Abstract
The genome is organized into large scale structures in the interphase nucleus. Pericentromeric heterochromatin represents one such compartment characterized by histones H3 and H4 tri-methylated at K9 and K20 respectively and with a correspondingly low level of histone acetylation. HP1 proteins are concentrated in pericentric heterochromatin and histone deacetylase inhibitors such as trichostatin A (TSA) promote hyperacetylation of heterochromatic nucleosomes and the dispersal of HP1 proteins. We observed that in mouse cells, which contain prominent heterochromatin, DNA topoisomerase IIβ (topoIIβ) is also concentrated in heterochromatic regions. Similarly, a detergent-resistant fraction of topoIIβ is associated with heterochromatin in human cell lines. Treatment with TSA displaced topoIIβ from the heterochromatin with similar kinetics to the displacement of HP1β. Topoisomerase II is the cellular target for a number of clinically important cytotoxic anti-cancer agents known collectively as topoisomerase poisons, and it has been previously reported that histone deacetylase inhibitors can sensitize cells to these drugs. While topoIIα appears to be the major target for most topoisomerase poisons, histone deacetylase-mediated potentiation of these drugs is dependent on topoIIβ. We find that while prior treatment with TSA did not increase the quantity of etoposide-mediated topoIIβ-DNA covalent complexes, it did result in a shift in their distribution from a largely heterochromatin-associated to a pannuclear pattern. We suggest that this redistribution of topoIIβ converts this isoform of topoII to a effective relevant target for topoisomerase poisons.
Key words: topoisomerase, histone deacetylase, histone acetyl transferase, chromatin remodelling, heterochromatin, euchromatin, etoposide, nucleolus, DNA damage
Introduction
Post-translational modifications of histone amino terminal tails are important for chromatin dynamics and organization. Methylation and acetylation of specific lysine residues, together with phosphorylation, polyADP-ribosylation and ubiquitylation modulate processes such as transcription, cell cycle progression, DNA damage responses, apoptosis and differentiation.1,2 Histone acetylation is associated with transcription. In contrast, constitutive heterochromatin, which is generally transcriptionally inert, contains low levels of histone acetylation and is characterized by trimethylated H3 lysine 9 and H4 lysine 20.3–5
Histone acetylation is governed by the opposing actions of histone acetyl transferases (HATs) and histone deacetylases (HDACs). Alterations in the structure or expression of HATs and HDACs are frequently reported in cancers and are linked to tumour development. Consequently, HDAC inhibitors (HDACIs) are undergoing trials as anticancer agents.6–9 HDACIs alone exert an antiproliferative effect and promote apoptosis in cancer cell-line models, but in addition HDACIs have been shown to sensitize cells to the cytotoxic effects of other anticancer drugs, particularly topoisomerase II poisons.10–16 Topoisomerase poisons such as etoposide and epirubicin are of great clinical importance and are widely used in cancer therapy.17,18
Topoisomerase II is an essential enzyme that allows the passage of one DNA duplex through a second. During its reaction cycle the dimeric enzyme introduces an enzyme-bridged DNA double-strand break (DSB) in one double helix through which a second duplex can be passed. At this stage each topoisomerase monomer is coupled to one free end of the first duplex through a 5′-phosphotyrosyl linkage. Topoisomerase poisons stabilize the enzyme-linked DSB, which is otherwise transient and rapidly re-ligated.19–21 The resultant topoisomerase II-DNA covalent complexes account for the cytotoxic properties of the drugs and are presumed to be converted to “frank” DSBs by repair processes and/or result in a damage response following collisions with transcriptional or replication machinery. Mammals possess two topoisomerase II isoforms, α and β, encoded by the TOP2A and TOP2B genes respectively.22,23 Although the isoforms have very similar enzymatic characteristics in vitro and share 70% amino acid identity, topoisomerase IIα and -β have different though possibly overlapping roles in mammalian cells. Topoisomerase IIα is essential for cell viability and is required for chromosome decatenation after S-phase,20 while top2β null mice develop to term but die perinatally with neural developmental defects.24 Recently, specific roles in transcription regulation and sperm chromatin remodelling have been uncovered for topoisomerase IIβ.25–27 The topoisomerase IIα isoform is the major target for most clinically-relevant topoisomerase poisons including etoposide and epirubicin12,28,29 although the -β isoform appears to contribute more for some topoisomerase poisons including mitoxantrone, mAMSA and XK469.28–30
Several mechanisms could explain the sensitization of cells to topoisomerase poisons by HDAC inhibitors13,15,31 but notably, it was recently reported that this potentiation is mediated specifically through topoisomerase IIβ.12 This was shown for etoposide and epirubicin as well as mitoxantrone. HDACIs cause chromatin decondensation, which may affect access of topoisomerase to DNA or access of topoisomerase poisons to the enzyme,14,32,33 resulting in elevated DNA damage and cytotoxicity. HDACI treatment leads to global changes in histone acetylation levels, including hyperacetylation of heterochromatin and redistribution of the non-histone heterochromatin protein HP1.34–37 It follows that this may affect the distribution of topoisomerase II in the interphase nucleus. Indeed, enzymatically active topoisomerase IIα is concentrated in heterochromatin in mid-late S-phase in HeLa cells and is delocalized by the HDACI TSA.38 We have studied the nuclear distribution of topoisomerase IIβ and show here that TSA leads to its redistribution in the nucleus and a change in the pattern of topoisomerase IIβ-DNA complexes induced by etoposide in favour of a more euchromatic pattern. We suggest that altering the distribution of topoisomerase IIβ in the nucleus and thus topoisomerase IIβ-DNA adducts across the genome converts topoisomerase IIβ into a more clinically useful target for topoisomerase poisons including etoposide.
Results
A fraction of topoisomerase IIβ is concentrated in heterochromatin.
