Abstract
In mammals actin contributes to transcription elongation by facilitating establishment of permissive chromatin. Here we report that the F-actin severing protein cofilin-1 is part of the same complex with actin and phosphorylated RNA polymerase (pol) II. In chromatin immunoprecipitation assays cofilin-1 was found selectively associated with transcribed regions of active genes, its occupancy being influenced by the polymerization state of actin. Cofilin-1 gene silencing led to a drop in FUrd incorporation into nascent transcripts. In cofilin-1 silenced cells chromatin immunoprecipitations showed that active genes were devoid of actin, phosphorylated pol II and displayed low histone H3 acetylation levels on K9. These findings suggest that cofilin-1 plays a major role in pol II transcription, facilitating association of elongating pol II and actin with active genes. We speculate that cofilin-1 performs its function in pol II transcription by regulating polymerization of gene-associated actin.
Key words: RNA polymerase II, transcription elongation, nascent mRNA, nuclear actin, cofilin
Introduction
In the cell nucleus, actin is an essential regulator of basal gene transcription. Actin associates with eukaryotic RNA polymerases, it occupies promoter and coding region of active genes and it is co-transcriptionally added to nascent ribonucleoprotein complexes.1,2 In mammalian cells, actin is required for both initiation and elongation phases of RNA polymerase II (pol II)-mediated transcription.3,4 During pre-mRNA elongation actin, hnRNP U and the histone acetyl transferase (HAT) PCAF associate with phosphorylated pol II carboxy-terminal domain (CTD).4,5 The actin-hnRNP U interaction mediates PCAF recruitment to active genes and contributes to pol II commitment to transcription elongation through chromatin-based mechanisms.6
Nuclear actin is known to assemble into dynamic higher order structures with a high turnover,7 suggesting that changes in actin polymerization may take place in the cell nucleus. Consistently there is evidence that both G-actin and F-actin binding proteins, including N-WASP and ARP2/3 complex which regulate nucleation and branching of actin filaments, are present in the cell nucleus.6,8–10 Overall their precise role in nuclear function is still unclear. In any case their engagement in controlling the state of actin polymerization and the finding that nuclear actin endures a continuous turnover has led to the hypothesis that there is a pool of G-actin monomers to feed growing polymeric actin structures presumably required for nuclear function.11,12
In living cells maintenance of a polymerization-competent G-actin pool depends on a set of F-actin depolymerising and severing proteins, such as cofilin.13 In mammals, there are three different cofilins/ADF (actin depolymerizing factors): cofilin-1, cofilin-2 and ADF. These proteins have distinct expression patterns: cofilin-1 is expressed in most embryonic and adult mouse cells, cofilin-2 is expressed in muscle cells and ADF is mainly found in epithelial and neuronal cells.14 Mechanistically, cofilin severs constrained actin filaments in a concentration-dependent manner and promotes in vivo cytoskeletal dynamics that are required for cytokinesis and cell motility.13,15–20 Cofilin has also been reported to be in the cell nucleus,21,22 but its potential role in nuclear function is not known.
In the present study we provide in vivo evidence that cofilin-1 is a novel key regulator of pol II transcription. Using run on assays performed in living cells we demonstrate that cofilin-1 is required for elongation of nascent transcripts. In chromatin immunoprecipitation experiments cofilin-1 gene silencing led to a drop in the levels of actin and pol II along active genes. Since cofilin-1 was found to associate with phosphorylated pol II, we suggest that cofilin-1 facilitates association of transcription machinery with actively transcribed genes presumably via its interaction with actin.
Results
In living cells cofilin-1 interacts with pol II-associated actin.
