which we assign the name “epichromatin”. Furthermore, we formulate an “epichromatin hypothesis”, suggesting that this chromatin may play a crucial role in organizing interphase nuclear architecture, justifying its conservation in the evolution of cell structure.
Results
Immunofluorescent analysis of a set of mouse anti-nucleosome autoantibodies.
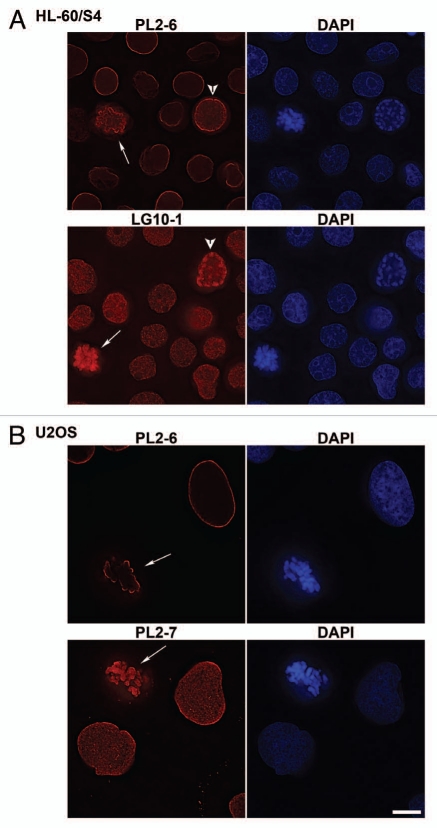
A number of mAbs derived from autoimmune mice were tested by immunostaining against rapidly growing HL-60/S4 (suspension cells) and U2OS (attached cells). The binding specificities of the mAbs (all IgGs) and verification that they are all anti-nucleosome antibodies have previously been determined by ELISA.11,13,14,17,18 Examples of the deconvolved slices from selected antibodies are shown in Figure 1: HL-60/S4 cells are displayed in A; U2OS cells in B. Monoclonal PL2-6 and PL2-7 were derived from the same mouse and are both anti-H2A-H2B-DNA;14,17 LG10-1 is from a different animal and has anti-H3-H4-DNA specificity.14,18 For the set of ten mAbs tested, the most typical immunostained images resembled those presented for LG10-1 and PL2-7; i.e., staining throughout interphase nuclei (except nucleoli) and throughout mitotic chromosomes, paralleling the DAPI staining of DNA. With one mAb, PL2-6, the images were quite different; i.e., staining was confined to the periphery of interphase nuclei and metaphase chromosomes, which we name “epichromatin”. By ELISA, PL2-6 resembled six other anti-H2A-H2B-DNA autoantibodies (including PL2-7) derived from the same animal.17 All displayed strong ELISA reactivity with the adsorbed complex of H2A-H2B-DNA, less reaction with H2A-H2B, and weak-to-negligible reactions with adsorbed separate histones or with an adsorbed H3-H4-DNA complex.17 One difference, observed by ELISA, is that the reactivity of PL2-6 to adsorbed H2A-H2B-DNA could be inhibited by preincubation with the H2A-H2B-DNA complex in buffer; which was not the case with the other tested mAbs.14 The unique staining pattern of PL2-6 (especially, the immunolocalization at the periphery of mitotic chromosomes) provoked us to examine accessibility of this epichromatin epitope throughout the cell cycle.
Figure 1.
Immunostaining of mammalian tissue culture cells with selected mouse monoclonal anti-nucleosome antibodies. Cell types: (A) HL-60/S4; (B) U2OS. Mouse mAbs (PL2-6, PL2-7 and LG10-1) staining are shown in red: DAPI staining in blue. The arrows denote mitotic cells. The arrowheads point to prophase nuclei. Each image is a single deconvolved optical slice. Bar equals 10 µm for both (A and B).
The epichromatin epitope is present throughout the cell cycle.
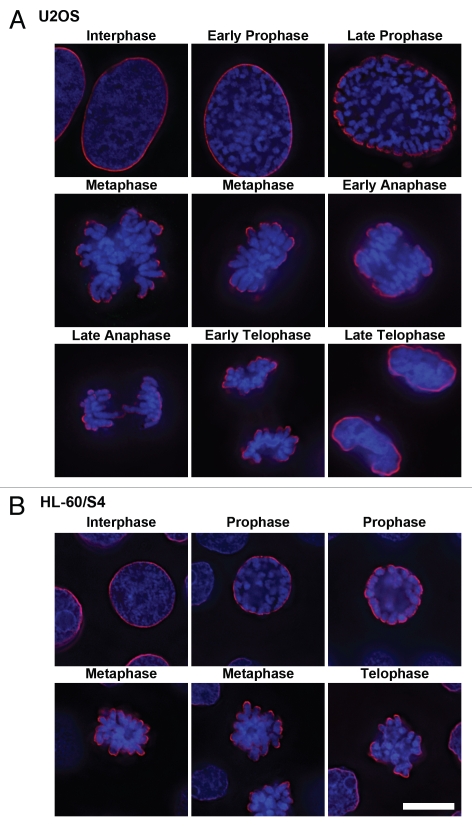
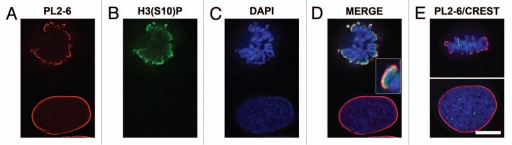
Immunostaining experiments clearly demonstrated that the epichromatin epitope recognized by PL2-6 is present throughout the cell cycle, even after nuclear envelope (NE) breakdown and before post-mitotic NE reformation. Immunostaining images throughout mitosis, plus interphase, are presented for U2OS (Fig. 2A and Sup. Vid. 1–5) and for HL-60/S4 cells (Fig. 2B). Identical results were obtained with formaldehyde (PFA) or with methanol/acetone fixation; only the PFA results are presented in Figure 2. During prophase the epichromatin epitope is accessible to antibody at peripheral regions of the condensing chromosomes adjacent to the dissociating nuclear envelope. During metaphase and early anaphase, the epitope outlines the exterior surface of the congressed chromosomes; more internal chromosome arms and ends do not exhibit the epitope. By late anaphase the epichromatin epitope becomes visible on the internal trailing arms and ends of some mitotic chromosomes. Finally, during telophase the epichromatin epitope “spreads” around the nuclear periphery, as the clustered mitotic chromosomes fuse and the NE reforms. By contrast, co-immunostaining with the “mitotic marker”, rabbit anti-H3(S10)p,19,20 clearly revealed the presence of this antigenic determinant during mitosis and its disappearance during interphase (Fig. 3B); whereas the epichromatin epitope persisted at every phase of the cell cycle (Fig. 3A). Examination of serial slices of mitotic figures (data not shown) demonstrated that the H3(S10)p antigenic determinant is present throughout the chromosome set, although staining is stronger near the periphery. This observation (i.e., stronger peripheral staining) is in agreement with previously published images (see figure 3 in ref. 19 and figure 1 in ref. 20). When the “merge” image is examined closely (insert, Fig. 3D), the epichromatin epitope appears to be slightly exterior to, and overlapping with H3(S10)p. An additional co-immunostaining experiment attempted to determine whether centromeric regions, as detected with autoimmune CREST antisera, revealed any special relationship to the epichromatin epitope. Figure 3E presents single optical slices of a metaphase plate and an interphase nucleus. All of the deconvolved slices were examined in this experiment (data not shown). Careful examination revealed that a few centromeres were close to the region of the epichromatin epitope, but the majority revealed no obvious proximity. The absence of PL2-6 staining in telophase chromosome “cores” is discussed in a later section.
Figure 2.
Immunostaining of the epichromatin epitope through mitosis in U2OS (A) and HL-60/S4 (B) cells. Mouse mAb PL2-6 staining is shown in red; DAPI staining in blue. Each image is a single deconvolved optical slice. Bar equals 10 µm for both (A and B). Projection videos for some of these images of U2OS can be found in the Supplementary Video Files, specifically: late prophase (Video 1); early anaphase (Video 2); late anaphase (Video 3); early telophase (Video 4); late telophase (Video 5).
Figure 3.
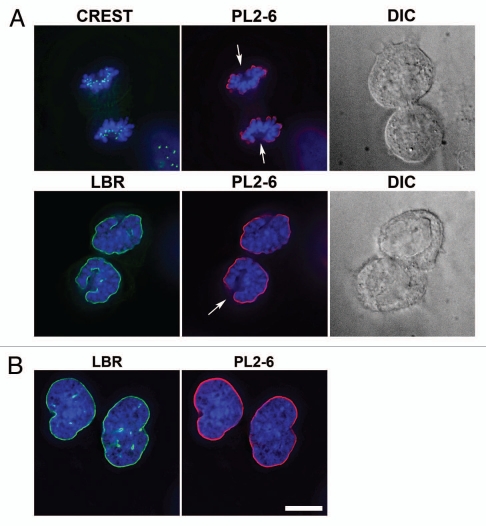
Co-immunostaining of U2OS interphase and metaphase cells with the epichromatin, mitotic marker and centromere (CREST) antibodies. The mitotic cell is on top of each part; the interphase cell is at the bottom. (A) mouse mAb PL2-6; (B) rabbit anti-H3 phosphorylated at serine 10, the mitotic marker H3(S10)p; (C) DAPI; (D) merge with an insert showing a 3-fold enlargement of the region denoted by an asterisk; (E) merged image of PL2-6, CREST and DAPI. Mouse mAb PL2-6 staining is shown in red, H3(S10)p and CREST in green, DAPI in blue. Each part is a single deconvolved optical slice of the same field. Bar equals 10 µm.
