Abstract
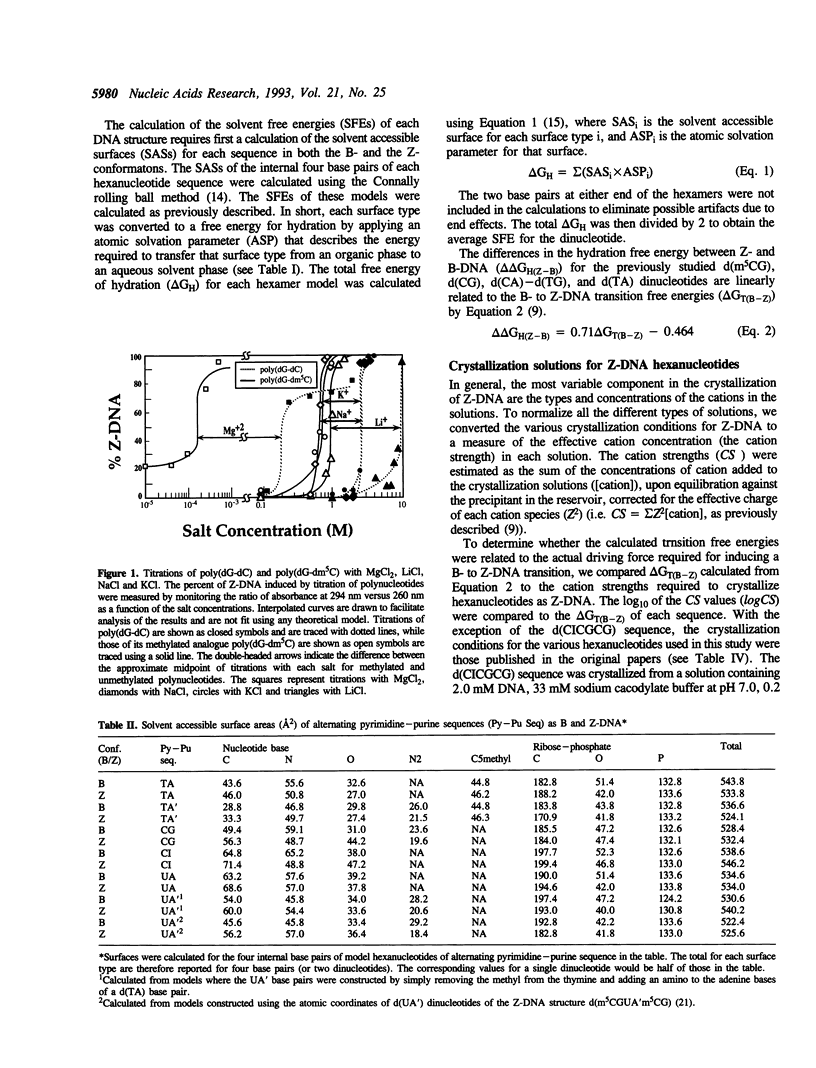
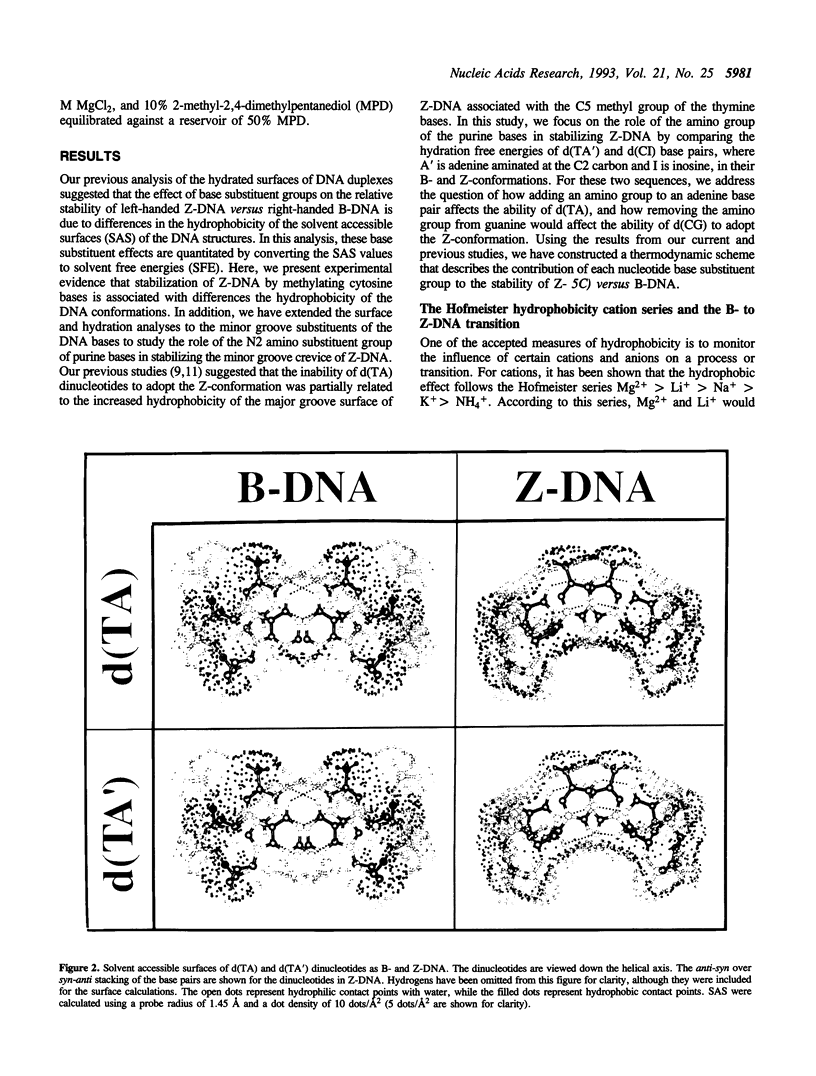
We present a study of how substituent groups of naturally occurring and modified nucleotide bases affect the degree of hydration of right-handed B-DNA and left-handed Z-DNA. A comparison of poly(dG-dC) and poly(dG-dm5C) titrations with the lipotropic salts of the Hofmeister series infers that the methyl stabilization of cytosines as Z-DNA is primarily a hydrophobic effect. The hydration free energies of various alternating pyrimidine-purine sequences in the two DNA conformations were calculated as solvent free energies from solvent accessible surfaces. Our analysis focused on the N2 amino group of purine bases that sits in the minor groove of the double helix. Removing this amino group from guanine to form inosine (I) destabilizes Z-DNA, while adding this group to adenines to form 2-aminoadenine (A') stabilizes Z-DNA. These predictions were tested by comparing the salt concentrations required to crystallize hexanucleotide sequences that incorporate d(CG), d(CI), d(TA) and d(TA') base pairs as Z-DNA. Combining the current results with our previous analysis of major groove substituents, we derived a thermodynamic cycle that relates the systematic addition, deletion, or substitution of each base substituent to the B- to Z-DNA transition free energy.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anshelevich V. V., Vologodskii A. V., Frank-Kamenetskii M. D. A theoretical study of formation of DNA noncanonical structures under negative superhelical stress. J Biomol Struct Dyn. 1988 Oct;6(2):247–259. doi: 10.1080/07391102.1988.10507711. [DOI] [PubMed] [Google Scholar]
- Behe M., Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc Natl Acad Sci U S A. 1981 Mar;78(3):1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll M., Fita I., Lloveras J., Subirana J. A., Bardella F., Huynh-Dinh T., Igolen J. Structure of d(CACGTG), a Z-DNA hexamer containing AT base pairs. Nucleic Acids Res. 1988 Sep 12;16(17):8695–8705. doi: 10.1093/nar/16.17.8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll M., Wang A. H., van der Marel G. A., van Boom J. H., Rich A. Crystal structure of a Z-DNA fragment containing thymine/2-aminoadenine base pairs. J Biomol Struct Dyn. 1986 Oct;4(2):157–172. doi: 10.1080/07391102.1986.10506337. [DOI] [PubMed] [Google Scholar]