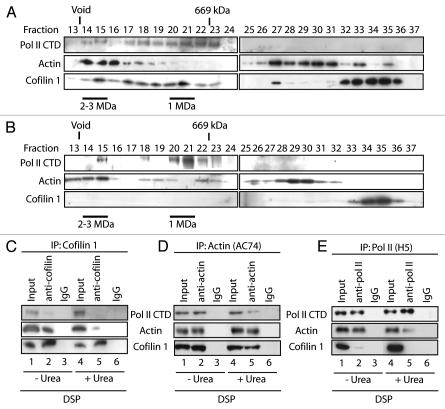
To test whether in the cell nucleus cofilin-1 interacts with actin and pol II, we treated HeLa cells with the cell-permeable and reversible short-range (11 Å) crosslinker dithiobis-succinimidylpropionate (DSP). Soluble HeLa nuclear extracts were fractionated by gel filtration chromatography on Superose 6HR and co-purifications of proteins monitored on immunoblots. We confirmed that actin and phosphorylated pol II coelute with apparent molecular mass of 2–3 MDa, independently of the DSP crosslinking (Fig. 1A and B).5 Furthermore we found that a fraction of nuclear cofilin-1 also coeluted with actin and phosphorylated pol II (Fig. 1A) but without crosslinking the cofilin signal was almost entirely detected towards low molecular weight fractions (Fig. 1B). DSP crosslinked nuclear protein extracts were next assayed by immunoprecipitation with antibodies against cofilin-1, actin (AC74) and phosphorylated pol II (H5). The immunoblots in Figure 1 show that cofilin-1 and actin were reciprocally co-immunoprecipitated even in denaturing conditions (Fig. 1C and D). Since in 8M urea DSP crosslinking is too inefficient to show interactions mediated by a third protein we conclude that cofilin-1 is crosslinked directly to actin and hence that cofilin-1 and actin interact directly in vivo. The immunoblots in Figure 1D and E also confirmed previous work that actin and phosphorylated pol II can be crosslinked under denaturing conditions indicating that actin interacts directly with phosphorylated pol II (Fig. 1A and cf lanes 1–4). Small fractions of cofilin-1 and phopshorylated pol II were reciprocally co-precipitated from DSP-crosslinked nuclear extracts only in non-denaturing conditions (Fig. 1C and E). Taken altogether these findings show that in living cells cofilin-1 interacts with pol II-associated actin and it is conceivably part of the same complex with the pol II machinery.
Figure 1.
Cofilin-1 is part of the same complex with actin and phosphorylated pol II. (A) Resolution of cofilin-1 containing nuclear complexes by gel filtration from DSP crosslinked HeLa cells. Fractions obtained from a Superose 6HR column were analyzed on immunoblots with the indicated antibodies. The position of the 670 kDa marker (thyroglobulin) is indicated. (B) Resolution of actin-containing nuclear complexes by Superose 6HR from non-crosslinked cells. (C and D) Nuclear extracts from DSP-treated cells were prepared and split into two equal portions; one was treated with urea. All samples were subjected to immunoprecipitation with (C) cofilin-1, (D) actin and (E) pol II (H5) antibodies. In all cases non-specific IgGs were used as control. Bound proteins were resolved by SDS-PAGE and analyzed on immunoblots with antibodies to pol II, actin and cofilin-1.
Cofilin-1 preferentially occupies transcribed region of active pol II genes.
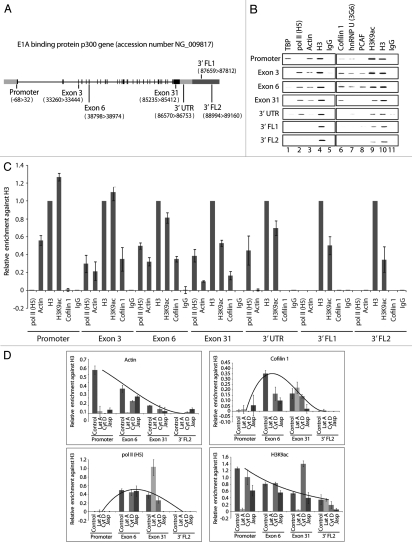
We next applied ChIP to determine whether cofilin-1 occupies gene promoter and coding region and whether its occupancy correlates with actin, phosphorylated pol II, hnRNP U and PCAF. Gene occupancy was monitored along the EP300 gene, which contains up to 31 exons and allows for a better resolution of the ChIP experiments. We studied occupancy at promoter, proximal and distal exons, 3′ untranslated regions (UTR) of the open reading frame as well as two 3′ gene flanking (FL) regions located outside the coding region (Fig. 2A). Chromatin from formaldehyde-crosslinked HeLa cells was subjected to immunoprecipitations with multiple antibodies. We used the anti-actin antibodies 1C7 (which recognizes an epitope that is not accessible in F-actin),23 and AC74.5 Furthermore we used antibodies to hnRNP U, PCAF, cofilin-1, TATA-box binding protein (TBP) and phosphorylated pol II (H5), as well as antibodies to histone H3, K9-acetylated H3 (H3K9ac) and non-specific IgGs. The precipitated chromatin was analyzed by PCR with primers amplifying EP300 gene promoter, exon 3, exon 6, exon 31, 3′ UTR, 3′ FL1 and 3′ FL2 (Fig. 2A). As previously reported gene promoter was co-precipitated using the anti-actin AC74 antibody5 (not shown) and interestingly, also with 1C7 (Fig. 2B and C). As expected gene promoter was precipitated with anti-TBP, H3 as well as H3K9ac antibodies. Weak signals were revealed with anti-cofilin, hnRNP U and PCAF antibodies