Exposure of cultured cells to HDAC inhibitors results in cell cycle arrest and apoptosis. Prolonged exposure to low doses leads to chromatin decondensation associated with hyperacetylation of histones H3 and H4. In mouse cells under these conditions, pericentromeric heterochromatin relocates from a largely perinucleolar to a more peripheral distribution and in mouse and human cells, HP1 proteins become dissociated from heterochromatic regions.34,36 In the light of this HDAC-mediated chromatin remodelling, and the role of topoisomerase IIβ in sensitization of cells to topoisomerase poisons by HDAC inhibitors, we hypothesized that HDAC inhibitors such as TSA or VPA may alter the subnuclear distribution of topoisomerase IIβ. This, either directly or through increased accessibility of decondensed chromatin could lead to increased cytotoxicity associated with topoisomerase poisons. Previous studies have reported differing subnuclear distributions for topoisomerase IIβ. Topoisomerase IIβ has variously been reported to be concentrated in the nucleoli or at the periphery of heterochromatic regions in fixed cells,43–45 and in live cells,45 while others have not found the protein to be specifically nucleolar.39,46 The reasons for these differences are not clear, but probably reflect, at least in part, species differences and differences in specimen preparation. Nucleoli of mouse cells for example are associated at their periphery with AT-rich pericentromeric heterochromatin, which clusters in so-called chromocentres around centromeres. These chromocentres can be readily visualized in fluorescence microscopy of Hoechst or DAPI stained cells or with antibodies against HP1 proteins (Fig. 1A and Sup. Fig. 1). Heterochromatin in human cells by contrast, is less easily visualized. We show here that in mouse epithelial cells, under standard paraformaldehyde fixation conditions and using a series of characterized antibodies, that topoisomerase IIβ has a nucleoplasmic distribution, with an increased concentration in chromocentres as identified by DAPI and anti-HP1β staining (Fig. 1A). Although not excluded from nucleoli, topoisomerase IIβ did not appear to be concentrated in these regions (see Figs. 1A and 2). By contrast, topoisomerase IIα was distributed more evenly throughout interphase nuclei (see Fig. 1B). Since chromocenters are largely clustered around nucleoli in mouse cells it was important to unequivocally distinguish between nucleolar and heterochromatic topoisomerase IIβ staining. This was achieved by co-staining cells with anti-topoisomerase IIβ and either anti-HP1β to identify heterochromatin or anti-fibrillarin to identify nucleoli. As shown in Figure 2A, regions of intense topoisomerase IIβ staining were largely non-overlapping with the fibrillarin signal, but almost coincident with the HP1 and DAPI signals, which largely clustered around the outside of nucleoli. Consistently, centromers, as visualized by CENP-B staining, were contained within the regions of intense topoisomerase IIβ (and HP1β and DAPI) staining (Fig. 2B).
Figure 1.
Topoisomerase IIβ staining co-localizes with DAPI-bright and HP1 staining regions. (A) Mouse C127I cells were fixed with paraformaldehyde and permeabilized (see Materials and Methods) and processed for immunofluorescence with the anti-topoisomerase IIβ rabbit antisera 3535β, 18513β, 30400β or anti-topoisomerase IIβ MAB H8, and rat MAB MAC353 and were counterstained with DAPI. (B) C127I cells were similarly processed using anti-topoisomerase IIα rabbit antiserum 18511. Z-stacks of fluorescence images were captured. Deconvolved equatorial sections are shown. Scale bar in main image, 10 mm; scale bar in insert 4 µm.
Figure 2.
Topoisomerase IIβ is concentrated in chromocentres often surrounding nucleoli in mouse epithelial cells. (A) Cells were fixed and processed for immunofluorescence with anti-topoisomerase IIβ (green) and anti-fibrillarin or MAC353 (red) and were counterstained with DAPI. Z-stacks of fluorescence images were captured. Deconvolved equatorial sections are shown for the non-merged images. Equatorial xy planes together with xz and yz sections are shown for the merged images. Scale bar 10 µm. (B) Cells were fixed and stained for topoisomerase IIβ or HP1β (green) and centromeres (CENP-B, red).
TSA mobilizes topoisomerase IIβ from heterochromatin.
As alluded to above, prolonged exposure of mouse cells to the HDAC inhibitor TSA results in histone hyperacetylation and movement of heterochromatin clusters from a largely perinucleolar to a more peripheral distribution and dissociation of heterochromatin protein HP1. Since like HP1, topoisomerase IIβ is concentrated in heterochromatin in C127I cells, we hypothesized that TSA treatment would also lead to its redistribution. C127I cells were treated with TSA concentrations from 2 to 80 nM for 2 or 5 days and then examined for topoisomerase IIβ and HP1β distribution by immunofluorescence. After 5 day's exposure to TSA, topoisomerase IIβ and HP1β both showed a progressive dissociation from heterochromatic regions (as visualized by DAPI staining) with dose of TSA (Fig. 3). After 5 day's exposure to 50 nM TSA, little or no heterochromatic concentration of either protein was apparent. After 2 days exposure to 50 nM TSA most topoisomerase IIβ and HP1β was displaced from heterochromatin. After 5 day's exposure to 50 or 80 nM TSA, most cells appeared larger and flatter then untreated cells. Median topoisomerase IIβ immunofluorescence per cell was increased almost two fold by TSA (Fig. 4A). This may reflect increased antigen accessibility, but notably median DNA content per nucleus was also increased, as assessed by quantitative DAPI fluorescence (Fig. 4C), consistent with accumulation of cells in G2 or with aberrant chromosome numbers resulting from TSA-induced mitotic defects and in addition, the nuclei of TSA treated cells are larger than those of untreated cells (see Fig. 3), Topoisomerase IIβ protein levels were not changed by TSA in western blot analysis where protein loading was standardized by micrograms of protein loaded (Fig. 4D). By comparison, it was previously noted that HDACIs did not significantly alter topoisomerase IIβ protein level in human MCF7 cell extracts.12 The level of HP1β immunofluorescence remained unchanged by TSA (Fig. 4B), consistent with previous observations.34 When topoisomerase IIβ dissociated from heterochromatin it appeared in a fine speckled pattern throughout the nucleus and was occasionally excluded from heterochromatic regions (Fig. 3 fourth and fifth rows).
Figure 3.
TSA-mediated disassociation of topoisomerase IIβ and HP1β from heterochromatin. C127I cells were cultured in media containing the indicated concentrations of TSA for two or five days before fixing and processing for immunofluorescence with the antibodies shown. Stacks of images were collected and deconvolved images were merged into maximum intensity projections.
Figure 4.