The epichromatin epitope persists in metaphase chromosomes even with nuclear envelope components dispersed into the cytoplasm.
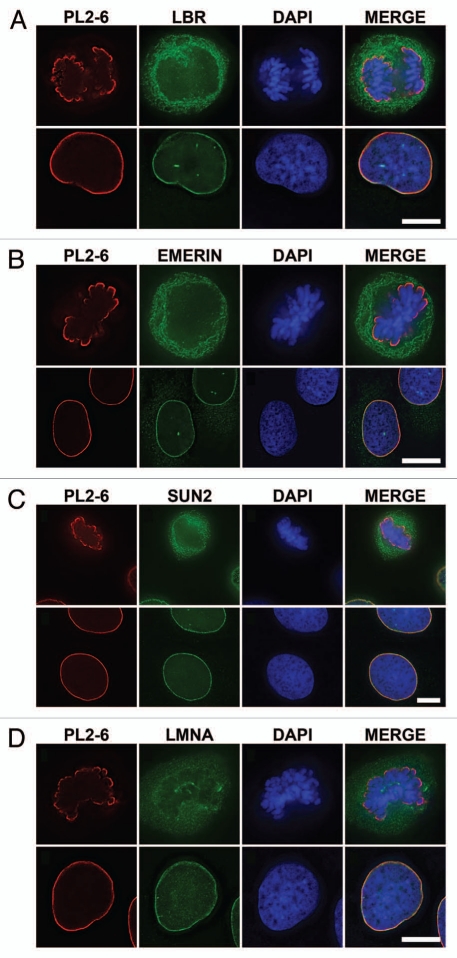
During the dissolution of the NE at mitosis, all of its components move into the cytoplasm.21 Integral membrane proteins are retained in the endoplasmic reticulum (ER) by virtue of their transmembrane domains (TM). Lamin B receptor (LBR) is an excellent example of an inner membrane protein that persists in the ER during mitosis, returning early to the post-mitotic reforming NE.22 Figure 4A demonstrates in U2OS cells that LBR is present in the interphase NE, but withdraws into the ER during mitosis; whereas PL2-6 stains both the interphase epichromatin and the exterior of the mitotic chromosome cluster. Similar sets of images were obtained for emerin (Fig. 4B) and SUN2 (Fig. 4C), both proteins possessing TM domains. Lamin A, which lacks direct attachment to the NE, is also dispersed into the mitotic cell cytoplasm (Fig. 4D). Similar image data for LBR and SUN2 have been obtained comparing interphase HL-60/S4 with metaphase cells (data not shown). It is clear from these experiments that “exposure” of the epichromatin epitope in mitotic chromosomes does not depend upon close proximity of the chromatin to an intact NE or to components of the NE. The epichromatin epitope appears to be an inherent property of chromatin throughout the cell cycle.
Figure 4.
Dispersion of LBR, emerin and SUN2 into the mitotic ER and of lamin A (LMNA) into the cytoplasm of U2OS cells, concurrent with persistence of the epichromatin epitope at the periphery of mitotic chromosomes. Mouse mAb PL2-6 staining is shown in red. Anti-LBR (A), anti-emerin (B), anti-SUN2 (C) and anti-LMNA (D) staining are indicated in green; DAPI in blue. In each part, the upper row of images is from the same mitotic cell; the bottom row is from the same interphase cell. Bar equals 10 µm.
The epichromatin epitope appears to be absent from several nuclear and chromosomal sites during the cell cycle.
As described earlier, PL2-6 does not stain the internal chromatin of interphase nuclei or the “internal” chromosome arms and ends of prophase, metaphase and early anaphase mitotic chromosomes. Another location that appears unreactive to the anti-epichromatin antibody is the mitotic chromosome “core”, a region facing the spindle pole, possessing centromeres, binding to the spindle microtubules, exhibiting proteins such as LAP2α and BAF and excluding LBR, LAP2β and lamin B.23,24 Examples of mitotic chromosome cores are presented in Figure 5A and Supplementary Figure 1. Note especially that CREST antibody stains centromere regions of the cores, while anti-LBR and PL2-6 primarily react with the telophase chromosome “periphery”. In contrast, SUN2 appears to stain the core and periphery of telophase chromosomes (Sup. Fig. 1). Eventually the daughter interphase nuclei do show LBR and epichromatin staining around the entire nuclear surface.
Figure 5.
Nuclear and chromosomal regions that demonstrate an absence of epichromatin staining. (A) presents telophase U2OS cells with discernable chromosome “core” regions. The left column of images pairs anti-centrosome CREST (top row) or anti-LBR (second row) with DAPI. The middle column of images pairs PL2-6 with DAPI. The right column presents a differential interference contrast (DIC) image of the separating daughter cells in the same field. Arrows point to mitotic chromosome “cores”. (B) displays interphase U2OS nuclei with intranuclear tubules. The left image pairs anti-LBR with DAPI. The right image pairs PL2-6 with DAPI. Intranuclear tubules are clearly stained by anti-LBR, but not by PL2-6. Bar equals 10 µm.
Within interphase nuclei of various types of attached cells (e.g., HeLa, NRK, 3T3 and CHO), intranuclear channels or tubules can be readily observed.25,26 These intranuclear tubules are generally oriented perpendicular to the cell attachment surface. Figure 5B and Supplementary Figure 2 present several examples of U2OS cells with intranuclear tubules. Since the tubules are primarily vertical in orientation, they usually present circular cross-sections. Figure 5B demonstrates that these tubules can be visualized by immunostaining with anti-LBR, but not with anti-epichromatin. Supplementary Figure 2 demonstrates that lamin A, emerin and SUN2 yield staining of the tubules, but PL2-6 does not. Current evidence supports that these tubules are extensions of the nuclear envelope, frequently adjacent to nucleoli and involved in Calcium release.27–29
There are numerous reasons why selected regions of interphase nuclei and mitotic chromosomes may not exhibit reactivity with the anti-epichromatin antibody. It may be that the local chromatin regions do not have the epitope or that it is blocked by bound molecules. Given that these regions can all be visualized by other IgG antibodies of the same molecular size, but different specificity, it is considered unlikely that there is a general penetration problem.
The epichromatin epitope exhibits a granular substructure at the nuclear envelope in cells and in isolated nuclei.
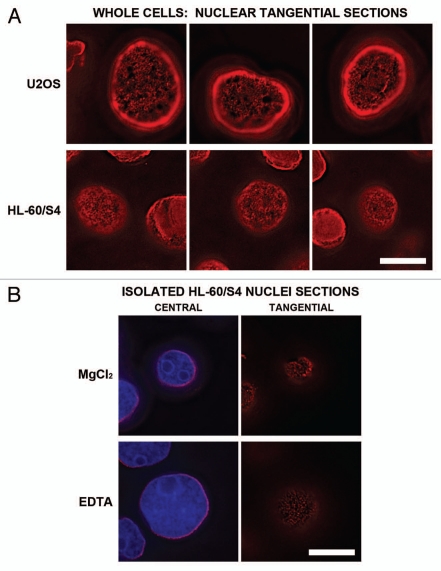
As shown in previous figures, mid-section optical slices of interphase nuclei suggest that the epichromatin epitope is continuous at the NE. However, Figure 6A presents evidence that is not an accurate impression. This Figure presents a gallery of tangential slices (i.e., just under the NE) stained with PL2-6, within interphase nuclei of U2OS and HL-60/S4 cells. It is evident that the epichromatin epitope exhibits a punctate/granular pattern at the NE. This granular substructure at the NE is preserved in detergent-buffer isolated nuclei from HL-60/S4 (Fig. 6B), washed with buffers containing divalent cations (1.5 mM MgCl2) or treated with combined chelators (EDTA and EGTA), prior to fixation and immunostaining. Because this granular substructure is seen at the NE of intact cells, it is unlikely to be an artifact of nuclear isolation. As with the regions of interphase chromatin and mitotic chromosomes that do not exhibit the epichromatin epitope, described earlier, the staining substructure at the NE might arise from a variety of possible causes which remain to be completely elucidated. But it is clear that since central optical sections in isolated and permeabilized nuclei show epichromatin staining only at the nuclear periphery, this characteristic staining pattern is not a consequence of inadequate antibody penetration.
Figure 6.