whereas no signal was detected with antibodies against phosphorylated pol II (H5) (Fig. 2B and C). The cofilin-1 antibody precipitated promoter-proximal and promoter-distal exons (Fig. 2B and C), similarly to antibodies against pol II (H5), AC74 (not shown),5 1C7, hnRNP U, PCAF, H3 and H3K9ac antibodies (Fig. 2B and C). When analyzing the 3′ end of the gene, anti-cofilin antibodies did not precipitate the 3′ UTR in contrast to anti-pol II (H5), H3 and H3K9ac antibodies (Fig. 2B and C). Similar distributions were revealed on other endogenous genes used as control (Sup. Fig. 1). Altogether these results were reproduced in at least 5 independent experiments. We conclude that cofilin-1 preferentially occupies gene coding region and occupancy correlates with actin and phosphorylated pol II.
Figure 2.
Cofilin-1 selectively occupies gene coding region. (A) The E1A binding protein p300 (EP300) gene. (B) ChIP assays with the indicated antibodies. Co-precipitated DNA was analyzed by PCR with primers for promoter, proximal and distal exons, 3′UTR and gene flanking regions (FL). (C) Quantification of relative occupancies at EP300 gene promoter, proximal and distal exons, 3′UTR, 3′ FL1 and FL2 of phosphorylated pol II (H5), actin, H3, H3K9ac, cofilin 1. The bar diagrams show relative amounts of gene promoter, exon 3, exon 6, exon 31, 3′UTR, FL1 and FL2 precipitated with antibodies against the above proteins and with a non-specific rabbit IgG used as control in the ChIP assay in part B. Calculations were made with respect to H3. Values were obtained by densitometric calculations of PCR products resolved by agarose gel electrophoresis (and revealed by ethidium bromide staining) in at least five independent experiments. Error bars represent standard deviations. (D) Cofilin-1 gene occupancy is sensitive to G-actin depletion. Densitometric quantification of relative occupancies determined by ChIP experiments on chromatin isolated from untreated cells or cells treated with the indicated drugs. Bars diagrams represent average values calculated from analysis of promoter, coding regions (exon 6 and exon 31) and 3′ FL2 over five independent experiments. All values are normalized against the signals obtained with an H3 antibody. See also supplemental information.
To evaluate whether gene occupancies of cofilin-1, phosphorylated pol II, actin and H3K9 acetylation are affected by changes in the state of actin polymerization we applied ChIP on chromatin isolated from latrunculin A-, cytochalasin D- and jasplakinolide-treated cells.24–27 Overall cofilin-1 was not found at EP300 gene promoter and a general drop in actin and H3K9ac signals was observed (Fig. 2D and Sup. Fig. 2). When further analyzing proximal exons, distal exons and flanking regions we found that in latrunculin A-treated cells where G-actin is depleted, the promoter-proximal exon 6 was devoid of actin, cofilin-1, phosphorylated pol II (H5) and displayed low H3K9ac levels. Terminal exon 31 was also devoid of actin and H3K9ac signal but a local increment in pol II (H5) and cofilin-1 occupancy was revealed (Fig. 2D and Sup. Fig. 2). With jasplakinolide which impairs filaments de-polymerization and induces abnormally organized filaments we found that actin, cofilin-1 and pol II (H5) antibodies precipitated proximal exon 6 with low efficiency and were not present at terminal exon 31, concomitantly with a general decrease in H3K9ac levels (Fig. 2D and Sup. Fig. 2). Finally, in cells treated with cytochalasin D which severs F-actin all antibodies behaved similarly to control cells even though we detected a drop in the immunoprecipitations efficiencies (Fig. 2D and Sup. Fig. 2). In this case a significant increase in H3K9ac was detected only at exon 31 (Fig. 2D and Sup. Fig. 2). We conclude that gene occupancies of cofilin-1 and phosphorylated pol II as well as global H3K9 acetylation levels are sensitive to changes in the polymerization state of actin. The finding that actin and cofilin-1 exhibit contrasting occupancies on exon 31 is somewhat surprising but it may reflect exon-specific differences in protein composition within the nuclear actin complex. In any case global drops in H3K9 acetylation levels and in the levels of actin as monitored by both 1C7 and AC74 antibodies suggest that G-actin depletion is not likely to be compatible with active transcription.