Quantification of topoisomerase IIβ, HP1β and DNA content in C127I cells exposed for 5 days to 50 nM TSA. (A–C) Quantitative immunofluorescence images were collected at 10x. Horizontal bars = median values. (D) Western blot analysis of topoisomerase IIβ and HP1β. 5, 2.5 or 1.25 µg of protein from whole cells extracts were loaded as shown. Con, no treatment; TSA, 50 nM TSA 5 days.
Effect of HDAC inhibition on etoposide-induced topoisomerase-DNA complexes.
The topoisomerase II reaction mechanism allows the passage of one DNA duplex through another by transiently cleaving the first DNA helix to create an enzyme-bridge DNA gate through which the second duplex is transported. The break is subsequently ligated and the DNA structure restored.19–21 During the cleavage reaction, a covalent enzyme-DNA intermediate is formed between a tyrosine residue of each topoisomerase II monomer and the 5′-phosphate group of the cleaved DNA. This enzyme-bridged double-strand break normally exists transiently, but the action of topoisomerase poisons such as etoposide is to stabilize these intermediates. Both topoisomerase IIα and β are targets for etoposide, and etoposide-induced topoisomerase II-DNA complexes can be visualized and quantified using a fluorescence microscopy based method (Trapped in AgaRose DNA ImmunoStaning or TARDIS).42 This method involves ionic detergent and salt extraction of agarose-embedded cells, resulting in removal of proteins not covalently coupled to the nuclear DNA, which is too bulky to elute from the agarose. Covalently coupled proteins are then detected by immunofluorescence. In a variation of this approach adherent cells are extracted in a similar fashion on glass coverslips under slightly less harsh conditions (DRT extraction) without agarose embedding.38 We used this latter technique to examine the effect of TSA treatment on both the quantity and subnuclear distribution of topoisomerase IIβ-DNA complexes. While very low levels of nuclear topoisomerase IIβ fluorescence were observed in DRT-extracted control cells, in cells treated with etoposide topoisomerase IIβ immunofluorescence was easily detectable (Figs. 5 and 6). The median immunofluorescent signal in DRT-extracted, etoposide-treated cells corresponded to approximately 20% of the total topoisomerase IIβ complement measured in normally fixed (i.e., non-extracted) cells (Sup. Fig. 2). From knowledge of the action of etoposide, and analogous experiments employing the more harsh extraction conditions of the TARDIS protocol,42,47,48 we conclude that the etoposide-induced topoisomerase IIβ signal in DRT-extracted cells corresponds to covalent topoisomerase-DNA complexes. Notably, prior exposure to TSA (5 days 50 nM) caused a small decrease in the median levels of etoposide-induced, extraction resistant topoisomerase IIβ immunofluorescence when antibodies 18513 or 30400 were used, but did not significantly affect the signal with antibody 3535 (Fig. 5).
Figure 5.
Effect of TSA pre-treatment on the quantity of etoposide induced topoisomerase IIβ-DNA complexes. Cells grown on glass coverslips were either cultured in control medium or in medium containing 50 nM TSA for 5 days after which etoposide (50 µM) was added to the medium. Two hours later cells were extracted in DRT buffer (see Materials and Methods). Quantitative immunofluorescence was carried out using the anti-topoisomerase IIβ antibodies indicated.
Figure 6.
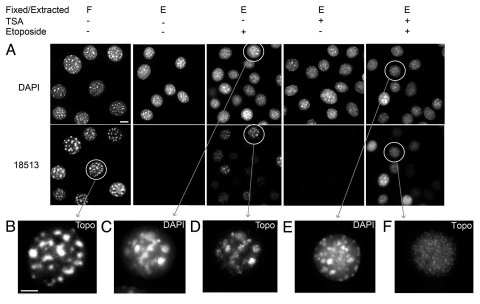
TSA alters the distribution of etoposide-induced topoisomerase IIβ-DNA complexes in the nucleus. (A) Cells were cultured either in control medium or in medium containing 50 nM TSA for 5 days, after which etoposide (50 µM) was added to the medium. Cells were extracted in situ 2 hours later in DRT buffer before fixing (shown as “E”) or were immediately fixed with paraformaldehyde and permeabilized (shown as “F”, see Materials and Methods). Immunofluorescence was carried out using the anti-topoisomerase IIβ antibody 18513. Image stacks were collected. Deconvolved equatorial sections are shown. Circled nuclei are enlarged and shown below. Bar = 10 µm. (B) Enlarged nucleus indicated by the circled nucleus in the first column of A, 18513-stained. Bar = 5 µm. (C and D) Enlarged image of nucleus shown in the third column of A, stained with DAPI and 18513 (TopoIIβ) respectively. (E and F) Enlarged image of nucleus shown in the fifth column of A, stained with DAPI and 18513 (TopoIIβ) respectively.
In cells treated with etoposide for 2 hours before DRT extraction, topoisomerase IIβ as detected by antibody 18513 had a clustered appearance, with bright areas coincident with the bright DAPI regions (Fig. 6A). This heterochromatic pattern was apparent in more than 70% of cells, while the remainder displayed a more diffuse, speckled pattern. The variation in pattern between cells may have been due to the short drug exposure in an asynchronous population of cells. In contrast, little if any heterochromatic concentration of extraction-resistant topoisomerase IIβ was apparent in cells pre-treated with TSA before exposure to etoposide (Fig. 6A and compare D and F). Results were similar with antibody 30400 and 3535. Thus, consistent with the TSA-induced redistribution of total topoisomerase IIβ (see Fig. 2), the nuclear distribution of etoposide-induced topoisomerase IIβ-DNA complexes is altered by TSA, to a more euchromatic pattern.
Notably, TSA treatment alone did not significantly affect the quantity of DRT extraction-resistant topoisomerase IIβ (Fig. S2), nor did it affect the magnitude of etoposide induced histone H2AX phosphorylation, a marker for DNA double-strand breaks (not shown). However, we did find that it had an unexpected effect on the background pattern of γH2AX. In control cells, γH2AX is present in a faint speckled pattern in the nucleus, typically with one or a small number of bright foci, presumably originating from spontaneous DNA double-strand breaks (DSBs). However, in cells exposed to TSA for 5 days, γH2AX was concentrated almost exclusively adjacent to large chromocentres (see Fig. 7). These TSA-induced γH2AX foci were not as intense as those induced by etoposide, which also form predominantly outside heterochromatin,49 but contributed to an overall increase in background γH2AX (Fig. 7A and B). Notably, it has recently been demonstrated that HDAC inhibition slows replication and leads to replication-associated DNA damage and H2AX phosphorylation,50 and we suspect that this is likely to be the cause of the TSA-induced peri-heterochromatic gH2AX seen here. Thus increased background H2AX phosphorylation and the redistribution of topoisomerase IIβ are likely to be independent effects of the TSA-mediated remodelling of heterochromatin.