Immunostaining of the epichromatin epitope in tangential optical sections of nuclei from U2OS and HL-60/S4 cells. Mouse mAb PL2-6 staining is shown in red; DAPI in blue. (A) presents tangential sections of nuclei within intact cells. The top row of three images displays sections of U2OS cells; the second row is from HL-60/S4 cells. (B) shows central and tangential sections of isolated HL-60/S4 cell nuclei, washed in different buffers prior to fixation and immunostaining. In the top row, the isolated nucleus was washed in 1.5 mM MgCl2, 0.2 mM EGTA, 50 mM HEPES (pH 7.0); bottom row, washed in 0.2 mM EDTA, 0.2 mM EGTA, 50 mM HEPES (pH 7.0). Each image is a single deconvolved optical slice. In order to visualize the low amount of epichromatin immunofluorescence at the tangent of the NE, the brightness of the PL2-6 red signal was greatly increased. Bar equals 10 µm.
Immunoelectron microscopy documents epichromatin epitope localization at the nuclear envelope and in envelope-limited chromatin sheets (ELCS) of granulocytic HL-60/S4 cells.
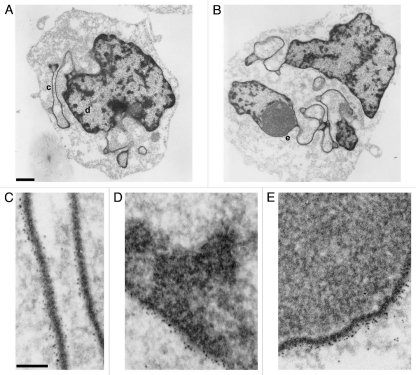
When human leukemic HL-60 cells are treated with all-trans retinoic acid (RA), they cease dividing and terminally differentiate into granulocytic forms. This differentiation process takes 3–4 days in the subline HL-60/S4, leading to nuclear lobulation and formation of extensive sheets containing a single layer of ∼30 nm chromatin fibers sandwiched between apposed nuclear envelopes.5,30 These envelope-limited chromatin sheets (ELCS) are an extension of the adjacent NE membranes, with the ∼30 nm chromatin layer in ELCS continuous to the most peripheral chromatin layer in the adjacent nuclear lobule. Employing pre-embedded thin section immunoelectron microscopy with a gold-labeled secondary antibody, the presence of the epichromatin epitope can be clearly visualized (Fig. 7). Two examples of HL-60/S4 granulocytic cells exhibiting nuclear irregularity and extensive ELCS formation are presented in Figure 7A and B. C shows an enlargement of region “c” in A, with two ELCS lying side-by-side; each is studded with gold grains. D presents an enlargement (region “d“ of A) at the interphase NE, again revealing gold-labeled antibody at the periphery. E shows a single chromatin layer sandwiched in between the nucleolus and the NE (region “e” of B) with clear labeling of the epichromatin epitope. These images demonstrate that the epichromatin epitope is present at the most peripheral single layer of ∼30 nm chromatin fibers and not detected within more interior chromatin.
Figure 7.
Immunoelectron microscopic labeling of the epichromatin epitope at the NE periphery and within ELCS of RA treated HL-60/S4 cells. (A and B) display two different cells which exhibit nuclear lobulation and extensive formation of ELCS. Enlarged regions taken from (A and B) are as follows: (C) taken from (A) (region “c”), presents two parallel ELCS; (D) taken from (A) (region “d”), displays a segment of the nuclear surface; (E) taken from (B) (region “e”), shows a single peripheral heterochromatin layer adjacent to a nucleolus. The NE and ELCS membranes can not be visualized because of the post-fixation detergent extraction and because the samples were not fixed with OsO4. Magnification bar values: (A and B) 1 µm; (C–E) 100 nm.
The NE and ELCS membranes can not be visualized in Figure 7 for several reasons: (1) the post-fixation extraction with 0.1% Triton X-100 removes most of the lipids; (2) the samples were not fixed with OsO4, (which enhances lipid contrast in the electron microscope). Evidence is presented in Supplementary Figure 3 to show that an integral membrane protein component of the NE (LBR) does remain in place, even though the membrane lipids can not be visualized. Immunoelectron microscopic staining of granulocytic HL-60/S4 cells for LBR clearly demonstrates the presence of gold particles along the ELCS and at the NE.
The epichromatin epitope is conserved among animals and plants.
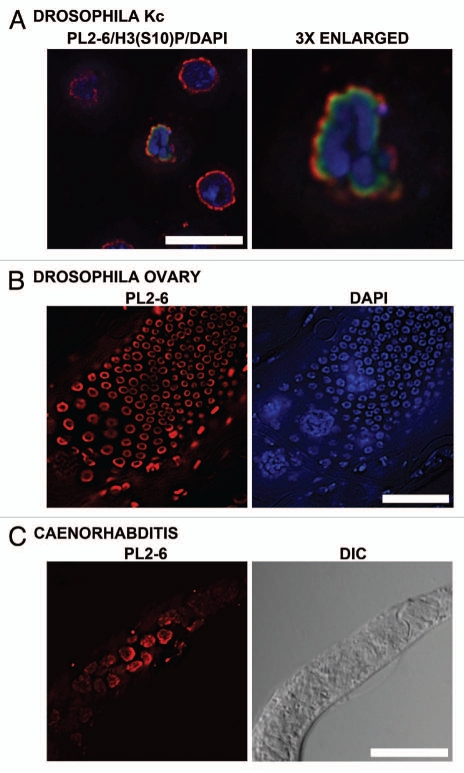
In order to explore the evolutionary conservation of the epichromatin epitope, studies were performed on a variety of eukaryotic organisms with well studied genetic systems. The invertebrates, Drosophila melanogaster and Caenorhabditis elegans, both have highly diverged homologs of vertebrate lamins in their NE.31 Figure 8 presents image data to demonstrate that the epichromatin epitope can be found at the periphery of interphase nuclei in both species, as well as at the surface of mitotic chromosomes in Drosophila Kc cells. This conclusion is clearly shown in Figure 8A, which presents a merged image of staining by mAb PL2-6 (red), rabbit anti-H3(S10)p (green) and DAPI (blue). The 3x enlarged cluster of mitotic chromosomes closely resembles a similar mitotic cluster shown previously (Fig. 3) for U2OS cells; the epichromatin epitope appears to be more exterior than the H3(S10)p determinant. Figure 8B shows a portion of a Drosophila larval ovary. The characteristic nuclear peripheral staining by PL2-6 can be readily observed in the surface cells of the ovary, demonstrating the expected location of this epitope. However, deeper cells within the ovary did not stain. We suspect that this is an antibody “penetration” problem due to the thickness of the whole ovary. Figure 8C presents a PL2-6 immunostained image and a DIC image of a C. elegans worm. The epichromatin epitope can be detected at the periphery of the interphase nuclei. As with the Drosophila larval ovary, antibody “penetration” into the whole organism may have prevented the staining of more internal cells. None-the-less, it is clear that the epichromatin epitope staining of peripheral chromatin within interphase nuclei exists in multicellular invertebrates, despite their highly divergent NE composition.31
Figure 8.
Immunostaining of the epichromatin epitope in Drosophila melanogaster and C. elegans cells. (A) Drosophila Kc cells immunostained with mAb PL2-6 (red), rabbit anti-H3 phosphorylated at serine10, the mitotic marker H3(S10)p (green) and DAPI (blue). (A) (right) is a 3-fold enlargement of the mitotic chromosomes in (A) (left). (B) (left) displays PL2-6 staining (red) of Drosophila ovary cells; DAPI (blue) of the same field is shown at (B) (right). (C): C. elegans worm: left, immunostaining with PL2-6; right, DIC image. Magnification bars equal 10 µm in (A and C); 5 µm, (B).
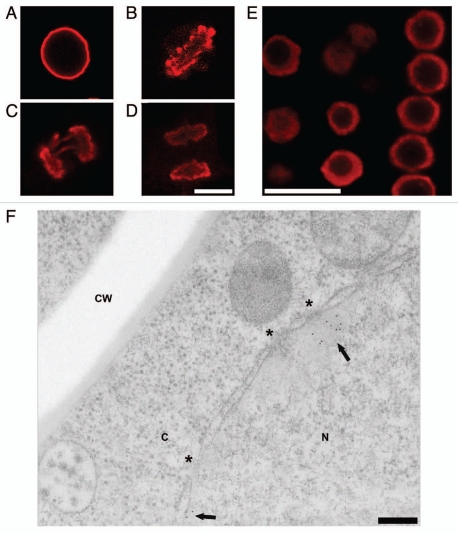
Plant cell NEs are even more divergent from higher metazoans than observed with the invertebrates cited above, exhibiting an absence of homologs to lamins, LBR and most other NE-associated proteins.32–34 None-the-less, Figure 9A–D convincingly demonstrates that the epichromatin epitope is present at the periphery of interphase nuclei and mitotic chromosomes in tobacco BY-2 tissue culture cells and in interphase nuclei of Arabidopsis thaliana root tips (Fig. 9E). Figure 9F displays an immunoelectron micrograph with gold-labeled antibody specifically localizing PL2-6 proximal to the NE in high pressure freezing/freeze substitution post-embedded samples of Arabidopsis thaliana root tips. Collectively, the immunostaining of invertebrate animal and plant cells strongly argues that the epichromatin epitope is highly conserved among very diverse species with vastly different NE composition and, likely, very different DNA sequences proximal to the NE.