Cofilin-1 posttranscriptional gene silencing leads to a transcriptional block.
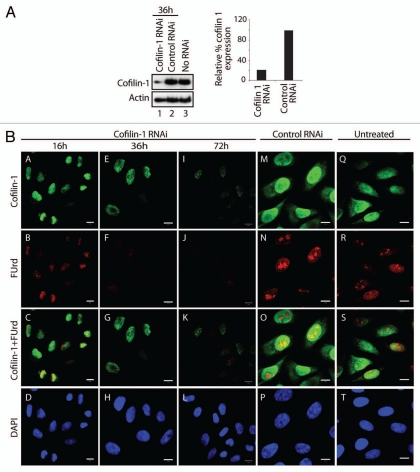
If cofilin-1 occupies active genes it may be involved in pol II transcription. To test this hypothesis HeLa cells were transfected with an RNA duplex against a target sequence within the open reading frame of the cofilin-1 gene and steady state protein expression was monitored on immunoblots of total cell lysates. An almost complete shutdown of the cofilin-1 gene resulting in very low endogenous cofilin-1 steady state expression was already observed after 36 h from transfection (Fig. 3A). To test whether silencing of the cofilin-1 gene induced abnormalities in transcription patterns we subjected cofilin-1 silenced HeLa cells to a short FUrd pulse chase. The incorporated FUrd was revealed by immunofluorescence and confocal microscopy. Concomitantly with decreased steady state expression of cofilin-1 the number of FUrd-rich foci was specifically downregulated (Fig. 3B). Quantification of nucleoplasmic FUrd-rich foci in three independent cofilin-1 gene silencing experiments showed a specific 80–90% reduction. Interestingly under the same conditions we found that a considerable proportion of cells displaying low steady state cofilin-1 expression revealed increased levels of nuclear polymeric actin foci detectable with Alexa568-phalloidin (Fig. 4A–C). These findings are consistent with the observation that post-transcriptional gene silencing of cofilin-1 induces changes in the G- to F-actin ratio, leading to formation of abnormal F-actin structures.16 We conclude that in the cell nucleus cofilin-1 is required for pre-mRNA synthesis and may be important to regulate actin polymerization.
Figure 3.
Cofilin-1 is required for in vivo FUrd incorporation. (A) Cofilin-1 steady state expression levels on immunoblots. Right part, quantification of cofilin-1 steady state protein expression relative to actin. (B) In cofilin-1 silenced HeLa cells, incorporated FUrd was monitored by immunostaining (red) at 16 h, 36 h and 72 h after transfection of the cofilin-1 specific siRNA oligos and overlaid with cofilin-1 (green). The DNA was detected by DAPI staining (Scale bar, 10 µm).
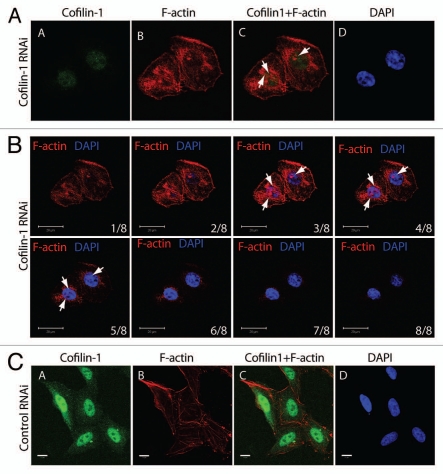
Figure 4.
Post-transcriptional gene silencing of cofilin-1 leads to nuclear accumulation of F-actin foci. (A) Cofilin-1 silenced HeLa cells were analyzed by confocal fluorescence microscopy, with an anti-cofilin 1 antibody (green) and Alexa568-conjugated phalloidin (red) at 36 h after transfection of the cofilin-1 specific siRNA oligos. The DNA was detected by DAPI staining (Scale bar, 10 µm). Arrows point towards examples of accumulation of nuclear polymeric actin structures stained with phalloidin. (B) Multiple confocal sections of the same experiment in (A) showing that the phalloidin-stainable foci are likely to be present in the cell nucleus (Scale bar, 20 µm, thickness of each section was 1 µm). (C) In control siRNA experiments cofilin-1 is present in the cell nucleus and phalloidin labels cytoplasmic F-actin (Scale bar 10 µm).