Figure 7.
TSA alters the background pattern of histone H2AX phosphorylation. (A) TSA-treated or control cells were plated onto coverslips and subsequently exposed to medium containing 0 or 1 µM etoposide. Cells were fixed and processed for immunofluorescence using anti-γH2AX and anti topoisomerase IIβ (18513). Images were collected with a 40x objective, maximum intensity projections derived from deconvolved images are shown. In the first two columns, greyscale image levels were adjusted such that background (signal present in the absence of a DSB-inducing agent) γH2AX was clearly visible. In the third column levels were adjusted as for the counting of induced γH2AX foci, such that etoposide-induced γH2AX foci are clearly visible but with minimal contribution from the background. (B) Quantitative immunofluorescent analysis of background γH2AX intensity in control cells and cells treated with 50 nM TSA for 5 days.
The effect of HDACIs on the distribution of topoisomerase IIβ in human cells.
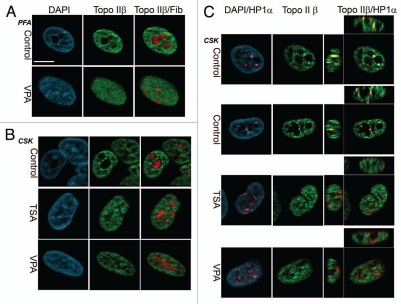
Since HDACIs have been shown to potentiate topoisomerase poisons in human cancer cell lines, we also examined the effect of TSA and valproic acid (VPA) on topoisomerase II distribution in the human lung cancer cell line A549. Topoisomerase IIβ was distributed in a nucleoplasmic pattern, with some concentration in the perinucleolar regions of most cells (Fig. 8A). The perinucleolar pattern was more pronounced when cells were permeabilized prior to fixation (Fig. 8B). Nucleoli of human cells are typically surrounded by clusters of heterochromatin identified by late replication timing, histone methylation and the presence of HP1 proteins.4,49 In human cells, HP1α gives the most robust pericentromeric staining pattern of the three HP1 species.36,51 When we compared the distribution of HP1α to that of topoisomerase IIβ, HP1 substantially coincided with the areas of brightest topoisomerase IIβ signal (Fig. 8C). When cells were treated with TSA or VPA, topoisomerase IIβ adopted a more pan-nuclear distribution, as was observed following HDACI treatment of mouse cells (Fig. 3). HP1α still presented a focal distribution under these conditions, as observed previously,36 but these foci were no longer associated with bright topoisomerase IIβ staining (Fig. 8C). Thus, as in mouse C127I cells, topoisomerase IIβ is partially concentrated in heterochromatic regions of the nucleus of A549 cells and is redistributed into a more uniform pattern upon HDACI treatment. We also investigated the distribution of topoisomerase IIβ in two human lymphoblastoid cell lines K562 and Nalm-6. Topoisomerase IIβ appeared nucleoplasmic in distribution in these cells, but after mild detergent extraction (CSK buffer) the remaining topoisomerase IIβ was concentrated in the DAPI-bright heterochromatic regions surrounding nucleoli or at the nucleolar periphery (Sup. Fig. 3), similar to the distribution in A549 cells.
Figure 8.
Topoisomerase IIβ distribution and HDACI-induced redistribution in A549 lung cancer cells. (A) A549 cells were fixed with paraformaldehyde, permeabilized and immunostained with rabbit anti-topoisomerase IIβ (green) and anti-fibrillarin MAB (red). In the lower row, cells were exposed to 2 mM VPA for 24 hours prior to fixation. (B) Control cells or cells treated with 2 mM VPA or 50 nM TSA for 48 hours were permeabilized prior to fixation as described in Materials and Methods (CSK extraction). Antibodies are as in (A). (C) Control, TSA treated or VPA treated cells were CSK extracted as in (B) and then fixed and immunostained with anti-rabbit topoisomerase II (18513β) and mouse anti HP1α. In all cases equatorial confocal planes are shown. In the right-hand column of part (C) x-z and y-z confocal sections are also displayed for the topoisomerase II β/HP1α merged images. Scale bar = 10 µm.
Topoisomerase IIβ mediates the potentiating effect of HDACI.
We investigated the ability of the HDACI valproic acid to potentiate the growth inhibitory/cytotoxic effects of topoisomerase poisons in A549 cells using a series of topoisomerase poisons with different selectivities for topoisomerase IIα and -β, see Table 1 and Sup. Table 1. The most robust potentiation was observed with mitoxantrone and mAMSA, while marginal potentiation was obtained for etoposide and no significant effect was seen for doxorubicin or epirubicin. Cells lacking topoisomerase IIβ have previously been shown to be resistant to mitoxantrone and mAMSA,28,29 indicating a significant role for this isozyme in the cytotoxicity of these drugs, whereas the degree of resistance to etoposide or doxorubicin in topoisomerase IIβ null cells was smaller. Similarly, we found the IC50 for mitoxantrone in growth inhibition assays is five times higher in topoisomerase IIβ null Nalm-6 cells than in their wild type counterparts while the ratio is only 2.8 for etoposide (data not shown). Thus, these data support previous work12 identifying a functional interaction between topoisomerase IIβ and HDAC inhibitors such as VPA in the synergistic action of histone deacetylase inhibitors with topoisomerase poisons.
Table 1.