Figure 9.
Immunostaining of the epichromatin epitope in tobacco and Arabidopsis thaliana cells. (A–D), confocal sections of mitotic stages seen in tobacco BY-2 cells immunostained with mAb PL2-6 (red): (A) interphase; (B) metaphase plate; (C) anaphase; (D) telophase. (E) confocal section of a whole mount of a Arabidopsis root tip stained with PL2-6 (red). (F) electron micrograph of a post-embedded immunogold stained thin section of a high pressure freezing/freeze substituted Arabidopsis root tip. The arrows point to the 5 nm gold near the NE. The astericks indicate the position of nuclear pores. CW, cell wall; C, cytoplasm; N, nucleus. Magnifications: (A–D), bar in (D) equals 10 µm; (E) bar equals 10 µm; (F) bar equals 200 nm.
Immunoblotting with PL2-6.
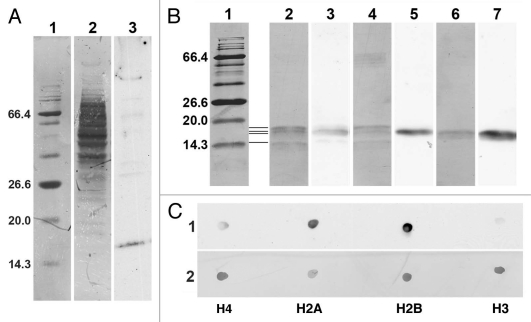
Most of our current knowledge about the binding specificity of the epichromatin antibody (PL2-6) is derived from ELISA studies.11,13,14,17,18 We know, based upon ELISA quantitation, that PL2-6 binds strongly to mononucleosomes and to a ternary complex of histones H2A + H2B + DNA, weakly to H2A + H2B and very weakly to H3 + H4 + DNA, individual histones or DNA alone. In the present study, we attempted to see whether PL2-6, PL2-7 and LG10-1 were capable of providing information by immunoblotting procedures. Figure 10A presents an immunoblot analysis of PL2-6 reacted with a total cell extract of U2OS cells (a similar experiment with PL2-7 and LG10-1 did not provide any ECL signals using the same extract of U2OS cells). Figure 10A reveals that most of the extracted proteins, when stained with Coomassie Blue (lane 2), migrated between ∼36 to ∼100 kDa. However, the major anti-epichromatin reactive band migrated at ∼18 kDa (lane 3), a region which includes the inner histones. A few very faint higher molecular weight bands were also detected with PL2-6. Figure 10B presents immunoblots of PL2-6 against several types of samples, including core mononucleosomes from HeLa cells and purified Xenopus recombinant core histones, individually or in various equimolar combinations. The image of the immunoblot shown in Figure 10B presents alternating lanes of the Coomassie Blue (CB) stained membrane (lanes 1, 2, 4 and 6) interspersed with carefully aligned ECL images from the same membrane, revealing PL2-6 reactivity (lanes 3, 5 and 7). Figure 10B lanes are as follows: lane 1, CB stained protein molecular weight markers; the region between lanes 1 and 2, representation of the positions of the core histones (from top to bottom, H3, H2B, H2A, and H4); lanes 2 and 3, HeLa mononucleosomes; lanes 4 and 5, equimolar mixture of recombinant Xenopus histones H4, H2A, H2B and H3; lanes 6 and 7, equimolar mixture of recombinant Xenopus histones H2A and H2B. Even with the limited resolution of this 17.5% polyacrylamide gel, the data clearly indicate that H4 and H3 show little reactivity with PL2-6; but H2A and H2B appear to exhibit significant reactivity. These results provoked us to perform dot blots with the purified individual recombinant Xenopus inner histones (Fig. 10C). Equimolar aliquots, based upon measurements of the histone concentrations using the molar extinction coefficients at 276 nm, were spotted onto Immobilon-P membranes, reacted with PL2-6, analyzed by ECL (Fig. 10C, strip 1) and CB stained (strip 2). The ECL results clearly show that, on a molar basis, PL2-6 reacts most strongly with recombinant Xenopus H2B and H2A and much more weakly with H4 and H3. Semi-quantitative estimates of reaction intensities were obtained from replicate dot arrays and different ECL exposure times, employing ImageJ for measurement of film densities (see Materials and Methods). The averaged results indicated that (setting the H2B reaction intensity at 100%) H2A exhibited ∼85% reaction intensity, H4 ∼22% and H3 ∼9%. In summary, these immunoblot results indicate that PL2-6 can recognize a homologous epitope within both histones H2A and H2B. Thus, the anti-epichromatin antibody (PL2-6), which has been extensively characterized by ELISA analyses, is capable of reacting by immunoblotting with the separate histones H2A and H2B, despite the fact that these separate histones give very weak reactions with PL2-6 by ELISA.
Figure 10.
Immunoblot and immunodot analysis of the reactivity of PL2-6. (A) 4–20% gradient SDS-PAGE immunoblot analysis of U2OS total cell extract. Lanes: 1, BioLab protein molecular weight (mol wt) standards, stained with Coomassie Blue (CB), indicating the mol wt (kDa) of several proteins; 2, total cell extract stained with CB; 3, ECL reaction with PL2-6. (B) 17.5% SDS-PAGE with the following lanes: 1, protein mol wt markers stained with CB; 2 and 3, HeLa core mononucleosomes; 4 and 5, equimolar mixture of recombinant Xenopus inner histones H4, H2A, H2B and H3; 6 and 7, equimolar mixture of recombinant Xenopus inner histones H2A and H2B. All lanes are from the same gel. Lanes 1, 2, 4 and 6, CB stained. Lanes 3, 5 and 7, ECL exposures carefully aligned to lanes 2, 4 and 6, respectively. Mol wt values (kDa) of the markers are indicated to the left of lane 1. The four thin horizontal lines between lanes 1 and 2 denote the positions of the four inner histones, starting with the lowest band (H4) and progressing upward, H4, H2A, H2B and H3. (C) Immunodot blots of equimolar aliquots of purified individual recombinant Xenopus inner histones (H4, H2A, H2B and H3). Strip 1, ECL reaction with PL2-6. Strip 2, identical membrane strip after CB staining.
Discussion
A chromatin epitope involving histones H2A and H2B and DNA, recognized by a mouse monoclonal autoimmune anti-nucleosome antibody (PL2-6), specifically localizes at the peripheral surface of interphase chromatin and mitotic chromosomes and is conserved by evolution in its location among various plant and animal cells. We suggest the name “epichromatin” to denote this unique chromatin surface conformation. To the best of our knowledge, no such epitope has been reported before in either interphase or mitotic cells, nor has there been a suggestion that surface chromatin conformation throughout the cell cycle differs from bulk chromatin conformation. The staining of interphase nuclei is reminiscent of the classical “rim” pattern, commonly seen with antibodies against lamins or nuclear envelope integral membrane proteins (see for example, ref. 35 and 36). During mitosis the lamina/nuclear envelope “rim” staining pattern disappears, being last observed in early prophase and again visible at telophase, the periods when the nuclear envelope is breaking down or reforming.21,37 By contrast, the epichromatin epitope persists throughout mitosis, including metaphase, being independent of the presence of a NE. In addition, we have shown that two closely related mouse monoclonal autoimmune anti-nucleosome antibodies, PL2-7 and LG10-1 stain throughout the entire interphase nuclei and the entire set of mitotic chromosomes (generally resembling DAPI staining), minimizing the possibility that the surface staining of PL2-6 is due to a penetration problem.
Published and present data imply some molecular characteristics about PL2-6 . ELISA data11,13,14,17,18 demonstrate that the chromatin epitope recognized by PL2-6 is a conformational one, involving a ternary complex of H2A, H2B and DNA. Even though the epitope is present in the binary complex of H2A and H2B, reactivity is augmented by complexing with (mixed sequence) DNA on a surface, as in ELISA plates, or in solution. Another observation (MM unpublished), using a protocol employed with a different antibody,38 is that trypsin removal of the histone basic tails in nucleosomes does not eliminate the ELISA reactivity of the epitope. This implies that the epichromatin epitope is present in the conserved globular regions of histones H2A and H2B, which primarily reside within the nucleosome cores. Even though the ELISA reactivity of PL2-6 with uncomplexed H2A or H2B is very weak,17 it is able to yield immunoblotting results with both native Hela and recombinant H2A and H2B. The fact that PL2-6 reacts with individual recombinant H2A and H2B argues that histone post-translational modifications are not a part of the epichromatin epitope.39 Furthermore, the data imply either that there are homologous sites in the histone globular domains for Xenopus H2A and H2B, similar enough to react with PL2-6 and/or sequences within H2A and H2B that are each part of the conformational epitope. Furthermore, these sequences should be present within the corresponding mouse histones (i.e., the presumed immunogens which induced the autoimmune antibody). A CLUSTAL multiple alignment of the major mouse and Xenopus histones (data not shown) does indicate scattered residue identities, conservative and neutral replacements within the histone globular regions which are candidates for homologous epitope regions. Although not yet established, it appears likely that most of the nucleosomes within interphase and metaphase chromatin possess the histone amino acid residues involved in the epichromatin epitope, but that these residues are not exposed (possibly due to binding interactions or chromatin higher order structures). In addition, there may be a subset of nucleosomes which have a different histone composition and lack the epichromatin epitope. For example, mammalian centromeric regions contain H2A.Z in many of the nucleosomes.40 Indeed, the present study documents (Fig. 5 and Sup. Fig. 1) that during late anaphase-telophase, the mitotic chromosome “core” (centromere) region is unreactive with PL2-6. There is increasing evidence that centromere regions in a variety of species differ considerably in histone composition and nucleosome conformation.41 Defining the molecular structure of the epichromatin epitope and possible binding interactions remains a worthy endeavor.