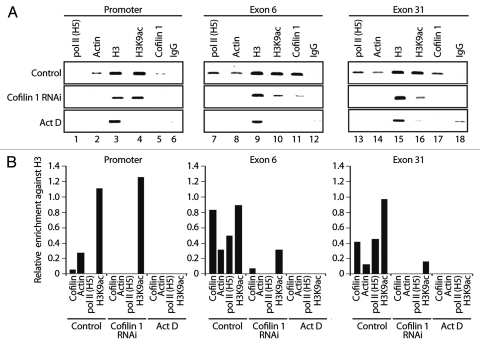
To get mechanistic insights we performed ChIP on cofilin 1-silenced cells with pol II (H5), actin, H3 and H3K9ac antibodies. In the absence of cofilin-1 actin occupancy at EP300 gene promoter dropped to very low levels (Fig. 5 and cf. lane 2). Cofilin 1 silencing also led to a moderate increase in the H3K9 acetylation levels at gene promoter, an observation which is not yet fully understood (Fig. 5 and cf. lane 4). When analyzing gene occupancy at exons 6 and 31, we found that neither pol II (H5) nor actin antibodies precipitated significant amounts of both exons (Fig. 5). Low amounts were also precipitated with the H3K9ac antibody; as expected, the cofilin-1 antibody did not precipitate any of the gene fragments analyzed (Fig. 5). Finally, in non-silenced cells, cofilin-1 and actin gene occupancies were found to be sensitive to transcription inhibition by actinomycin D, correlating with drops in pol II and H3K9 acetylation (Fig. 5). We conclude that cofilin-1 is necessary for association of actin and phosphorylated pol II with active genes presumably through a mechanism that controls and maintains polymerization of gene-associated actin.
Figure 5.
Cofilin-1 facilitates association of actin and elongating pol II with active genes. ChIP on cofilin-1 silenced cells or control siRNA as well as actinomycin D treated cells, with the indicated antibodies. PCR analysis was with primers amplifying promoter, exons 6 and exon 31 of the EP300 gene. Bottom parts, densitometric quantification of relative occupancies. Bars diagrams represent values calculated from analysis of promoter, exon 6 and exon 31 in duplicates. All values are normalized against the signals obtained with an H3 antibody.
Discussion
Overall, our results suggest that cofilin-1 plays a key role in pol II transcription. When studying cofilin 1 gene occupancy we discovered that cofilin-1 selectively occupies transcribed regions of constitutively expressed genes while being mostly excluded from promoters. These observations and evidence that cofilin 1 is part of the same complex with actin and phosphorylated pol II suggest that cofilin-1 is not likely to be involved in the initial phases of pol II transcription. In fact as a consequence of cofilin-1 gene silencing run on assays revealed a global decrease in nascent mRNA levels. Furthermore under the same conditions ChIP assays showed that promoter-proximal and promoter-distal exons were specifically devoid of actin and phosphorylated pol II and displayed low levels of H3K9 acetylation. These findings suggest a functional link between cofilin-1 and elongating pol II. We favor the idea that cofilin-1 is primarily involved in transcription elongation, where cofilin-1 may facilitate association of phosphorylated pol II and actin with active genes.
Cofilin-1 gene silencing also resulted in actin depletion from proximal and distal exons and in turn, G-actin depletion led to a general decrease in gene occupancies by cofilin-1, actin, pol II and H3K9, suggesting that G-actin is a requisite for active transcription. As mentioned above the local increment in the levels of cofilin-1 and pol II occupancy in contrast with a drop in actin and H3K9 at promoter-distal exon 31 upon latrunculin A treatment is surprising. One possible explanation is that the composition of actin complexes may vary dynamically along different exonic regions and the cofilin-actin interaction may be temporally precluded. In any case given that cofilin-1 posttranscriptional gene silencing inhibited incorporation of FUrd in nascent transcripts, our findings suggest that there is a fundamental link between the role of cofilin-1 in pol II transcription elongation and polymerization of gene-associated actin. Consistent with its F-actin severing activity,13,18,28 cofilin-1 silencing led to accumulation of abnormal polymeric nuclear actin structures detectable by phalloidin. While it is unlikely that gene-associated actin is in a classical F-actin form, evidence that gene-associated actin can be coprecipitated with both AC74 (data not shown; reviewed in ref. 5) and 1C7 antibody specific for an epitope not accessible in F-actin23 suggests that gene-associated actin is present in different forms, monomeric as well as polymeric. Based on our findings, it is likely that a fraction of gene-associated actin is competent for cofilin binding. Therefore we speculate that cofilin-1 may function in transcription elongation by contributing to a local pool of monomeric G-actin molecules that feed growing actin polymers along active genes.
