Abstract
Peptide-based hydrogels are an important class of biomaterials finding use in food industry and potential use in tissue engineering, drug delivery and microfluidics. A primary experimental method to explore the physical properties of these hydrogels is rheology. A fundamental understanding of peptide hydrogel mechanical properties and underlying molecular mechanisms is crucial for determining whether these biomaterials are potentially suitable for biotechnological uses. In this critical review, we cover the literature containing rheological characterization of the physical properties of peptide and polypeptide-based hydrogels including hydrogel bulk mechanical properties, gelation mechanisms, and the behavior of hydrogels during and after flow.
I. Introduction
Highly hydrated and porous materials, biopolymer hydrogels have been widely used in the food and pharmaceutical industries1 in addition to current and future applications in tissue engineering,2–4 drug delivery5–7 and microfluidics.8,9 A large number of these hydrogels were made from either natural occurring proteins, like collagen and gelatin, or synthetic polypeptides. The use of amino acids as material building blocks allows the designer to easily incorporate possible biofunctionality, biocompatibility and biodegradability into the hydrogel.
While these biologically relevant properties are certainly important for potential biomaterial applications, the mechanical and viscoelastic properties of peptide and polypeptide-based hydrogels are of growing interest relative to their uses in new technologies. Herein we specifically focus on rheological characterization of peptide and polypeptide-based hydrogels relative to aspects of bulk mechanical and viscoelastic properties, observations of gelation mechanisms, and the behavior of hydrogels during and after flow. Fundamental principles and applications of hydrogels in general have already been discussed thoroughly in a number of comprehensive reviews,2–7,10,11 and so this material won’t be repeated here. Mechanical properties are criteria growing in importance as to judge the feasibility of a hydrogel for a specific biological application.3,12,13 For example, the gel has to be rigid enough to sustain itself as a scaffold for cell growth while appropriate mechanical stiffness is essential for regulation of cell phenotype, cell adhesion and cell gene expression.14 Proper flow properties allow hydrogels to be excellent candidates for injectable therapeutic delivery vehicles if they shear-thin upon the application of a proper shear stress and rapidly self-heal into solids once the stress is removed. In this case, solid, preformed gels are capable of being delivered to a targeted in vivo site by simple syringe injection. Upon removal of injection shear, immediate recovery of gel stiffness enables payloads (e.g. therapeutics) that were encapsulated during hydrogel formation to remain localized against in vivo biological forces.13,15 Finally, fundamental understanding of gelation mechanisms helps unveil the pathways of forming a network, inspiring further development of hydrogels for tissue repair and drug delivery.16
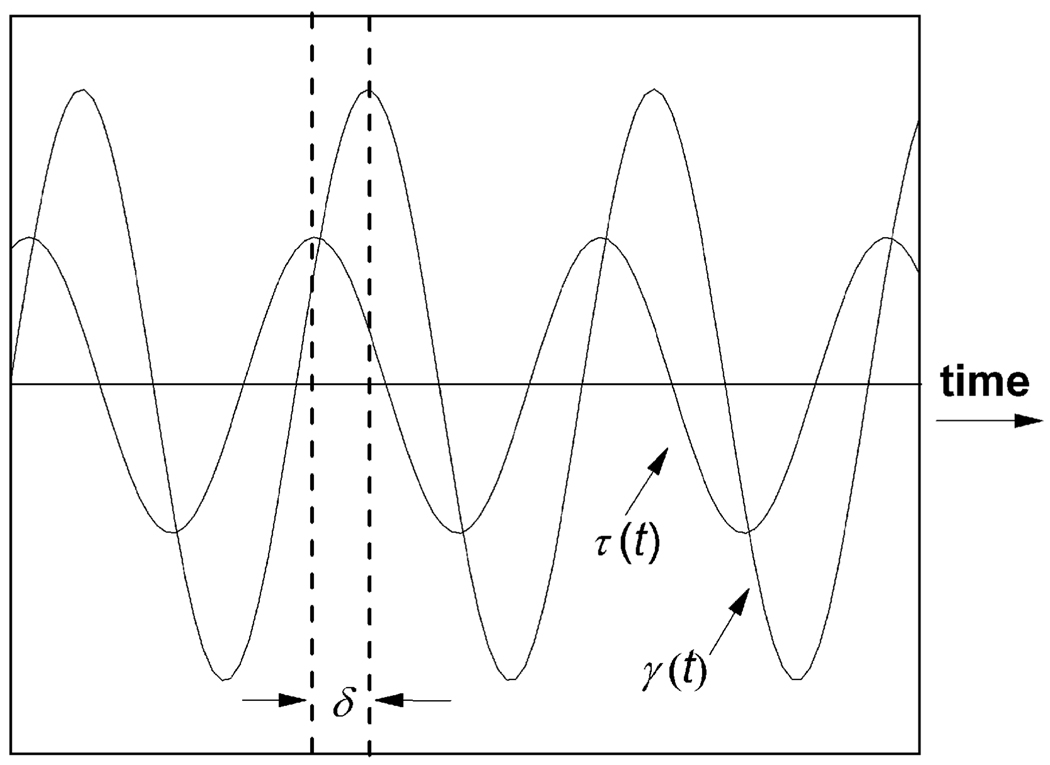