It is tempting to speculate whether epichromatin may have a functional significance, and if so, what it might be. At the onset of such speculative thinking, it should be explicitly stated that epichromatin may have no functional significance. It may simply be that the epichromatin nucleosomal conformation results from the lack of a complete surrounding by adjacent chromatin with consequent exposure to cytoplasm (mitosis) or the nuclear envelope (interphase). A physical analogy might be the surface tension of liquid water. From this point-of-view, the epichromatin nucleosomal conformation is the consequence of a different macromolecular environment. Extending this analogy, it is conceivable that epichromatin has a general function; i.e., separating or protecting chromatin, creating a barrier without membranes.
An alternative view is that epichromatin has a more specific functional significance, of sufficient importance to be highly conserved in evolution. We describe this view as the “epichromatin hypothesis”. This hypothesis postulates that a unique peripheral chromatin conformation plays an active role in ensuring the continuity of nuclear architecture throughout the cell cycle, especially during post-mitotic nuclear envelope reformation. Furthermore, it is suggested that this function (maintaining nuclear architecture throughout the cell cycle) has been highly conserved by evolution. Epichromatin could affect post-mitotic nuclear reformation by presenting preferred interaction sites for early-binding nuclear envelope integral membrane proteins (e.g., LBR and LAP2β21,22,37). In a “deterministic” version of the epichromatin hypothesis, this attractive chromatin conformation is always located at the same regions of specific chromosomes, ensuring that these chromosome regions have preferred positions adjacent to the nuclear envelope during post-mitotic nuclear reformation. In a “stochastic” version of the hypothesis, the epichromatin conformation does not have precise chromosomal locations, but dynamically fluctuates around chromosome surfaces. It is also conceivable that the epichromatin regions exhibit tissue specificity, possibly reflecting which regions of the genome are heterochromatic or modified. We do not know whether the epichromatin epitope is associated with specific DNA sequences; however, its similar localization in divergent plant and animal cell nuclei strongly suggests that neither specific DNA sequences nor CpG DNA methylation42 are required.
There is at least one other nuclear structure, where specific DNA sequences do not directly dictate the formation and location of an essential chromosomal feature; i.e., centromeres, which may represent a provocative comparison to epichromatin. Centromeres are ubiquitous structures, observed in plant and animal cells, involved in holding sister chromatids together, being a platform for kinetochores and ensuring proper segregation of homologous chromosomes during mitosis.41,43–47 The underlying DNA sequence is variable, comparing different species; although proximity to repetitive DNA is a frequent motif. The current view is that centromeres are epigenetic structures built around a centromere-specific highly conserved histone H3 variant CENH3, which is critical for the binding of other centromere-specific proteins and the establishment of unique nucleosomal structures. CENH3 deposition on chromatin does not occur during S phase; but, rather, at the end of mitosis. Current studies are exploring the structural basis of the epigenetic maintenance of centromere localization through cell division. Clearly, we need to know whether epichromatin histones include specific variants that might preserve a unique chromatin conformation by epigenetic mechanisms.
In many respects, our view of interphase epichromatin resembles that of van Steensel and co-workers.2,9,10,48 This group has examined the interphase nuclear chromatin regions that are proximal to the nuclear envelope in Drosophila Kc cells and in human fibroblasts, employing transfection with a chimeric gene containing lamin B fused to DNA methylase, followed by subsequent analysis of the methylated DNA fragments. These lamina-associated domains (LADs) of chromatin do not display specific DNA sequences. They vary in size (0.1–10 Mb), are largely “gene-poor”, with those genes present being transcriptionally repressed. Furthermore, there is no simple correlation with epigenetic repressive histone markers or with active histone markers; i.e., H3 and H4 acetylation and H3K4 methylation are largely depleted. Perhaps the most revealing characteristic of LADs occurs at their borders, which appear to be relatively sharp, containing binding sites for an “insulator” protein (CTCF), CpG islands and promoters of genes with transcription directed away from the nuclear envelope. The authors suggest a model with active gene loops interspersed between the LADs and directed away from the repressive nuclear envelope environment. It is possible that epichromatin includes (or is equivalent to) the LADs within the interphase nucleus. Unfortunately, since the nuclear envelope breaks down during mitosis, LADs can not be analyzed from this stage of the cell cycle.
The observation that interphase CTs sometimes exhibit a non-random radial distribution within interphase nuclei6,7,49 suggests that for some CTs, epichromatin regions may participate in directing chromosomes to proximity with the nuclear envelope. However, our images from late anaphase and telophase cells (Fig. 2 and Suppl. Videos 3 and 4) suggest that late in mitosis most chromosomes exhibit the surface epichromatin epitope. From the point-of-view of our epichromatin hypothesis, this implies that other factors might restrict the chromatin surfaces involved in post-mitotic nuclear envelope reformation. The complexities of measurements and interpretations of CT arrangements within interphase nuclei have been discussed.7 Radial and neighboring arrangements among CT are not generally fixed or simple, which may signify that epichromatin can not have a strictly deterministic influence upon interphase nuclear architecture. Many factors, including nuclear envelope shape, flexibility and composition and interactions with cytoskeletal elements must play a role in defining nuclear architecture.50–52 To cite one example, pericentromeric heterochromatin regions of mouse granulocytes frequently cluster adjacent to the nuclear envelope making nodules into the cytoplasm.53 In this situation, it is evident that the nuclear envelope integral membrane protein LBR is necessary for the heterochromatin to exhibit a peripheral localization. In another remarkable variation of interphase nuclear architecture, heterochromatin in the retinal rod cells of nocturnal (but not diurnal) mammals is “inverted” (clustered) into the middle of nuclei, apparently to function in channeling the light.54 A somewhat similar rearranged heterochromatin nuclear structure phenotype is observed in the mutant blood granulocytes of humans (Pelger-Huet anomaly) and mice (Ichthyosis) due to a deficiency of LBR.55 All of these nuclear variations suggest that the epichromatin conformation may be only one of many factors involved in specifying post-mitotic nuclear architecture.
Our observation of an epichromatin epitope in interphase and metaphase cells provokes a new question: what is the relationship between epichromatin and heterochromatin or euchromatin? The immunogold electron microscopy of RA treated (granulocytic) HL-60/S4 cells (Fig. 7) clearly demonstrates the presence of the epichromatin epitope within the condensed heterochromatic region adjacent to the nuclear envelope, as well as within the single heterochromatic layer of ELCS.5 These images prove that the epichromatin epitope does include peripheral heterochromatin. However, it is becoming increasingly clear that some transcriptionally-active euchromatin can be localized near the nuclear envelope.2,56 Experiments should be devised to test whether such euchromatin regions also exhibit the epichromatin epitope. Sites of DNA synthesis occur in different regions of the interphase nucleus during progression through S phase.57–59 The most peripheral chromatin of the interphase nucleus appears to replicate during late S, yielding immunofluorescent images of incorporated nucleotides very similar to those of epichromatin staining, and containing the “G band” gene-poor heterochromatin. However, in our studies epichromatin staining of metaphase chromosomes does not resemble the alternating patterns of R (gene-rich) and G bands, traditionally employed for chromosome karyotyping. The basis for our more uniform staining of the exposed surfaces of mitotic chromosomes remains to be explored. It is important to mention that mitotic chromosomes are surrounded by a complex mixture of proteins and RNPs, called the “perichromosomal layer”,60,61 which may influence the exposure of the epichromatin epitope and the existence of an epichromatin conformation. It is of particular interest that the cell proliferation associated nuclear antigen (Ki-67),62 which localizes within interphase nucleoli, appears to bind to the surface of metaphase and anaphase chromosomes. It may be that epichromatin can act as a “platform” for the binding of proteins or particles which require distribution to both daughter nuclei.
Much remains to be determined about epichromatin, for example: the molecular structure of the epitope; the nature of the chromatin conformation; correlations with underlying or surrounding DNA; correlations with epigenetic markers; characterization of binding partners. Furthermore, it remains to be established whether the concept of epichromatin and the suggested “epichromatin hypothesis” will be useful in advancing our understanding of the structural and functional “memory” of the interphase nucleus.
Materials and Methods
Reagents and antibodies.