- Connolly M. L. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983 Aug 19;221(4612):709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- Dang L. X., Pearlman D. A., Kollman P. A. Why do A.T base pairs inhibit Z-DNA formation? Proc Natl Acad Sci U S A. 1990 Jun;87(12):4630–4634. doi: 10.1073/pnas.87.12.4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D., McLachlan A. D. Solvation energy in protein folding and binding. Nature. 1986 Jan 16;319(6050):199–203. doi: 10.1038/319199a0. [DOI] [PubMed] [Google Scholar]
- Ellison M. J., Feigon J., Kelleher R. J., 3rd, Wang A. H., Habener J. F., Rich A. An assessment of the Z-DNA forming potential of alternating dA-dT stretches in supercoiled plasmids. Biochemistry. 1986 Jun 17;25(12):3648–3655. doi: 10.1021/bi00360a026. [DOI] [PubMed] [Google Scholar]
- Fujii S., Wang A. H., van der Marel G., van Boom J. H., Rich A. Molecular structure of (m5 dC-dG)3: the role of the methyl group on 5-methyl cytosine in stabilizing Z-DNA. Nucleic Acids Res. 1982 Dec 11;10(23):7879–7892. doi: 10.1093/nar/10.23.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho P. S., Kagawa T. F., Tseng K. H., Schroth G. P., Zhou G. W. Prediction of a crystallization pathway for Z-DNA hexanucleotides. Science. 1991 Nov 15;254(5034):1003–1006. doi: 10.1126/science.1948069. [DOI] [PubMed] [Google Scholar]
- Kagawa T. F., Stoddard D., Zhou G. W., Ho P. S. Quantitative analysis of DNA secondary structure from solvent-accessible surfaces: the B- to Z-DNA transition as a model. Biochemistry. 1989 Aug 8;28(16):6642–6651. doi: 10.1021/bi00442a017. [DOI] [PubMed] [Google Scholar]
- Kollman P., Weiner P., Quigley G., Wang A. Molecular-mechanical studies of Z-DNA: a comparison of the structural and energetic properties of Z- and B-DNA. Biopolymers. 1982 Oct;21(10):1945–1969. doi: 10.1002/bip.360211003. [DOI] [PubMed] [Google Scholar]