Mechanistically, our observations suggest that controlled actin polymerization may be a requirement for pol II transcription. The findings that different actin forms are likely to associate with active genes and that G-actin depletion affects gene occupancy of the transcription machinery as well as actin point to the possibility that dynamic actin polymerization occurs along active genes as a primary requisite for basal transcription elongation. Transcription occurs against a mixture of complex histone modifications, which probably have several roles.29 Gene silencing of cofilin-1 induced low levels of acetylated H3K9 and prevented elongating pol II as well as actin from associating with chromatin. Actin interacts with hnRNP U to recruit the HAT PCAF that acetylates H3K9.5 Since the actin-hnRNP U interaction is established outside the gene promoter,5 hnRNP U is likely to interact with polymeric actin. Changes in the polymerization state of actin may accompany the transition of the polymerase machinery from promoter to transcribed region, providing a platform for recruitment of pol II transcriptional coactivators such as PCAF. Whether other changes in epigenetic marks are induced in an actin-dependent manner is not known. In either case, we suggest that in the cell nucleus cofilin-1-mediated F-actin severing activity is important for polymerization of gene-associated actin. We speculate that this mechanism provides enhanced processivity and directionality to pol II while synchronizing transcription machinery with chromatin modification for efficient pre-mRNA elongation.
Materials and Methods
Antibodies and reagents.
For chromatin immunoprecipitation experiments mouse monoclonal antibodies to phosphorylated pol II (H5), TBP (clone 1TBP18) and hnRNP U (3G6) as well as the rabbit polyclonal antibodies to cofilin-1, non-acetylated histone H3 and acetylated H3K9 were purchased from AbCam (Cambridge, UK). Mouse monoclonal antibody against PCAF (E-8) was from Santa Cruz Biotechnology Inc., (Santa Cruz, CA USA). The anti-actin antibody AC74 was from Sigma whereas the 1C7 monoclonal antibody to actin was a gift of Cora-Ann Schoenenberger (Biocentrum, University of Basel).23 The non specific rabbit IgG was from DIANOVA (Hamburg, Germany). The monoclonal antibody to bromouridine triphosphate (BrUTP) used to monitor FUrd incorporation in nascent transcripts was from Roche. For F-actin staining we used Alexa568-conjugated phalloidin (Molecular Probes).
Cells and drugs treatment.
HeLa cells were cultured with antibiotics in Dulbecco's minimum essential medium as described by Obrdlik et al. (2008).5 Living HeLa cells were incubated with 2 mM latrunculin A (Sigma), 2 mM cytochalasin D (Sigma) and 1 mM jasplakinolide (Calbiochem) at 37°C diluted in culture medium.
Cofilin-1 gene silencing.
siRNA duplexes against the human cofilin-1 gene (accession number NM_005507, CCU CUA UGA UGC AAC CUA U) or the control eGFP gene (Accell eGFP siRNA, D-001940-01-05) were transfected into HeLa cells according to the manufacturer's instructions (Dharmacon Research, Boulder, CO). siRNA duplexes against cofilin-1 and eGFP were applied by transfection with Lipofectamine 2000 (Invitrogen) at a final concentration of 20 nM.
FUrd incorporation in living cells and immunofluorescence microscopy.
For FUrd incorporation, the method was adapted from Mitchell and Fraser (2008).30 Briefly HeLa cells harvested 12, 36 and 72 h after transfection with cofilin-1 and eGFP siRNAs were incubated with FUrd supplemented medium for 25 min at 37°C to allow incorporation into nascent RNA transcripts. In all cases FUrd was added to a final concentration of 2 mM. In all cases the run on reactions were stopped by replacing the cell culture medium with ice-cold PBS. Incorporated FUrd was detected on fixed and permeabilized cells with a polyclonal antibody to BrUTP by immunofluorescence and confocal microscopy, using a LSM 510 Laser Scanning Microscope (Zeiss, Jena, Germany). These experiments were repeated three times and on average as much as 70% of those cells with decreased expression of cofilin-1 showed decreased FUrd incorporation levels. Quantification of the number of nucleoplasmic FUrd-rich foci was performed using the ImageJ software.