The primary experimental method with which researchers explore the viscoelastic properties of hydrogels is rheology. Importantly, equipment to perform the basic rheological exploration of hydrogel materials is inexpensive (<$100k) and fits on the bench top of any biomaterials laboratory. The field of rheology theory, measurements and equipment is well introduced in books on the subject, e.g. by Macosko17 and Mezger.18 However, we would like to briefly describe the very basics of what hydrogel properties can be measured with rheology as these properties are frequently discussed in the remainder of the article. To assess mechanical properties quantitatively, primarily small deformation rheology experiments are performed on biopolymeric hydrogels.19 By small deformation, the measurement is meant to be carried out within linear viscoelastic region of a material, ensuring the measured hydrogel properties are independent of the magnitude of imposed strain or stress.17,18 Typical small deformation tests are small amplitude oscillatory shear (SAOS) measurement as well as creep and creep recovery tests. The principle of SAOS is illustrated in Fig. 1.
Fig. 1.
Principle of a small amplitude oscillatory shear measurement.
For controlled-strain rheometers, shear strain is applied to the sample in a sinusoidal oscillation, γ(t) = γ0(sin ωt), and the measured shear stress is a phase-shifted sine wave with τ(t) = τ0(sin ωt + δ) in which ω is the applied angular frequency and δ is the phase difference between the two waves. For stress-controlled rheometers, the shear stress is applied as τ(t) = τ0(sin ωt) and the resulting shear strain is measured as γ(t) = γ0(sin ωt + δ). For a purely elastic material, the strain and stress waves are in phase (δ = 0°) while a purely viscous response has the two waves out of phase by 90°(δ = 90°). Viscoelastic materials give rise to a phase-angle somewhere in between.18,20
In small amplitude oscillatory shear measurements, the shear storage modulus, G′, loss modulus, G″ and loss factor, tan δ, are critical hydrogel properties monitored against time, frequency and strain. If complex notation is used to describe an applied sinusoidal strain, g = g0 exp(iwt), then the complex modulus of the tested material is G*(w) = s*/g* = G′ + iG″ with G′ and G″ as the real (i.e. elastic or in phase) and imaginary (i.e. viscous or loss or out of phase) components of G*, respectively.21 The loss factor, tan δ, is defined as G″/G′. To re-emphasize, G′ measures the deformation energy stored during shear process of a test material (i.e. the stiffness of the material) and G″ is representative of the energy dissipated during shear (i.e. the flow or liquid-like response of the material). If G″ > G′ (tan δ > 1), the sample behaves more like a viscous liquid while, conversely, when G′ > G″, and, thus, tan δ < 1, the sample behaves more like an elastic solid.17,18
For gel samples, these parameters are often measured as a function of time, strain and frequency. By monitoring the temporal evolution of G′ and G″ one can actively observe gelation. By monitoring the moduli vs. strain one can determine the linear viscoelastic region within which G′ and G″ are independent of shear strain. By measurement of the moduli vs. frequency one can easily observe the behavior of the hydrogel at short vs. long time scales. The frequency dependence of the moduli is a critical hydrogel characteristic to observe and appreciate since a single material can look quite solid-like (G′ ≫ G″) at a high frequency/fast timescale but behave much more liquid-like (G″ > G′) at low frequency/long time scales. Gelation kinetics and final gel stiffness are critical hydrogel material properties that directly impact final uses of the materials such as successful, homogeneous encapsulation of a mammalian cell payload and the desired behavior of a cultured cell population, respectively. In addition, the appropriate strain and frequency range for linear visco-elastic behavior is critical to be assessed initially.