Paraformaldehyde (PFA) was prepared as an 8% solution in distilled water (pH 7–8) and stored in aliquots at −20°C. Poly-L-lysine (MW 150–300 KD, Catalogue # P-1399) and Nuclei EZ Prep (Catalogue # NUC-101) were purchased from Sigma-Aldrich (St. Louis MO). Complete Mini protease inhibitor was obtained from Roche Diagnostics (Germany). All buffers were made from reagent grade components, with stocks sterilized by autoclaving or membrane filtration. Fluorescent secondary goat antibodies, Alexa 568 and Alexa 488 conjugates were obtained from Invitrogen GmbH (Darmstadt, Germany).
Cells, tissues and organisms.
Animal materials. Human HL-60/S4 suspension cells were maintained in RMPI 1640 medium, plus 10% heated fetal calf serum, 1% Pen/Strep and 2 mM L-glutamine, as described earlier.30 Human U2OS cells were cultivated in DMEM medium, plus 20% fetal calf serum and 2 mM L-glutamine. Drosophila melanogaster Kc 167 cells were maintained at 25°C, in Schneider's Drosophila Medium supplemented with 10% heated fetal calf serum, in the presence of Pen/Strep. HL-60/S4 cells were harvested for microscopy at a concentration of ∼106/mL; coverslip attached U2OS and Drosophila Kc cells were used prior to confluence. Caenorhabditis elegans was cultivated on nematode growth medium (NGM) agar at 20°C as described previously.63
Plant materials. A stably transformed Arabidopsis thaliana line64 was grown on Murashige and Skoog medium containing 0.5% agar. Roots were harvested 5 days after germination for whole mount immunofluorescence. Suspension cultures of BY-2 tobacco (Nicotiana tabacum) were cultivated as previously described65 and analysed 3 days after subculturing.
Immunostaining and fluorescence microscopy.
HL-60/S4 cells were allowed to settle onto fresh polylysine-coated microscope slides for 30–60 min at RT in a moist chamber. The attached HL-60/S4, U2OS and Drosophila Kc cells were fixed with fresh 4% PFA/PBS for 15 min at RT. The following steps included: 50 mM NH 4Cl (1 min), PBS washes (2 × 1 min), 0.1% Triton X-100 in PBS (20 min); PBS (3 × 5 min); blocking with 10% normal goat serum/PBS (30 min, RT). For some experiments, HL-60/S4 and U2OS cells were fixed with anhydrous methanol (−20°C, 10 min), followed by acetone (−20°C, 10 min) and subsequent washes with PBS, prior to blocking.
Primary antibody dilutions were as follows: PL2-6, PL2-7 and most other mAbs, 1:200 dilution (∼8 µg/ml); LG10-1, 1:400 (∼3 µg/ml). For some experiments, the “mitotic marker” rabbit anti-H3(S10)p (Millipore, Billerica MA) was employed at a 1:200 dilution. Co-immunostaining was also performed with human auto-immune anti-centromere (CREST) antisera, as described previously.35 Secondary antibody dilutions (Alexa 568 and 488) were all at 1:100. DAPI was included during incubation with the secondary antibodies. Incubations were in a moist chamber (1 hr, 37°C). Slides were mounted in SloFade Antifade Kit (Life Technologies Co., Carlsbad CA) using a #1.5 thickness square coverslip and sealed with clear nail polish. Optical sections on all animal cells were collected on a DeltaVision Core microscope (Applied Precision Inc., Issaquah WA) using either a 40x, 60x or a 100x objective as “RGB” images. Images were deconvolved using the built in SoftWoRx software. Adobe Photoshop was used to assemble figures, adjust the size of individual images and bring color levels of individual panels to comparable intensities, no changes in gamma were made and then “RGB” was changed to “CMYK”. Videos of selected mitotic U2OS cells stained with PL2-6 were calculated from 12 projections between ±30° in 10° intervals around the vertical axis, employing Imaris software (Bitplane Inc., Saint Paul MN). The movies should be opened with a video player in the “loop” mode.
Ovaries were dissected in PBS from 4-day-old Drosophila melanogaster wild-type Oregon R females, grown at 25°C on standard food. They were fixed in 4% PFA/PBS on ice for 30 min and washed in PBS. After blocking with 2% BSA/0.1% Triton X-100/PBS for 1 hour, the samples were incubated with PL2-6 (1:200) overnight at 4°C in blocking buffer. Subsequently, the ovaries were washed for 1 hr in blocking buffer and incubated 1 hr with Alexa 568 anti-mouse in blocking buffer and washed again for 1 hr. The nuclei were stained with DAPI in PBS for 10 min. After staining, the ovaries were spread out on a glass slide in mounting medium (80% glycerol/0.4% N-propyl-gallate/PBS), a coverslip was applied and sealed with nail polish.
The immunostaining method employed with C. elegans had the following modifications. The worms were pelleted from M9 buffer and fixed overnight at 4°C in 4% PFA/PBS. Following washes in 0.5% Triton X-100/PBS, the pellet was permeabilized with 0.5% Triton X-100/1% DTT/PBS for 2 hrs at 37°C and repeated washes as before. The worms were incubated in blocking buffer (1% BSA/0.5% Triton X-100/PBS) for 1 hr at RT, followed by the primary antibody (PL2-6, 1:200) in blocking buffer overnight at RT. Following additional washes, the collected worms were incubated with secondary antibody (Alexa 568 anti-mouse, 1:100) in 0.1% BSA/0.5% Triton X-100/PBS for 4 hrs at 37°C. After final washes in 0.5% Triton X-100/PBS, the stained worms were placed on microscope slides and embedded in Vectashield Hard Set Mounting Medium for Fluorescence.
Arabidopsis thaliana plants were fixed and processed as previously described.66 Tobacco BY-2 suspension culture cells were fixed and processed as described67 with minor changes. One batch of cells was fixed with 4% PFA for 1 h; the other one with 1% glutaraldehyde for 15 min. Samples were washed with Sörensen buffer instead of PBS. Samples were incubated at 4°C with first antibody (PL2-6, 1:200). ALEXA-FLUORR conjugate 546 (Life Technologies Co.,) was used as secondary antibody. Imaging was performed with a Zeiss Axiovert LSM510 Meta CLSM using a C-Apochromat 63x/1.2 W corr water immersion objective. For the Metadetector, the main beam splitter (HFT) 488/543 was used. Pinholes were adjusted to 1 Airy Unit.
Immunoelectron microscopy.
Two types of gold-labeled immunoelectron microscopy were employed: (1) pre-embedded immunostaining of fixed and detergent extracted HL-60/S4; (2) post-embedded immunostaining on thin sections of high pressure freezing/freeze-substituted preparations of Arabidopsis root tips.
For the pre-embedded staining reaction, the procedure largely resembled that used for immunofluorescent staining, with the following modifications: (1) permeabilization of the fixed cells with 0.1% Triton X-100/PBS was extended to 30 min at RT; (2) 6 nm gold-labeled secondary antibody was incubated for 4 hrs in a moist chamber at 37°C, followed by three 5 min washes with PBS; (3) after the antibody reactions, the coverslips were fixed with 2.5% glutaraldehyde in 50 mM cacodylate buffer for 5 min at RT and 25 min at 4°C; (4) coverslips were given three washes in 50 mM cacodylate prior to dehydration and embedding in epon, by standard procedures; (5) coverslips were removed from the epon in liquid N2.
For the post-embedded immunostaining on high pressure freezing/freeze substituted preparations of Arabidopsis thaliana root tips, six-day-old root tips were cut from the seedling, submerged in 140 mM sucrose, 7 mM trehalose and 7 mM Tris buffer (pH 6.6). Four to five submerged root tips were collected, transferred to planchettes (Wohlwend GmbH, Sennwald, Switzerland; type 241 and 242) and frozen in a high pressure freezer (Bal-Tec HPM010, Lichtenstein). Freeze substitution was performed in a Leica EM AFS2 freeze substitution unit (Leica, Germany) in 9.9 ml dry acetone supplemented with 100–200 µl 20% uranyl acetate in methanol (0.2–0.4% final) at −85°C for 16 hrs before gradually warming up to −50°C over a 5 hr period. After washing with 100% ethanol for 60 min, the roots were infiltrated and embedded in Lowicryl HM20 at −50°C (intermediate steps of 30, 50, 75% HM20 in ethanol, 1 hr each), and polymerized for 3 days with ultraviolet (UV) light in the freeze substitution apparatus. To increase sectioning quality, the blocks were then hardened with UV light for another 4 hr at RT. Ultrathin sections were cut on a Leica Ultracut S and incubated with the primary antibody (PL2-6, 1:3) followed by incubation with 5 nm gold-coupled secondary antibody (BioCell GAR5, 1:50) in 1% BSA/PBS. Sections were examined in a JEM1400 transmission electron microscope (JEOL, Japan) operating at 80 kV. Micrographs were recorded with a FastScan F214 digital camera (TVIPS, Germany).
Immunoblotting and dot blotting.