Potentiation data obtained in A549 lung cancer cells for mitoxantrone, mAMSA, etoposide, doxorubicin and epirubicin, and the effect of the HDAC inhibitor VPA
| TopoII poison | Topoisomerase II preference | VPA pre-treatment (1.6 mM) | Potentiation |
| Mitoxantrone | α and β | + | YES |
| − | YES1.6,8 | ||
| mAMSA | α and β | + | YES |
| − | YES8 | ||
| Etoposide | α > β | + | YES |
| − | YES8 | ||
| Doxorubicin | α > β | + | NO |
| − | NO | ||
| Epirubicin | N/D | + | NO |
| − | NO |
Cells were either plated directly or pre-incubated with 1.6 mM VPA before plating into 96 well plates in media containing a range of topoisomerase poisons and either 0, 1.6 or 8 mM VPA. Growth inhibition assays were performed and IC50 values were determined for each topoisomerase poison/VPA combination. Experiments were carried out at least in triplicate and potentiation values (PF50) were calculated (see Table S1). Where potentiation is scored as “YES”, IC50 values obtained with VPA were significantly lower than that obtained with topoisomerase poison alone (p < 0.05 one tailed paired t-test). Topoisomerase II preference pertains to evidence for the relative involvement of topoisomerase IIα or –β in mediating the cytotoxic effects in cell line systems (Errington, et al. 1999; Toyoda, et al. 2008).
What causes topoisomerase IIβ to be retained in heterochromatin?
The very similar subnuclear localization of topoisomerase IIβ and HP1β and their parallel dispersal from heterochromatin following TSA treatment suggests that the two proteins might interact in a protein-protein complex. Another piece of evidence that suggests this is the very similar phenotype of mice null for TOP2b and CBX1, the genes encoding topoisomerase IIβ and HP1β respectively. Both TOP2b and CBX1 null mice develop apparently normally in utero, but die perinatally from respiratory failure due to failure to innervate the diaphragm.24,52 We carried out a co-immunoprecipitation experiment to determine whether HP1β and topoisomerase IIβ exist in a stable protein complex. While antibodies 18513 and MAC353 efficiently precipitated their cognate antigens (topoisomerase IIβ and HP1β respectively), 18513 immunoprecipitates did not contain detectable HP1β and MAC353 did not specifically precipitate topoisomerase IIβ (data not shown). Thus, topoisomerase IIβ and HP1β do not exist in a protein complex, at least not one that is stable under the conditions in which the cell extracts were prepared. So the heterochromatic accumulation of topoisomerase IIβ can probably not be explained by association with HP1β. Other candidates include interaction with another heterochromatin protein such as HP1α or KAP1 or with a modified histone such as H3K9me3 (involved in targeting HP1β)3 or H4K20me3. Whether any of these features are involved remains to be elucidated. Notably, topoisomerase II has previously been shown by photo bleaching studies to be quite mobile within the nucleus,45 thus interactions resulting in the observed heterochromatic concentration of topoisomerase IIβ are presumable not of high affinity.
Discussion
Several mechanisms have been proposed to account for the sensitization of cells to topoisomerase poisons by HDACIs. These include modulation of apoptotic pathways15 and the general decondensation of chromatin following histone hyperacetylation, leading to increased DNA damage. For example, SAHA and VPA cause chromatin decondensation in MCF7 cells as viewed by EM and DNase sensitivity.11,12 This decondensation may allow greater access or accumulation of topoisomerase poisons in chromatin. Similarly, prolonged VPA treatment (48 hr) has been shown to result in reduction in HP1, DNMT1, SMC1 and SMC3 protein levels in MCF7 cells, though this was not observed in mouse cells for any of the HP1 isotypes.34 Our findings add to this chromatin-mediated mechanism by demonstrating a redistribution of topoisomerase IIβ within the nucleus following HDAC inhibition analogous to that previously observed for HP1β. Specifically we find that in mouse C127I cells, which possess easily identifiable heterochromatic foci, topoisomerase IIβ is concentrated in heterochromatin and redistributes to a speckled pan-nuclear pattern upon TSA-treatment (Figs. 1–3 and S1). Previous photo-bleaching studies have shown that topoisomerase IIβ is relatively mobile in the nucleus,45 thus association with heterochromatic or other structures is likely to be transient with exchange between compartments the equilibrium of which is altered by hyperacetylation of heterochromatin during HDACI treatment.
Human cells do not contain the prominent chromocentres that are present in mouse cells, but topoisomerase IIβ was concentrated in the perinucleolar region and in foci co-localizing with HP1α in A549 human lung carcinoma cells. Notably though, topoisomerase IIβ was not found in the barr-body in female human cells (E. Heard, personal communication), suggesting that its observed accumulation is limited to constitutive heterochromatin. As was observed in mouse cells, topoisomerase IIβ was redistributed following TSA or VPA treatment (Fig. 8). TSA also altered the distribution of etoposide-induced topoisomerase IIβ-DNA complexes. In non-TSA treated C127I cells, adducts were concentrated in heterochromatic regions, reflecting the distribution of the enzyme in these cells, while after TSA treatment, adducts were distributed more evenly throughout the nucleus.
We have not addressed the HDAC specificity of the observed topoisomerase II redistribution, but we note that HDAC2 was found to be the predominant target in MCF7 cells for HDACIs with regards to chromatin decondensation, downregulation of HP1 and other chromatin proteins and potentiation of epirubicin cytotoxicity.33 We have employed TSA and VPA in the work reported here. Both inhibit HDAC2, although VPA but not TSA selectively induces proteasomal degradation of HDAC2.53 In addition, topoisomerase IIα and IIβ physically interact with HDAC1 and HDAC2 and topoisomerase IIβ is associated with HDAC1 in the NuRD complex.16,54 Previous descriptions of the intranuclear distribution of topoisomerase IIβ have varied in the literature.39,43–46 This may reflect the dynamic nature of topoisomerase IIβ. Topoisomerase IIβ is recruited to the promoters of certain genes during transcriptional activation25,26 and it is possible that the HDACI-induced redistribution of topoisomerase IIβ reported here reflects the normal dynamic behaviour of the protein during the execution of transcriptional programs, though this possibility remains to be explored. We have consistently observed topoisomerase IIβ to be concentrated in chromocentres in untreated fixed mouse cells (Figs. 1–3 and S1). Notably, this distribution is mirrored in the pattern of topoisomerase IIβ-DNA adducts formed in living cells by etoposide (Fig. 6), arguing against the observed pattern being an artifact of fixation.