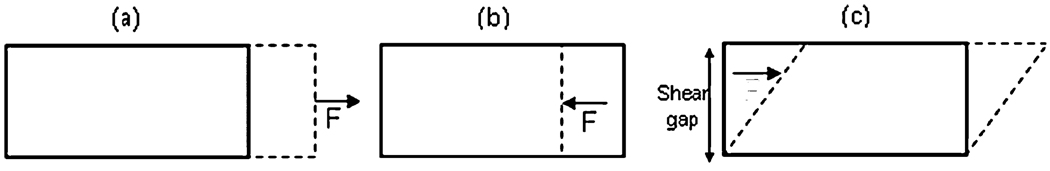
Measuring principles of tensile tests and compression tests resemble those of oscillatory shear tests except that the gels undergo uniaxial deformation during elongation or compression rather than a displacement gradient across a shear gap, as illustrated in Fig. 2.
Fig. 2.
(a) Elongational deformation in a tensile test. (b) Compressional deformation in a compression test. (c) Displacement gradient across the shear gap in an oscillatory shear test.
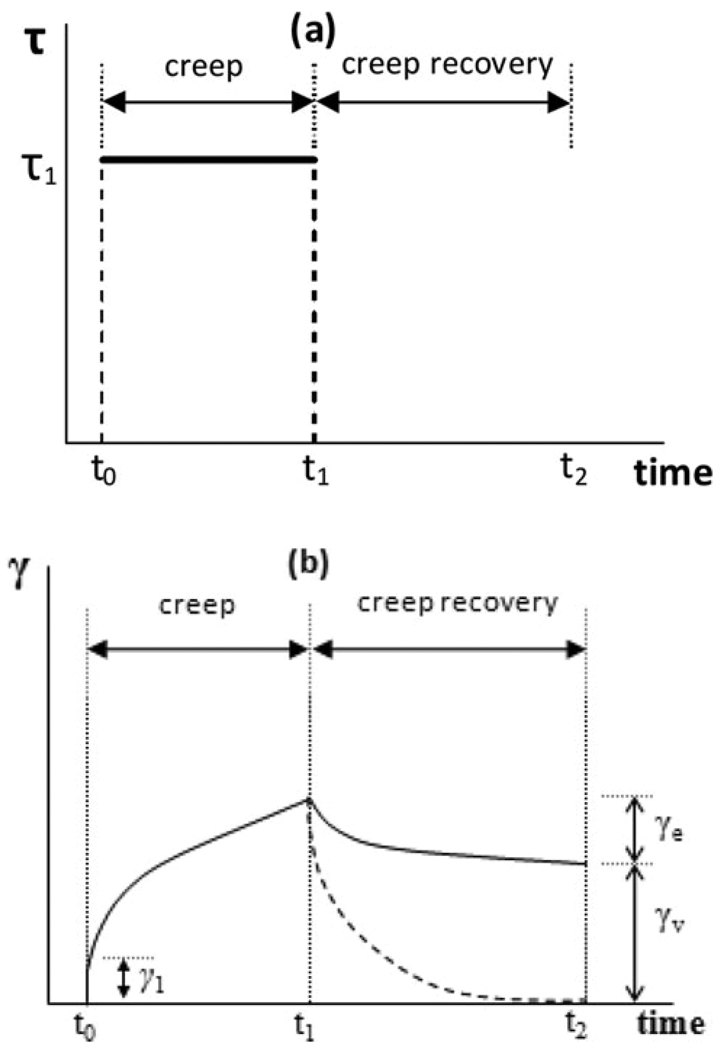
As an alternative to SAOS measurements, creep and creep recovery tests17 are also adopted to investigate the time-dependent evolution of compliance which helps understanding long-term viscoelastic behavior of hydrogels.22,23 The characterization of compliance is important since various mammalian cell types exert stress on hydrogel scaffolds and exhibit different behaviors in response to the compliance of the gel scaffolds.24,25 In principle, creep and creep recovery tests are often performed in consecutive order (Fig. 3a); there is an instantaneous step raise of stress from 0 to τ1 that is then kept constant from t0 to t1 in the creep phase. Then the stress is completely removed in the following recovery phase. The resulting strain is recorded as a function of time (t0 < t < t2) in both tests.
Fig. 3.
(a) Creep test—stress is suddenly raised to τ1 which lasts from t0 to t1; creep recovery—stress is removed completely at t1 and τ = 0 between t1 to t2. (b) Resulting strain as a function of time. γ(t) is different at creep recovery phase for a viscoelastic material (solid line) and an viscoelastic solid (dash line).