In order to immunoblot a U2OS total extract, cells were washed in 0.1 mM EDTA/PBS, trypsinized, counted with a hemocytometer and washed in PBS containing both Complete (Roche Diagnostics) and PMSF (Sigma Aldrich) protease inhibitors. Cells (3 × 106) were lysed in 300 µL of 1x Laemmli sample buffer (10% glycerol, 3% SDS, 62.5 mM Tris-HCl, 50 mM DTT and 0.05% bromphenol blue). The cells were heated to 95° C for 5 min, allowed to cool and sheared by repeated passage through a 26G × 1/2″ hypodermic needle. 10 µL of the cell lysate was loaded per lane on a BioRad 4–20% precast gradient SDS-PAGE gel. Protein transfer, blocking, immunology and the ECL reaction were exactly as described previously,68 employing PL2-6 at a 1:2,000 dilution.
For an immunoblotting experiment with histones, three samples were examined by electrophoresis in a 17.5% SDS-PAGE: (1) native HeLa core mononucleosomes; (2) an equimolar mixture of Xenopus recombinant core histones H4, H2A, H2B and H3; (3) an equimolar mixture of Xenopus recombinant core histones H2A and H2B. Different loads of each sample were run on the gel, spanning 1–5 µg histones per lane. Protein transfer, blocking, immunology and the ECL reaction were exactly as described previously,68 employing PL2-6 at a 1:2,000 dilution.
The dot blotting procedure followed a protocol from Millipore describing its use on Immobilon-P membranes. Concentrations of the individual recombinant Xenopus inner histones H4, H2A, H2B and H3 were measured by absorbance at 276 nm in guanidine HCl buffer and calculated from known molar extinction coefficients. These histone stock solutions were diluted to 100 µM with the immunoblotting transfer buffer (20 mM sodium borate, 1 mM EDTA, pH 8.8). Several loads (3, 6 and 9 µl) of each histone were applied to the Immobilon-P membrane in a grid-like pattern on top of a filter paper stack. The spots were allowed to dry, than wetted again with methanol prior to reaction with PL2-6 (1:2,000) and ECL, as employed for immunoblotting. For a semi-quantitative analysis of the ECL reaction intensities of the dots, three replicate dot arrays were established onto the Immobilon-P membrane and several ECL exposure times were obtained. Employing ImageJ software (rsb.info.nih.gov/ij/), integrated spot densities were measured, corrected for adjacent blank areas, normalized to the strongest reacting histone (H2B, set to 1.0) and averaged to yield relative reaction strengths.
Acknowledgements
A.L.O. and D.E.O. wish to express their gratitude to Harald Herrmann and Peter Lichter for generously hosting us in their laboratories at the German Cancer Research Center (DKFZ, Heidelberg), during which time this work was performed. Gratitude is also expressed to the Department of Biology, Bowdoin College for encouraging us to continue scientific research. Cristina Pallares-Cartes (DKFZ) kindly provided the Drosophila samples. In addition, A.L.O. and D.E.O. thank Kati Toth and Jörg Langowski (DKFZ) and Andreas Ladurner (EMBL, Heidelberg) for gifts of HeLa mononucleosomal histones and recombinant Xenopus inner histones, and to Thorsten Kolb (DKFZ) for the gift of guinea pig anti-lamin A. Thanks are also extended to the ever-helpful team in the Herrmann group “Functional Architecture of the Cell”; Michaela Hergt, Helga Kleiner, Tanja Lichtenstern, Monika Mauermann and Dorothee Möller.
Abbreviations
- H2A, H2B, H3, H4
nucleosome inner histones
- CREST
auto immune antibody against centromere proteins
- CENH3
centromere H3 variant
- H3(S10)p
anti-phosphorylated serine 10 in H3
- mAb
monoclonal antibody
- SLE
systemic lupus erythematosus
- ELISA
enzyme-linked immunosorbent assay
- CT
chromosome territory
- LBR
lamin B receptor
- ELCS
nuclear envelope-limited chromatin sheets
- DIC
differential interference contrast
- NE
nuclear envelope
- CB
Coomassie blue
- PAGE
polyacrylamide gel electrophoresis
- ECL
enhanced chemiluminescence
- LADs
lamina-associated domains
- TM
transmembrane domain
- ER
endoplasmic reticulum
- PFA
paraformaldehyde
- DTT
dithiothreitol
Supplementary Material
References
- 1.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 2.Kalverda B, Roling MD, Fornerod M. Chromatin organization in relation to the nuclear periphery. FEBS Lett. 2008;582:2017–2022. doi: 10.1016/j.febslet.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 3.Towbin BD, Meister P, Gasser SM. The nuclear envelope —a scaffold for silencing? Curr Opin Genet Dev. 2009;19:180–186. doi: 10.1016/j.gde.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev. 2009;19:172–179. doi: 10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olins DE, Olins AL. Nuclear envelope-limited chromatin sheets (ELCS) and heterochromatin higher order structure. Chromosoma. 2009;118:537–548. doi: 10.1007/s00412-009-0219-3. [DOI] [PubMed] [Google Scholar]
- 6.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 7.Cremer T, Cremer M. Chromosome Territories. Cold Spring Harb Perspect Biol. 2010;2:22. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 10.de Wit E, van Steensel B. Chromatin domains in higher eukaryotes: insights from genome-wide mapping studies. Chromosoma. 2009;118:25–36. doi: 10.1007/s00412-008-0186-0. [DOI] [PubMed] [Google Scholar]
- 11.Monestier M. Autoantibodies to nucleosomes and histone-DNA complexes. Methods. 1997;11:36–43. doi: 10.1006/meth.1996.0385. [DOI] [PubMed] [Google Scholar]
- 12.Ballestar E, Esteller M, Richardson BC. The epigenetic face of systemic lupus erythematosus. J Immunol. 2006;176:7143–7147. doi: 10.4049/jimmunol.176.12.7143. [DOI] [PubMed] [Google Scholar]
- 13.Kramers C, Hylkema MN, van Bruggen MC, van de Lagemaat R, Dijkman HB, Assmann KJ, et al. Anti-nucleosome antibodies complexed to nucleosomal antigens show anti-DNA reactivity and bind to rat glomerular basement membrane in vivo. J Clin Invest. 1994;94:568–577. doi: 10.1172/JCI117371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kramers K, Stemmer C, Monestier M, van Bruggen MC, Rijke-Schilder TP, Hylkema MN, et al. Specificity of monoclonal anti-nucleosome auto-antibodies derived from lupus mice. J Autoimmun. 1996;9:723–729. doi: 10.1006/jaut.1996.0094. [DOI] [PubMed] [Google Scholar]
- 15.Van Hooser AA, Mancini MA, Allis CD, Sullivan KF, Brinkley BR. The mammalian centromere: structural domains and the attenuation of chromatin modeling. FASEB J. 1999;13:216–220. doi: 10.1096/fasebj.13.9002.s216. [DOI] [PubMed] [Google Scholar]
- 16.Moroi Y, Peebles C, Fritzler MJ, Steigerwald J, Tan EM. Autoantibody to centromere (kinetochore) in scleroderma sera. Proc Natl Acad Sci USA. 1980;77:1627–1631. doi: 10.1073/pnas.77.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Losman MJ, Fasy TM, Novick KE, Monestier M. Monoclonal autoantibodies to subnucleosomes from a MRL/Mp(-)+/+ mouse. Oligoclonality of the antibody response and recognition of a determinant composed of histones H2A, H2B and DNA. J Immunol. 1992;148:1561–1569. [PubMed] [Google Scholar]
- 18.Monestier M, Novick KE. Specificities and genetic characteristics of nucleosome-reactive antibodies from autoimmune mice. Mol Immunol. 1996;33:89–99. doi: 10.1016/0161-5890(95)00115-8. [DOI] [PubMed] [Google Scholar]
- 19.Hake SB, Garcia BA, Kauer M, Baker SP, Shabanowitz J, Hunt DF, et al. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc Natl Acad Sci USA. 2005;102:6344–6349. doi: 10.1073/pnas.0502413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McManus KJ, Hendzel MJ. The relationship between histone H3 phosphorylation and acetylation throughout the mammalian cell cycle. Biochem Cell Biol. 2006;84:640–657. doi: 10.1139/o06-086. [DOI] [PubMed] [Google Scholar]
- 21.Guttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10:178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- 22.Olins AL, Rhodes G, Welch DBM, Zwerger M, Olins DE. Lamin B Receptor. Nucleus. 2010;1:1–18. doi: 10.4161/nucl.1.1.10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foisner R. Cell cycle dynamics of the nuclear envelope. The Scientific World Journal. 2003;3:20. doi: 10.1100/tsw.2003.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haraguchi T, Kojidani T, Koujin T, Shimi T, Osakada H, Mori C, et al. Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J Cell Sci. 2008;121:2540–2554. doi: 10.1242/jcs.033597. [DOI] [PubMed] [Google Scholar]