The cytotoxic potential of topoisomerase poisons such as etoposide is mediated predominantly through topoisomerase IIα.12,28,29 HDACIs sensitize cells to topoisomerase poisons including etoposide, but the sensitization is reported to be dependent upon topoisomerase IIβ.12 This apparent contradiction can be resolved if HDACIs somehow recruit topoisomerase IIβ as a target for topoisomerase poisons. A possible mechanism for this recruitment is provided by the chromatin decondensation, release of topoisomerase IIβ from heterochromatic sites and a shift in the distribution of resulting topoisomerase IIβ adducts towards the euchromatic portion of the nucleus that is induced by HDACIs.
Materials and Methods
Cell culture and reagents.
Mouse C127I cells and A549 human lung carcinoma cells were acquired from European Collection of Cell Cultures (ECACC). Both cell lines were cultured in DMEM containing 10% FCS and antibiotics. Trichostatin A (Sigma) was dissolved in water and was added to cell cultures at between 2 nM and 80 nM. VPA (Sigma) was added to cultures at the concentrations indicated in the text. Rabbit polyclonal antibodies 18511, 18513,39 3535 and 30400 were generated in house and their specificity was confirmed by western blotting with whole cell extracts and purified recombinant topoisomerase IIα and β. Anti topoisomerase IIβ mab H-8 was from Santa Cruz, mouse MAB anti-fibrillarin 18380 and anti-HP1β rat MAB MAC353 were from Abcam, mouse anti-γH2AX MAB was from Millipore.
Growth inhibition assays.
Control (untreated) cells or cells incubated in medium containing VPA at 1.6 or 8 mM (10% and 50% of IC50 respectively) for 72 hours were trypsinized and plated into 96 well plates containing combinations of VPA (0, 1.6 or 8 mM) and either vehicle or a dose range of topoisomerase II poison. Plates were incubated for 72 hours under normal conditions, after which relative cell growth was determined by XTT assay (Roche) according to the manufacturer's instructions. Optimum cell plating density was determined from growth curves. Data was normalized to the growth obtained with VPA alone. Assays were performed at least in triplicate and IC50 and PF50 values were calculated. Potentiation factors (PF50) are the ratio of the concentration of topoisomerase poison required to achieve a 50% growth inhibition (IC50) in the absence of VPA to that in the presence of VPA.
Immunofluorescence analysis.
Cells were grown on glass cover slips. For standard paraformaldehyde fixation, coverslips were briefly washed in PBS, immersed in 4% paraformaldehyde in PBS for 10 minutes at room temperature and then permeabilized by incubation in KCM+T [120 mM KCl, 20 mM NaCl, 10 mM TRIS-HCl, pH 7.5, 0.5 mM EDTA, 0.1% (v/v) Triton X-100] for 5 minutes. Coverslips were then blocked for 1 hour at room temperature with 10% dried milk powder, 5% BSA in KCM+T. First and second antibody incubations were in blocking buffer for 1 hour each at room temperature. Washes were carried out using KCM+T. Second antibodies were Alexa Fluor-488 or Alexa Fluor-594 conjugated (Invitrogen). After second antibody incubation and washing, coverslips were mounted on slides with Vectashield containing DAPI and were sealed with nail varnish. For CSK extraction, coverslips were incubated on ice for 5 minutes in CSK buffer [10 mM Pipes (pH 7.0), 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA 0.5% Triton-X100] containing protease inhibitors, before paraformaldehyde fixation and immunostaining. For DRT extraction, coverslips were extracted once on ice in [30 mM HEPES, 65 mM Pipes, 10 mM EGTA, 2 mM MgCl2, pH 6.9 with 350 mM NaCl, 0.5% Triton X-100] containing protease inhibitors for two minutes with occasional gentle agitation. Cells were then fixed for 10 minutes in 4% paraformaldehyde in PBS before immunostaining as above. Images were obtained using an Olympus IX81 motorized microscope fitted with a Hamamatsu Orca-AG monochrome camera. Z-stacks were collected using a 40x objective (PlanS Apo, NA 0.95) at 0.2 µm intervals or using a 60x objective and OptiGrid confocal (Qioptiq) at 0.3 µm intervals. Iterative deconvolution was performed using Volocity software (Improvision).
Western blotting.
Cells were grown in 9 cm tissue culture flasks. Whole cell extracts were prepared, separated by SDS PAGE and blotted onto nitrocellulose membranes as described previously in reference 40.
Quantitative immunofluorescence.
Cells were grown on glass coverslips and immunostained as above. Topoisomerase II was detected using the primary antibodies indicated and Alexa Fluor 594-conjugated second antibodies (Invitrogen). Microscopy was performed using an Olympus IX81 motorized microscope with a 10x objective (UPlanFLN, NA 0.3) and employing a 120 W X-Cite illumination system. Greyscale images were collected with a Hamamatsu Orca-AG camera with 2 × 2 binning. Images were background and shade corrected as described previously in reference 41 and 42. Fluorescence intensity (integrated fluorescence) per nucleus was determined for DAPI and Alexa Fluor 488 channels using Volocity Quantitation software (Improvision). Data was plotted using Graphpad Prism.
Acknowledgements
This work was supported by a grant (05-179) from the Association for International Cancer Research.