In Fig. 3b, temporal evolution of strains for a viscoelastic material and an elastic solid is illustrated. In the creep phase, the strain curves basically overlap, displaying an immediate strain jump of γ1 due to pure elastic response to sudden application of τ1 at t0 and a subsequent time-dependent increase of strain. When steady state is reached at later stages of creep, the strain versus time curve is linear and the slope equals the applied stress τ1 over zero-shear viscosity η0. However, in the following recovery phase, the elastic solid manages to recover completely while the viscoelastic material only recovers part of the deformation γe (here we assume the interval between t1 and t2 is sufficiently long). Hence, the permanent deformation, γv, represents the viscous portion of a material and γe the elastic portion. As practical examples, polymer melts are observed to behave like a viscoelastic material with some solution-like properties while different types of crosslinked gels can exhibit elastic, solid-like behavior.18,22 Hydrogels from peptidic molecules can display behavior anywhere between solid-like and solution-like behavior depending on the hydrogel design.
The creep compliance is defined as J(t) = γ(t)/τ0 which has a unit of reciprocal modulus (1/Pa). Within the linear viscoelastic region, like all other measured parameters, the creep compliance is independent of applied stress and therefore all J(t) curves obtained under various stresses should overlap with each other. Sometimes creep compliance is compared to reciprocal shear modulus measured in small amplitude oscillatory shear tests in order to judge if the sample displays pure elastic behavior.22,23
In addition to the measurements described above, it is essential to assess the properties of hydrogels during flow as well as their abilities to retain or recover their solid form morphology and rigidity after experiencing shear flow or large strain. As stated previously, shear-thinning and self-healing hydrogels can be excellent candidates for injectable therapeutic delivery vehicles. Monitoring rheological behavior and structural evolution of these gels during and after flow can help evaluate encapsulated therapy retention and delivery during syringe injection and the ability of the material to stay localized after injection against possible biological forces in vivo.14,15
In the literature to be discussed in this review, peptide and polypeptide molecule chemistry as well as sample conditions are varied in order to examine whether gel rheological behavior is dependent on factors like peptide sequence, peptide concentration, temperature, pH and so on. For various gel systems, research efforts have been devoted to establish models derived from theories in order to explain the relation among these experimental parameters. These theories and modeling will only be discussed here in detail if necessary. Instead, we present recent progress on the design and observation of peptide and polypeptide hydrogel rheological behavior. While we will briefly discuss the properties of peptidic hydrogels during and after flow, the review will not focus generally on the broader field of non-linear viscoelasticity that measures and models properties such as shear thinning and strain hardening in polymeric systems. We will focus on linear viscoelastic properties observed with rheology. In addition, while we’re focusing on bulk rheology characterization of peptidic materials, there is also the growing field of manipulation and observation of probe particles embedded in desired gels or solutions to determine microrheological properties. While not the focus of this review, there are other reviews that thoroughly discuss the uses of micro-rheology.26,27 We primarily focus on recent peptide-based gelling systems but also discuss several classic studies. For organizational purposes throughout the remainder of the review, peptidic and polypeptidic hydrogels are categorized with respect to their molecular origin and design. First, natural protein-based hydrogels are discussed. Next synthetic peptide and polypeptide-based gelling systems are discussed by class, e.g. hydrogels stabilized by interactions between helices, coiled coils or β-sheet; elastin-like polypeptides; peptide amphiphiles; and, lastly, short peptides.
II. Gel systems based on natural polypeptides
Natural polypeptide-based hydrogels are potentially good candidates for tissue engineering and drug delivery as they meet a majority of the design criteria for biomaterials.3,10 Some of these molecules, such as gelatin, are derived from natural protein sources,28 therefore displaying possible biocompatibility and biodegradability. Other natural polypeptides, such as silk sequences, have the mechanical properties desired for scaffolds to support tissue constructs.29 Although relevant studies have been ongoing for decades, these natural gel-forming materials are not fully understood. For instance, the exact kinetic pathway of gelation, as well as the precise gel point, for many natural proteins remains unclear. However, the significant efforts being made to understand the rheological properties of natural protein hydrogels clearly reveal the potential of these natural materials as biomaterials in the future.