Protein-protein interaction assays.
In vivo protein to protein crosslinking with dithiobis-succinimidylpropionate (DSP, Sigma) and nuclear extracts preparation were done essentially as described in reference 4 and 5. Where indicated nuclear extracts from DSP-crosslinked HeLa cells were prepared in 8 M urea. The extracts were diluted 10 times to reduce the urea concentration and used immediately for immunoprecipitation with anti-cofilin-1, actin (AC74) and pol II (H5) antibodies or IgGs. Immunoprecipitations were done following standard procedures.
Chromatin immunoprecipitations.
ChIP was performed as previously described in reference 5. Briefly, formaldehyde cross-linked chromatin was obtained from HeLa cells untreated or treated with latrunculin A, cytochalasin D, jasplakinolide, from HeLa cells subjected to trasfection with cofilin-1 siRNA oligonucleotides, control eGFP oligonucleotides or subjected to actinomycin D treatment. Cross-linked chromatin isolated from each condition was immunoprecipitated with antibodies to TBP, actin (AC74 and 1C7), cofilin-1, hnRNP U (3G6), PCAF, non acetylated H3, acetylated H3K4 and non-specific rabbit IgGs as indicated in the figures accompanying main text and supplementary information. DNA-protein complexes were analyzed by PCR with specific primers for amplifying multiple regions of the EP300 gene, including promoter, exons 3, 6, 21, 31, 3′ UTR, 3′ gene flanking regions 1 and 2 as well as S19, β-tubulin and GAPDH genes (Sup. Fig. 1). About 1/10 of the precipitated DNA was used for each PCR reaction. PCR products were separated by electrophoresis on 2% agarose gels and visualized with ethidium bromide. Quantification of amplified signals was performed with ImageJ and Excell packages. All ChIP experiments were successfully reproduced in multiple rounds, three to five times. All sequences of the primers used in the ChIP experiments to amplify multiple regions of the EP300 gene are listed in the Supplemental Table 1. Sequences of primers to amplify S19, β-tubulin and GAPDH gene promoters and coding regions were previously published in reference 5.
Acknowledgements
This work was supported by grants from the Swedish Research Council (Vetenskapsrådet) and the Swedish Cancer Society (Cancerfonden) to P.P.
Supplementary Material
References
- 1.Hofmann WA. Cell and molecular biology of nuclear actin. Int Rev Cell Mol Biol. 2009;273:219–263. doi: 10.1016/S1937-6448(08)01806-6. [DOI] [PubMed] [Google Scholar]
- 2.Louvet E, Percipalle P. Actin and myosin in gene transcription. Int Rev Cell Mol Biol. 2009;272:107–147. doi: 10.1016/S1937-6448(08)01603-1. [DOI] [PubMed] [Google Scholar]
- 3.Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol. 2004;6:1094–1101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- 4.Kukalev A, Nord Y, Palmberg C, Bergman T, Percipalle P. Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat Struct Mol Biol. 2005;12:238–244. doi: 10.1038/nsmb904. [DOI] [PubMed] [Google Scholar]
- 5.Obrdlik A, Kukalev A, Louvet E, Farrants AK, Caputo L, Percipalle P. The histone acetyltransferase PCAF associates with actin and hnRNP U for RNA polymerase II transcription. Mol Cell Biol. 2008;28:6342–6357. doi: 10.1128/MCB.00766-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Percipalle P. The long journey of actin and actin-associated proteins from gene to polysomes. Cell Mol Life Sci. 2009;66:2151–2165. doi: 10.1007/s00018-009-0012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MG. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol. 2006;172:541–552. doi: 10.1083/jcb.200507101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X, Yoo Y, Okuhama NN, Tucker PW, Liu G, Guan JL. Regulation of RNApolymerase-II-dependent transcription by N-WASP and its nuclear-binding partners. Nat Cell Biol. 2006;8:756–763. doi: 10.1038/ncb1433. [DOI] [PubMed] [Google Scholar]
- 9.Yoo Y, Wu X, Guan JL. A novel role of the actin-nucleating Arp2/3 complex in the regulation of RNA polymerase II-dependent transcription. J Biol Chem. 2007;282:7616–7623. doi: 10.1074/jbc.M607596200. [DOI] [PubMed] [Google Scholar]