Supplementary Material
References
- 1.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Corpet A, Almouzni G. A histone code for the DNA damage response in mammalian cells? EMBO J. 2009;28:1828–1830. doi: 10.1038/emboj.2009.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cowell IG, Aucott R, Mahadevaiah SK, Burgoyne PS, Huskisson N, Bongiorni S, et al. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma. 2002;111:22–36. doi: 10.1007/s00412-002-0182-8. [DOI] [PubMed] [Google Scholar]
- 4.Wu R, Terry AV, Singh PB, Gilbert DM. Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Mol Biol Cell. 2005;16:2872–2881. doi: 10.1091/mbc.E04-11-0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kourmouli N, Jeppesen P, Mahadevhaiah S, Burgoyne P, Wu R, Gilbert DM, et al. Heterochromatin and trimethylated lysine 20 of histone H4 in animals. J Cell Sci. 2004;117:2491–2501. doi: 10.1242/jcs.01238. [DOI] [PubMed] [Google Scholar]
- 6.Duenas-Gonzalez A, Candelaria M, Perez-Plascencia C, Perez-Cardenas E, de la Cruz-Hernandez E, Herrera LA. Valproic acid as epigenetic cancer drug: preclinical, clinical and transcriptional effects on solid tumors. Cancer Treat Rev. 2008;34:206–222. doi: 10.1016/j.ctrv.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Molecular Oncology. 2007;1:19–25. doi: 10.1016/j.molonc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganesan A, Nolan L, Crabb SJ, Packham G. Epigenetic therapy: Histone acetylation, DNA methylation and anti-cancer drug discovery. Curr Cancer Drug Targets. 2009;9:963–981. doi: 10.2174/156800909790192428. [DOI] [PubMed] [Google Scholar]
- 9.Carey N, La Thangue NB. Histone deacetylase inhibitors: Gathering pace. Curr Opin Pharmacol. 2006;6:369–375. doi: 10.1016/j.coph.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 11.Marchion DC, Bicaku E, Daud AI, Richon V, Sullivan DM, Munster PN. Sequence-specific potentiation of topoisomerase II inhibitors by the histone deacetylase inhibitor suberoylanilide hydroxamic acid. J Cell Biochem. 2004;92:223–237. doi: 10.1002/jcb.20045. [DOI] [PubMed] [Google Scholar]
- 12.Marchion DC, Bicaku E, Turner JG, Daud AI, Sullivan DM, Munster PN. Synergistic interaction between histone deacetylase and topoisomerase II inhibitors is mediated through topoisomerase IIβ. Clin Cancer Res. 2005;11:8467–8475. doi: 10.1158/1078-0432.CCR-05-1073. [DOI] [PubMed] [Google Scholar]
- 13.Kurz EU, Wilson SE, Leader KB, Sampey BP, Allan WP, Yalowich JC, et al. The histone deacetylase inhibitor sodium butyrate induces DNA topoisomerase II alpha expression and confers hypersensitivity to etoposide in human leukemic cell lines. Mol Cancer Ther. 2001;1:121–131. [PubMed] [Google Scholar]
- 14.Hajji N, Wallenborg K, Vlachos P, Fullgrabe J, Hermanson O, Joseph B. Opposing effects of hMOF and SIRT1 on H4K16 acetylation and the sensitivity to the topoisomerase II inhibitor etoposide. Oncogene. 2010;29:2192–2204. doi: 10.1038/onc.2009.505. [DOI] [PubMed] [Google Scholar]
- 15.Hajji N, Wallenborg K, Vlachos P, Nyman U, Hermanson O, Joseph B. Combinatorial action of the HDAC inhibitor trichostatin A and etoposide induces caspase-mediated AIF-dependent apoptotic cell death in non-small cell lung carcinoma cells. Oncogene. 2008;27:3134–3144. doi: 10.1038/sj.onc.1210976. [DOI] [PubMed] [Google Scholar]
- 16.Tsai SC, Valkov N, Yang WM, Gump J, Sullivan D, Seto E. Histone deacetylase interacts directly with DNA topoisomerase II. Nat Genet. 2000;26:349–353. doi: 10.1038/81671. [DOI] [PubMed] [Google Scholar]
- 17.Chen AY, Liu LF. DNA topoisomerases: essential enzymes and lethal targets. Annu Rev Pharmacol Toxicol. 1994;34:191–218. doi: 10.1146/annurev.pa.34.040194.001203. [DOI] [PubMed] [Google Scholar]
- 18.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu LF, Liu CC, Alberts BM. Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell. 1980;19:697–707. doi: 10.1016/s0092-8674(80)80046-8. [DOI] [PubMed] [Google Scholar]
- 20.Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 21.Austin CA, Marsh KL. Eukaryotic DNA topoisomerase IIβ. Bioessays. 1998;20:215–226. doi: 10.1002/(SICI)1521-1878(199803)20:3<215::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 22.Austin CA, Fisher LM. Isolation and characterization of a human cDNA clone encoding a novel DNA topoisomerase II homologue from HeLa cells. FEBS Lett. 1990;266:115–117. doi: 10.1016/0014-5793(90)81520-x. [DOI] [PubMed] [Google Scholar]
- 23.Tan KB, Dorman TE, Falls KM, Chung TDY, Mirabelli CK, Crooke ST, et al. Topoisomerase IIa and topoisomerase IIβ genes—characterization and mapping to human chromosome-17 and chromosome-3, respectively. Cancer Res. 1992;52:231–234. [PubMed] [Google Scholar]
- 24.Yang X, Li W, Prescott ED, Burden SJ, Wang JC. DNA topoisomerase IIβ and neural development. Science. 2000;287:131–134. doi: 10.1126/science.287.5450.131. [DOI] [PubMed] [Google Scholar]
- 25.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, et al. A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 26.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 27.Leduc F, Maquennehan V, Nkoma GB, Boissonneault G. DNA damage response during chromatin remodeling in elongating spermatids of mice. Biol Reprod. 2008;78:324–332. doi: 10.1095/biolreprod.107.064162. [DOI] [PubMed] [Google Scholar]
- 28.Errington F, Willmore E, Tilby MJ, Li L, Li G, Li W, et al. Murine transgenic cells lacking DNA topoisomerase IIβ are resistant to acridines and mitoxantrone: analysis of cytotoxicity and cleavable complex formation. Mol Pharmacol. 1999;56:1309–1316. doi: 10.1124/mol.56.6.1309. [DOI] [PubMed] [Google Scholar]