(a) Gelatin gels
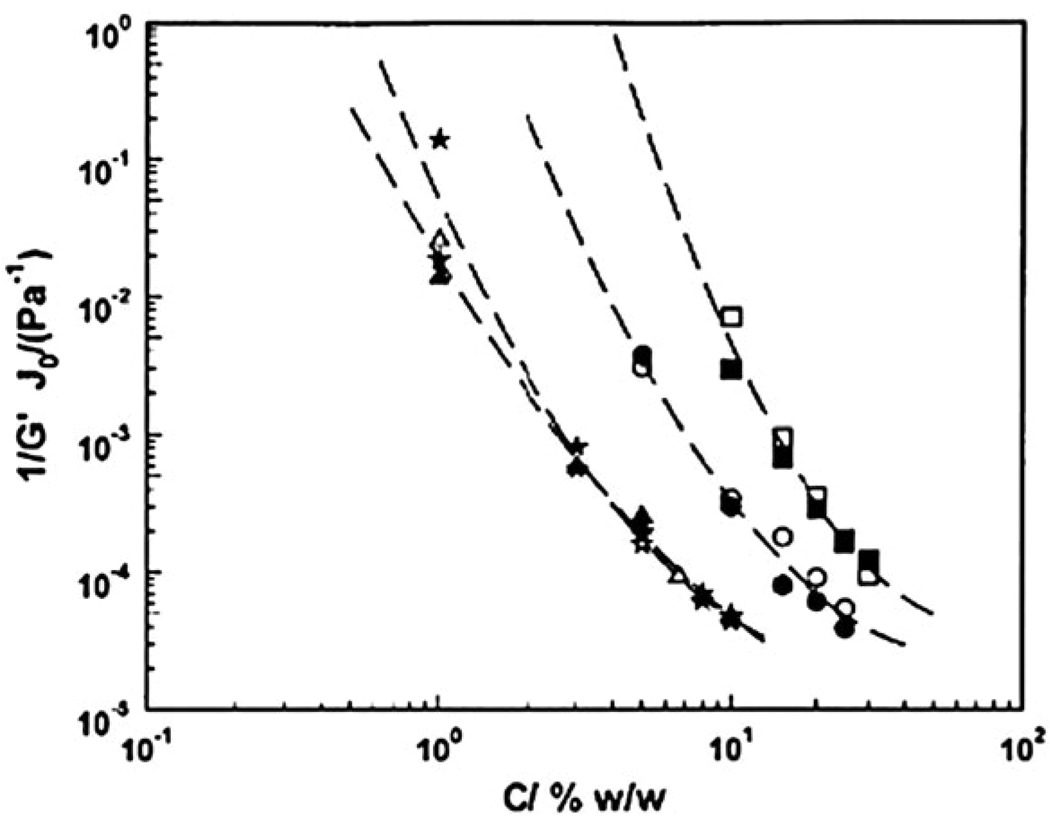
In addition to commercial applications in food, pharmaceutical and photographic industries,1 gelatin gels have been utilized for biomedical applications such as protein delivery30,31 due to their biocompatibility and thermo-responsive gelation. Gelatin is usually produced by denaturing naturally derived collagen in solution through either acidic or alkaline process,28 during which the triple-helical tropocollagen is separated into three single-strand gelatin molecules. When dissolved in warm water, these molecules have a random-coil conformation but undergo a coil–helix transformation when cooled.32,33 On formation of helices, gelation takes place leading to a thermo-reversible physical gel if the protein concentration is above a threshold value, the critical gelling concentration, C0.32–34 It is now generally accepted that the thermal stability of gelatin is closely relevant to the amount of the different amino acids proline, hydroxyproline and glycine in the original collagen source.35–37 A lower content of these amino acids produces a lower propensity of intermolecular helix formation, therefore leading to a higher C0 and a lower temperature of sol–gel transition.36 This coincides with observed gelling behavior of fish gelatins in contrast to that of mammalian gelatins due to different content of the three amino acids.22,38,39 Through small amplitude oscillatory shear measurements, Gilsenan and Ross-Murphy investigated how gelatin concentration and molecular weight affected gel melting temperature and the effects of gelatin concentration on equilibrium gel modulus. The rheological results were modeled in order to summarize the relation between these parameters.38,39 In addition, creep and creep recovery measurements were performed to observe long-term viscoelastic behavior of gelatin gels.22 The initial creep compliance obtained at the beginning of creep measurements (representative of pure elastic response) was compared to the inverse of the equilibrium gel modulus in dynamic oscillatory measurements, as illustrated in Fig. 4.22 The two parameters were similar in value and almost overlapped at higher gelatin concentrations, a strong indication that these physical gels possibly behave like elastic solids. However, there was a part of the deformation unrecovered at the end of recovery phase that indicates the gelatin gels possibly behave like a viscoelastic solution.17 To date, this contradiction of displaying both elastic solid and polymer liquid behavior remains and further attempts are needed to properly model this particular gel system.
Fig. 4.