- 25.Fricker M, Hollinshead M, White N, Vaux D. Interphase nuclei of many mammalian cell types contain deep, dynamic, tubular membrane-bound invaginations of the nuclear envelope. J Cell Biol. 1997;136:531–544. doi: 10.1083/jcb.136.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broers JL, Machiels BM, van Eys GJ, Kuijpers HJ, Manders EM, van Driel R, et al. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J Cell Sci. 1999;112:3463–3475. doi: 10.1242/jcs.112.20.3463. [DOI] [PubMed] [Google Scholar]
- 27.Echevarria W, Leite MF, Guerra MT, Zipfel WR, Nathanson MH. Regulation of calcium signals in the nucleus by a nucleoplasmic reticulum. Nat Cell Biol. 2003;5:440–446. doi: 10.1038/ncb980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lui PP, Chan FL, Suen YK, Kwok TT, Kong SK. The nucleus of HeLa cells contains tubular structures for Ca2+ signaling with the involvement of mitochondria. Biochem Biophys Res Commun. 2003;308:826–833. doi: 10.1016/s0006-291x(03)01469-4. [DOI] [PubMed] [Google Scholar]
- 29.Lee RK, Lui PP, Ngan EK, Lui JC, Suen YK, Chan F, et al. The nuclear tubular invaginations are dynamic structures inside the nucleus of HeLa cells. Can J Physiol Pharmacol. 2006;84:477–486. doi: 10.1139/y05-110. [DOI] [PubMed] [Google Scholar]
- 30.Olins AL, Buendia B, Herrmann H, Lichter P, Olins DE. Retinoic acid induction of nuclear envelope-limited chromatin sheets in HL-60. Exp Cell Res. 1998;245:91–104. doi: 10.1006/excr.1998.4210. [DOI] [PubMed] [Google Scholar]
- 31.Melcer S, Gruenbaum Y, Krohne G. Invertebrate lamins. Exp Cell Res. 2007;313:2157–2166. doi: 10.1016/j.yexcr.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 32.Meier I. Composition of the plant nuclear envelope: theme and variations. J Exp Bot. 2007;58:27–34. doi: 10.1093/jxb/erl009. [DOI] [PubMed] [Google Scholar]
- 33.Graumann K, Evans DE. The plant nuclear envelope in focus. Biochem Soc Trans. 2010;38:307–311. doi: 10.1042/BST0380307. [DOI] [PubMed] [Google Scholar]
- 34.Meier I, Brkljacic J. Adding pieces to the puzzling plant nuclear envelope. Curr Opin Plant Biol. 2009;12:752–759. doi: 10.1016/j.pbi.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 35.Olins AL, Olins DE. Cytoskeletal influences on nuclear shape in granulocytic HL-60 cells. BMC Cell Biol. 2004;5:30. doi: 10.1186/1471-2121-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olins AL, Hoang TV, Zwerger M, Herrmann H, Zentgraf H, Noegel AA, et al. The LINC-less granulocyte nucleus. Eur J Cell Biol. 2009;88:203–214. doi: 10.1016/j.ejcb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Margalit A, Vlcek S, Gruenbaum Y, Foisner R. Breaking and making of the nuclear envelope. J Cell Biochem. 2005;95:454–465. doi: 10.1002/jcb.20433. [DOI] [PubMed] [Google Scholar]
- 38.Losman JA, Fasy TM, Novick KE, Massa M, Monestier M. Nucleosome-specific antibody from an autoimmune MRL/Mp-lpr/lpr mouse. Arthritis Rheum. 1993;36:552–560. doi: 10.1002/art.1780360417. [DOI] [PubMed] [Google Scholar]
- 39.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 40.Greaves IK, Rangasamy D, Ridgway P, Tremethick DJ. H2A.Z contributes to the unique 3D structure of the centromere. Proc Natl Acad Sci USA. 2007;104:525–530. doi: 10.1073/pnas.0607870104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dalal Y, Bui M. Down the rabbit hole of centromere assembly and dynamics. Curr Opin Cell Biol. 2010;22:11. doi: 10.1016/j.ceb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kunert N, Marhold J, Stanke J, Stach D, Lyko F. A Dnmt2-like protein mediates DNA methylation in Drosophila. Development. 2003;130:5083–5090. doi: 10.1242/dev.00716. [DOI] [PubMed] [Google Scholar]
- 43.Mellone BG. Structural and temporal regulation of centromeric chromatin. Biochem Cell Biol. 2009;87:255–264. doi: 10.1139/O08-121. [DOI] [PubMed] [Google Scholar]
- 44.Gieni RS, Chan GK, Hendzel MJ. Epigenetics regulate centromere formation and kinetochore function. J Cell Biochem. 2008;104:2027–2039. doi: 10.1002/jcb.21767. [DOI] [PubMed] [Google Scholar]
- 45.Silva MC, Jansen LE. At the right place at the right time: novel CENP-A binding proteins shed light on centromere assembly. Chromosoma. 2009;118:567–574. doi: 10.1007/s00412-009-0227-3. [DOI] [PubMed] [Google Scholar]
- 46.Przewloka MR, Glover DM. The kinetochore and the centromere: a working long distance relationship. Annu Rev Genet. 2009;43:439–465. doi: 10.1146/annurev-genet-102108-134310. [DOI] [PubMed] [Google Scholar]
- 47.Dalal Y. Epigenetic specification of centromeres. Biochem Cell Biol. 2009;87:273–282. doi: 10.1139/O08-135. [DOI] [PubMed] [Google Scholar]
- 48.Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- 49.Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Webster M, Witkin KL, Cohen-Fix O. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J Cell Sci. 2009;122:1477–1486. doi: 10.1242/jcs.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burke B, Roux KJ. Nuclei take a position: managing nuclear location. Dev Cell. 2009;17:587–597. doi: 10.1016/j.devcel.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 52.Mejat A, Misteli T. LINC complexes in health and disease. Nucleus. 2009;1:13. doi: 10.4161/nucl.1.1.10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zwerger M, Herrmann H, Gaines P, Olins AL, Olins DE. Granulocytic nuclear differentiation of lamin B receptor-deficient mouse EPRO cells. Exp Hematol. 2008;36:977–987. doi: 10.1016/j.exphem.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, et al. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 55.Hoffmann K, Sperling K, Olins AL, Olins DE. The granulocyte nucleus and lamin B receptor: avoiding the ovoid. Chromosoma. 2007;116:227–235. doi: 10.1007/s00412-007-0094-8. [DOI] [PubMed] [Google Scholar]
- 56.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 57.O'Keefe RT, Henderson SC, Spector DL. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferreira J, Paolella G, Ramos C, Lamond AI. Spatial organization of large-scale chromatin domains in the nucleus: a magnified view of single chromosome territories. J Cell Biol. 1997;139:1597–1610. doi: 10.1083/jcb.139.7.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takebayashi S, Sugimura K, Saito T, Sato C, Fukushima Y, Taguchi H, et al. Regulation of replication at the R/G chromosomal band boundary and pericentromeric heterochromatin of mammalian cells. Exp Cell Res. 2005;304:162–174. doi: 10.1016/j.yexcr.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 60.Hernandez-Verdun D, Gautier T. The chromosome periphery during mitosis. Bioessays. 1994;16:179–185. doi: 10.1002/bies.950160308. [DOI] [PubMed] [Google Scholar]
- 61.Van Hooser AA, Yuh P, Heald R. The perichromosomal layer. Chromosoma. 2005;114:377–388. doi: 10.1007/s00412-005-0021-9. [DOI] [PubMed] [Google Scholar]
- 62.Endl E, Gerdes J. The Ki-67 protein: fascinating forms and an unknown function. Exp Cell Res. 2000;257:231–237. doi: 10.1006/excr.2000.4888. [DOI] [PubMed] [Google Scholar]
- 63.Morcos M, Du X, Pfisterer F, Hutter H, Sayed AA, Thornalley P, et al. Glyoxalase-1 prevents mitochondrial protein modification and enhances lifespan in Caenorhabditis elegans. Aging Cell. 2008;7:260–269. doi: 10.1111/j.1474-9726.2008.00371.x. [DOI] [PubMed] [Google Scholar]
- 64.Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell. 2006;18:715–730. doi: 10.1105/tpc.105.037978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bubeck J, Scheuring D, Hummel E, Langhans M, Viotti C, Foresti O, et al. The syntaxins SYP31 and SYP81 control ER-Golgi trafficking in the plant secretory pathway. Traffic. 2008;9:1629–1652. doi: 10.1111/j.1600-0854.2008.00803.x. [DOI] [PubMed] [Google Scholar]
- 66.Friml J, Benkova E, Mayer U, Palme K, Muster G. Automated whole mount localisation techniques for plant seedlings. Plant J. 2003;34:115–124. doi: 10.1046/j.1365-313x.2003.01705.x. [DOI] [PubMed] [Google Scholar]
- 67.Ritzenthaler C, Nebenfuhr A, Movafeghi A, Stussi-Garaud C, Behnia L, Pimpl P, et al. Reevaluation of the effects of brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. Plant Cell. 2002;14:237–261. doi: 10.1105/tpc.010237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Olins AL, Herrmann H, Lichter P, Olins DE. Retinoic acid differentiation of HL-60 cells promotes cytoskeletal polarization. Exp Cell Res. 2000;254:130–142. doi: 10.1006/excr.1999.4727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.