- 29.Toyoda E, Kagaya S, Cowell IG, Kurosawa A, Kamoshita K, Nishikawa K, et al. NK314, a topoisomerase II inhibitor that specifically targets the alpha isoform. J Biol Chem. 2008;283:23711–23720. doi: 10.1074/jbc.M803936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao H, Huang KC, Yamasaki EF, Chan KK, Chohan L, Snapka RM. XK469, a selective topoisomerase IIβ poison. Proc Natl Acad Sci USA J1—PNAS. 1999;96:12168–12173. doi: 10.1073/pnas.96.21.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Catalano MG, Fortunati N, Pugliese M, Poli R, Bosco O, Mastrocola R, et al. Valproic acid, a histone deacetylase inhibitor, enhances sensitivity to doxorubicin in anaplastic thyroid cancer cells. J Endocrinol. 2006;191:465–472. doi: 10.1677/joe.1.06970. [DOI] [PubMed] [Google Scholar]
- 32.Kim MS, Blake M, Baek JH, Kohlhagen G, Pommier Y, Carrier F. Inhibition of histone deacetylase increases cytotoxicity to anticancer drugs targeting DNA. Cancer Res. 2003;63:7291–7300. [PubMed] [Google Scholar]
- 33.Marchion DC, Bicaku E, Turner JG, Schmitt ML, Morelli DR, Munster PN. HDAC2 regulates chromatin plasticity and enhances DNA vulnerability. Mol Cancer Ther. 2009;8:794–801. doi: 10.1158/1535-7163.MCT-08-0985. [DOI] [PubMed] [Google Scholar]
- 34.Taddei A, Maison C, Roche D, Almouzni G. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat Cell Biol. 2001;3:114–120. doi: 10.1038/35055010. [DOI] [PubMed] [Google Scholar]
- 35.Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 36.Bartova E, Pachernik J, Harnicarova A, Kovarik A, Kovarikova M, Hofmanova J, et al. Nuclear levels and patterns of histone H3 modification and HP1 proteins after inhibition of histone deacetylases. J Cell Sci. 2005;118:5035–5046. doi: 10.1242/jcs.02621. [DOI] [PubMed] [Google Scholar]
- 37.Toth KF, Knoch TA, Wachsmuth M, Frank-Stohr M, Stohr M, Bacher CP, et al. Trichostatin A-induced histone acetylation causes decondensation of interphase chromatin. J Cell Sci. 2004;117:4277–4287. doi: 10.1242/jcs.01293. [DOI] [PubMed] [Google Scholar]
- 38.Agostinho M, Rino J, Braga J, Ferreira F, Steffensen S, Ferreira J. Human topoisomerase IIa: targeting to subchromosomal sites of activity during interphase and mitosis. Mol Biol Cell. 2004;15:2388–2400. doi: 10.1091/mbc.E03-08-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cowell IG, Willmore E, Chalton D, Marsh KL, Jazrawi E, Fisher LM, et al. Nuclear distribution of DNA topoisomerase IIβ: a potent nuclear targeting signal resides in the C-terminal 116 amino acids. Exp Cell Res. 1998;243:232–240. doi: 10.1006/excr.1998.4150. [DOI] [PubMed] [Google Scholar]
- 40.Cowell IG, Durkacz BW, Tilby MJ. Sensitization of breast carcinoma cells to ionizing radiation by small molecule inhibitors of DNA-dependent protein kinase and ataxia telangiectsia mutated. Biochem Pharmacol. 2005;71:13–20. doi: 10.1016/j.bcp.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 41.Frank AJ, Proctor SJ, Tilby MJ. Detection and quantification of melphalan-DNA adducts at the single cell level in hematopoietic tumor cells. Blood. 1996;88:977–984. [PubMed] [Google Scholar]
- 42.Willmore E, Frank AJ, Padget K, Tilby MJ, Austin CA. Etoposide targets topoisomerase IIa and IIβ in leukemic cells: Isoform-specific cleavable complexes visualized and quantified in situ by a novel immunofluorescence technique. Mol Pharmacol. 1998;54:78–85. doi: 10.1124/mol.54.1.78. [DOI] [PubMed] [Google Scholar]
- 43.Petrov P, Drake FH, Loranger A, Huang W, Hancock R. Localization of DNA topoisomerase II in Chinese hamster fibroblasts by confocal and electron microscopy. Exp Cell Res. 1993;204:73–81. doi: 10.1006/excr.1993.1010. [DOI] [PubMed] [Google Scholar]
- 44.Zini N, Santi S, Ognibene A, Bavelloni A, Neri LM, Valmori A, et al. Discrete localization of different DNA topoisomerases in HeLa and K562 cell nuclei and subnuclear fractions. Exp Cell Res. 1994;210:336–348. doi: 10.1006/excr.1994.1046. [DOI] [PubMed] [Google Scholar]
- 45.Christensen MO, Larsen MK, Barthelmes HU, Hock R, Andersen CL, Kjeldsen E, et al. Dynamics of human DNA topoisomerases IIa and IIβ in living cells. J Cell Biol. 2002;157:31–44. doi: 10.1083/jcb.200112023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaly N, Brown DL. Is DNA topoisomerase IIβ a nucleolar protein? J Cell Biochem. 1996;63:162–173. doi: 10.1002/(sici)1097-4644(19961101)63:2<162::aid-jcb4>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 47.Willmore E, Errington F, Tilby MJ, Austin CA. Formation and longevity of idarubicin-induced DNA topoisomerase II cleavable complexes in K562 human leukaemia cells. Biochem Pharmacol. 2002;63:1807–1815. doi: 10.1016/s0006-2952(02)00920-6. [DOI] [PubMed] [Google Scholar]
- 48.Errington F, Willmore E, Leontiou C, Tilby MJ, Austin CA. Differences in the longevity of topo IIa and topo IIβ drug-stabilized cleavable complexes and the relationship to drug sensitivity. Cancer Chemother Pharmacol. 2004;53:155–162. doi: 10.1007/s00280-003-0701-1. [DOI] [PubMed] [Google Scholar]
- 49.Cowell IG, Sunter NJ, Singh PB, Austin CA, Durkacz BW, Tilby MJ. γ H2AX foci form preferentially in euchromatin after ionising-radiation. PLoS ONE. 2007;2:1057. doi: 10.1371/journal.pone.0001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conti C, Leo E, Eichler GS, Sordet O, Martin MM, Fan A, et al. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins and induces DNA damage. Cancer Res. 2010;70:4470–4480. doi: 10.1158/0008-5472.CAN-09-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minc E, Allory Y, Worman HJ, Courvalin JC, Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- 52.Aucott R, Bullwinkel J, Yu Y, Shi W, Billur M, Brown JP, et al. HP1β is required for development of the cerebral neocortex and neuromuscular junctions. J Cell Biol. 2008;183:597–606. doi: 10.1083/jcb.200804041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kramer OH, Zhu P, Ostendorff HP, Golebiewski M, Tiefenbach J, Peters MA, et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003;22:3411. doi: 10.1093/emboj/cdg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson CA, Padget K, Austin CA, Turner BM. Deacetylase activity associates with topoisomerase II and is necessary for etoposide-induced apoptosis. J Biol Chem. 2001;276:4539–4542. doi: 10.1074/jbc.C000824200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.