Log J0 (closed symbols) and log 1/G′ (open symbols) plotted against log concentration tilapia-derived gelatin (triangles), bovine gelatin OC1 (stars), and two cod gelatin samples, IC (circles) and 2747 (squares).22 (Reprinted from G. M. Gilsenan et al., Shear creep of gelatin gels from mammalian and piscine collagens,22 permission from Elsevier).
(b) Globular protein-based hydrogels
Gelation of globular proteins is of practical industrial importance to structure fluids. To reduce problems arising in commercial production of foods and pharmaceuticals, it is essential to understand the fundamental mechanisms of gelation.40
Heat-induced gelation of globular proteins is often adopted in the production of food. During heat denaturation, globular proteins unfold allowing individual protein molecules to aggregate due to interactions including hydrogen bonding, hydrophobic effects, etc. Aggregation will lead to formation of a gel if the protein concentration equals or exceeds the critical gelling concentration. Generally there are two types of heat-set globular protein-based gels: particulate gels and fine-stranded gels. The former is often formed in a solution where the pH is close to isoelectric point (pI) which causes proteins to aggregate, ultimately forming a turbid gel composed of large aggregates.41–44 If the solution pH is far from pI, heat-induced gelation can give rise to a fine-stranded gel42,45–51 comprised of either curved, flexible strands or rigid, linear fibrils42,45,52–54 sometimes termed as “amyloid” fibrils due to their resemblance of amyloid protein fibrils.53,55
Over the years, comprehensive studies have been performed on a number of globular proteins such as bovine serum albumin, β-lactoglobulin and ovalbumin to correlate their rheological properties with structural and molecular characteristics.41,52,56–59 Based on experimental results, extensive efforts were taken to develop gelation models to further explain gelation processes. Most models try to summarize and predict effects of protein concentration, pH, temperature and ionic strength on gelation time, critical gelation concentration and equilibrium gel modulus. In the work of Clark, Ross-Murphy and related scholars, several models were applied to describe the gelling behavior of fibrillar β-lactoglobulin gels formed at both acidic and basic pH.60–66 Through comparison, these models were judged in their ability to describe experimental data.65,66 van der Linden et al. demonstrated an adjusted random contact model that described well the dependence of critical percolation concentration on ionic strength for a couple of fine-stranded gels.54,55,67,68 Other comprehensive studies involved establishing models that consider data on the aggregation process and hydrogel rheology but that also adopted techniques complementary to rheology such as light scattering.52,69–73
Lysozyme is a small globular protein existing in hen egg white and an interesting, recently considered, biomaterial candidate. The uniqueness of this protein is that lysozyme-based hydrogels are cytocompatible to living fibroblast cells, suggesting that globular protein-based hydrogels may be useful as scaffolds for tissue engineering.74,75 Miller and colleagues primarily focused on thermoreversible lysozyme-based gels formed in mixtures of water and dithiothreitol. They reported that gelation of lysozyme was achieved by heating the protein solution up to 85 °C and then slowly cooling back to room temperature, during which lysozyme proteins were denatured and formed β-sheet-rich fibrils that further entangled into a gel network. The resulting lysozyme-based hydrogel was reversed to the solution state upon heating.74–76 Gelation behavior and mechanical properties were monitored in dynamic oscillatory measurements. It was found that the critical gelation concentration of lysozyme was around 3 mM and gel of higher lysozyme concentration had a higher melting temperature.75,76 The plateau elastic modulus was found to be dependent on lysozyme concentration in a power law relation.75,76
(c) Fibrous protein-based hydrogels
Silks are extensively studied fibrous proteins that can produce fibers with mechanical properties superior to almost any other biopolymer or synthetic polymer in aspects of higher Young’s modulus and ultimate tensile strength.77–79 In addition, silk proteins are biocompatible and degradable,80–82 motivating various studies to explore silk gels as scaffolds for tissue engineering83–85 or blends with gelatin for purposes of drug delivery.86,87
Studies on natural silk protein usually focus on the fibroin material in silkworm cocoon and spidroin in dragline silk produced by spiders.88,89 Silk fibroin is the major protein found in natural silkworm fibers.90 At high enough concentration, silk fibroin starts to assemble in solution leading to a physical hydrogel of β-sheet-rich fibrils.91 Extensive studies have validated that silk fibroin hydrogel is cytocompatible and biodegradable.29,83,84,92–95 It was found that raising silk fibroin concentration or temperature resulted in gels with higher compressive strength and moduli.96 Gelation kinetics of fibroin was observed to be faster at low pH (<5), high temperature (>60 °C), or at appropriate ionic strength.83,84,91,96–98 Research by Kang et al. and Yoo et al. showed that stability99 and rigidity100 of silk fibroin hydrogel were affected by the amount of the polymer, poloxamer 407.99,100 Recently, Wang et al. successfully speeded up gelation of silk fibroin under physiological condition (37 °C and pH 7.4), enabling homogeneous encapsulation of living mesenchymal stem cells in three-dimensions.101 Yucel et al. showed that at a lower fibroin concentration, the vortexing of the protein solution also reduced the time needed for complete sol–gel transition. Moreover, these vortex-induced silk hydrogels shear-thinned upon syringe delivery and recovered their rigidity immediately after removal of injection shear.102 Synthetic silk–elastin-like hydrogels will be discussed later together with elastin-like polypeptide-based hydrogels.