- Melander W., Horváth C. Salt effect on hydrophobic interactions in precipitation and chromatography of proteins: an interpretation of the lyotropic series. Arch Biochem Biophys. 1977 Sep;183(1):200–215. doi: 10.1016/0003-9861(77)90434-9. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Energetics of B-to-Z transition in DNA. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6206–6210. doi: 10.1073/pnas.80.20.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Schneider B., Ginell S. L., Jones R., Gaffney B., Berman H. M. Crystal and molecular structure of a DNA fragment containing a 2-aminoadenine modification: the relationship between conformation, packing, and hydration in Z-DNA hexamers. Biochemistry. 1992 Oct 13;31(40):9622–9628. doi: 10.1021/bi00155a014. [DOI] [PubMed] [Google Scholar]
- Sinden R. R., Wells R. D. DNA structure, mutations, and human genetic disease. Curr Opin Biotechnol. 1992 Dec;3(6):612–622. doi: 10.1016/0958-1669(92)90005-4. [DOI] [PubMed] [Google Scholar]
- Soumpasis D. M., Robert-Nicoud M., Jovin T. M. B-Z DNA conformational transition in 1:1 electrolytes: dependence upon counterion size. FEBS Lett. 1987 Mar 23;213(2):341–344. doi: 10.1016/0014-5793(87)81519-3. [DOI] [PubMed] [Google Scholar]
- Vologodskii A. V., Frank-Kamenetskii M. D. Left-handed Z form in superhelical DNA: a theoretical study. J Biomol Struct Dyn. 1984 Jun;1(6):1325–1333. doi: 10.1080/07391102.1984.10507523. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Hakoshima T., van der Marel G., van Boom J. H., Rich A. AT base pairs are less stable than GC base pairs in Z-DNA: the crystal structure of d(m5CGTAm5CG). Cell. 1984 May;37(1):321–331. doi: 10.1016/0092-8674(84)90328-3. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Zhou G. W., Ho P. S. Stabilization of Z-DNA by demethylation of thymine bases: 1.3-A single-crystal structure of d(m5CGUAm5CG). Biochemistry. 1990 Aug 7;29(31):7229–7236. doi: 10.1021/bi00483a010. [DOI] [PubMed] [Google Scholar]