- 10.Jockusch BM, Schoenenberger CA, Stetefeld J, Aebi U. Tracking down the different forms of nuclear actin. Trends Cell Biol. 2006;16:391–396. doi: 10.1016/j.tcb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Gieni RS, Hendzel MJ. Actin dynamics and functions in the interphase nucleus: moving toward an understanding of nuclear polymeric actin. Biochem Cell Biol. 2009;87:283–306. doi: 10.1139/O08-133. [DOI] [PubMed] [Google Scholar]
- 12.Visa N, Percipalle P. Nuclear functions of actin. Cold Spring Harb Perspect Biol. 2010 doi: 10.1101/csh-perspect.a000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vartiainen MK, Mustonen T, Mattila PK, Ojala PJ, Thesleff I, Partanen J, et al. The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Mol Biol Cell. 2002;13:183. doi: 10.1091/mbc.01-07-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dedova IV, Nikolaeva OP, Mikhailova VV, dos Remedios CG, Levitsky DI. Two opposite effects of cofilin on the thermal unfolding of F-actin: A differential scanning calorimetric study. Biophys Chem. 2004;110:119–128. doi: 10.1016/j.bpc.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Hotulainen P, Paunola E, Vartiainen MK, Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian non-muscle cells. Mol Biol Cell. 2005;16:649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yap CT, Simpson TI, Pratt T, Price DJ, Maciver SK. The motility of glioblastoma tumour cells is modulated by intracellular cofilin expression in a concentration-dependent manner. Cell Motil Cytoskeleton. 2005;60:153–165. doi: 10.1002/cm.20053. [DOI] [PubMed] [Google Scholar]
- 18.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Bobkov AA, Muhlrad A, Pavlov DA, Kokabi K, Yilmaz A, Reisler E. Cooperative effects of cofilin (ADF) on actin structure suggest allosteric mechanism of cofilin function. J Mol Biol. 2006;356:325–334. doi: 10.1016/j.jmb.2005.11.072. [DOI] [PubMed] [Google Scholar]
- 20.Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Actin filament severing by cofilin. J Mol Biol. 2007;365:1350–1358. doi: 10.1016/j.jmb.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pendleton A, Pope B, Weeds A, Koffer A. Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J Biol Chem. 2003;278:14394–14400. doi: 10.1074/jbc.M206393200. [DOI] [PubMed] [Google Scholar]
- 22.Ye J, Zhao J, Hoffmann-Rohrer U, Grummt I. Nuclear myosin 1 acts in concert with polymeric actin to drive RNA polymerase I transcription. Genes & Dev. 2008;22:322–330. doi: 10.1101/gad.455908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoenenberger CA, Buchmeier S, Boerries M, Sütterlin R, Aebi U, Jockusch BM. Conformation-specific antibodies reveal distinct actin structures in the nucleus and the cytoplasm. J Struct Biol. 2005;152:157–168. doi: 10.1016/j.jsb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Goddette DW, Frieden C. Actin polymerization. The mechanism of action of cytochalasin D. J Biol Chem. 1986;261:15974–15980. [PubMed] [Google Scholar]
- 25.Cooper JA. Effects of cytochalasin and phalloidin on actin. J Cell Biol. 1987;105:1473–1478. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bubb MR, Spector I, Beyer BB, Fosen KM. Effects of jasplakinolide on the kinetics of actin polymerization. J Biol Chem. 2000;275:5163–5170. doi: 10.1074/jbc.275.7.5163. [DOI] [PubMed] [Google Scholar]
- 27.Morton WM, Ayscough KR, McLaughlin PJ. Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat Cell Biol. 2000;2:376–378. doi: 10.1038/35014075. [DOI] [PubMed] [Google Scholar]
- 28.Paavilainen VO, Bertling E, Falck S, Lappalainen P. Regulation of cytoskeletal dynamics by actin-monomer-binding proteins. Trends Cell Biol. 2004;14:386–394. doi: 10.1016/j.tcb.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 29.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;44:7407–7412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell JA, Fraser P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 2008;22:20–25. doi: 10.1101/gad.454008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.