Vollrath et al. reported that gelation of spidroin proteins was initiated at pH 5.5 and the resulting hydrogel was reverted to a solution at pH 7.103 Synthetic spidroins can also undergo a sol–gel transition and form fibrillar networks stabilized by chemical or physical cross-links. The physical spidroin gels were reported to be easily disrupted due to the fibril entanglement nature of crosslinks104–106 while the chemically crosslinked gels were much stiffer with elastic moduli up to around 1000 Pa.106
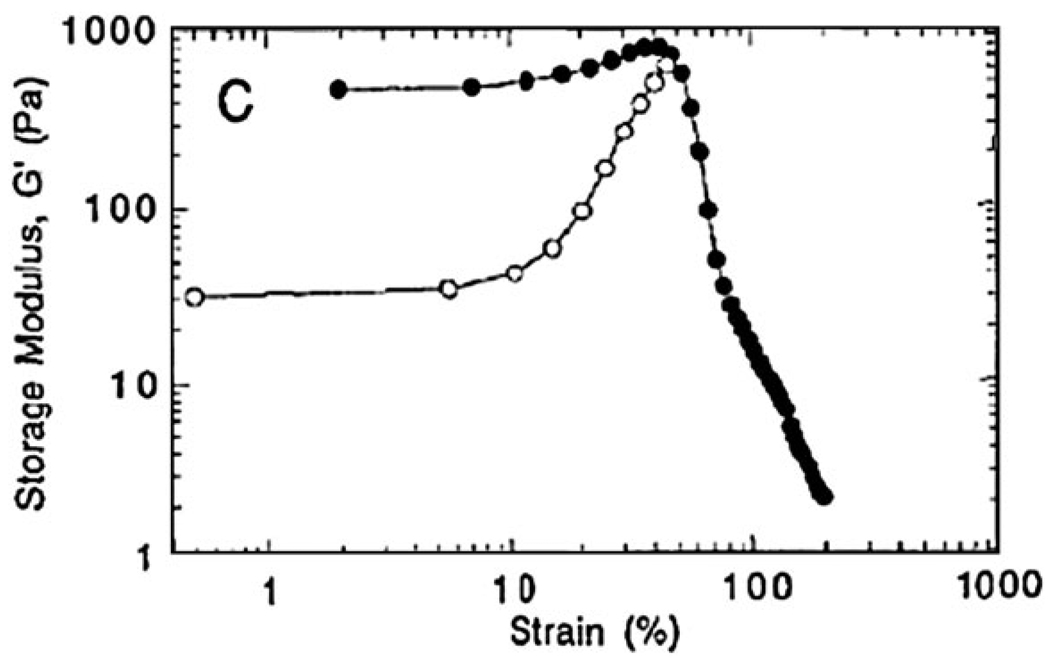
Fibrin is another natural, fibrous protein that has been shown to be useful as a biomaterial for wound healing and potentially as a vehicle for growth factor delivery.107,108 When an injury occurs to a blood vessel, fibrinogen is covalently crosslinked by thrombin, forming fibrin, which is further crosslinked by factor XIII into a network (fibrin clot) that coagulates the blood.109 As early as 1970’s, Fukada and Kaibara adopted various techniques110,111 to study dynamic rheological properties of fibrin clots as well as mechanism of blood coagulation.112–118 Ryan et al. performed a detailed investigation on the fundamental rheological behavior of fibrin clots and related that to structural characteristics, discovering that mechanical rigidity of fibrin clots was dependent on the concentration of thrombin, calcium, fibrinogen and, therefore, network and crosslink densities.119 By adopting inhibitors that prevent certain types of cross-links, the same group found that some species of crosslinked fibrin chains helped stiffen fibrin clots and proposed a mechanism to explain how this happened.120 Urech et al. reported that a high concentration of the ligand L1Ig6 incorporated into a fibrin matrix lowered the crosslink density of the resulting fibrin clot. However, this effect was neutralized by adding more factor XIII at a low concentration of the ligand.121 One unique property of fibrin hydrogel is strain stiffening.108 Shah and Janmey observed that with increasing shear strain the G′ of fibrin clots remained constant first but then started to grow sharply (Fig. 5). G′ at 50% strain was almost 20 times higher than that at small strain (<10%). However, the strain stiffening behavior of fibrin gels was obscured by the addition of platelets that raised the storage modulus at small strain.122 Parallel theoretical work was also carried out to yield rational explanations responsible for the strain hardening behavior of fibrin gels.123
Fig. 5.
Strain dependent storage modulus of fibrin gels with platelets (filled circles) and without platelets (open circles).122 (Reprinted from J. V. Shah et al., Strain hardening of fibrin gels and plasma clots,122 permission from Springer).
III. Synthetic biomimetic-polypeptide hydrogels
Although hydrogels from natural polypeptides can display excellent biocompatibility and biodegradability, limitations of their use as biomaterials do exist. For instance, these self-assembling physical gels are generally weak and lack batch-to-batch consistency regarding gel properties since it is difficult to maintain identical sample composition from natural sources.32 As an alternative approach, sequences of synthetic polypeptides can include segments mimicking those of natural polypeptides so that resultant materials inherit natural biofunctionality. For example, functional epitopes can be included to provide a biological function as well as to promote the formation of secondary bonds, thereby enhancing both mechanical and biological performance of resulting hydrogels. Hydrogels based on synthetic polypeptides are, therefore, promising since structural features and functionalities of the gel network can be manipulated as desired simply by further engineering the peptide sequence.
(a) Hydrogels based on coiled coil structures
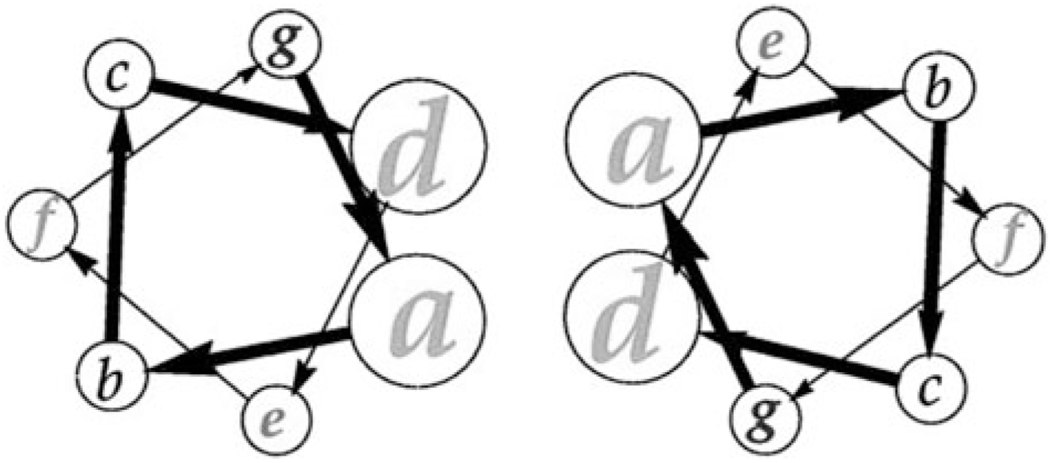
The coiled-coil is one of the major structural motifs for proteins and normally involves a seven-residue heptad periodic unit (abcdefg), as illustrated in Fig. 6.124 Positions a and d are generally occupied by hydrophobic residues responsible of interhelical hydrophobic interaction. Putting responsive residues in positions a, d, e and g results in peptide molecules responsive to various environmental stimuli and that can self-assemble into supramolecular nanostructures.124
Fig. 6.
A coiled-coil dimer based on heptad sequence repeat (abcdefg).124 (Reprinted from B. Ciani et al., A designed system for assessing how sequence affects α to β conformational transitions in proteins,124 with permission from the American Society for Biochemistry and Molecular Biology).
Hydrogelation of many synthetic peptidic systems can be driven by coiled-coil formation.125 A major approach involves putting coiled-coil-forming segments like leucine zippers into block copolymer polypeptides.125–133 Tirrell and colleagues designed a multidomain protein AC10A composed of a middle random coil block (C) flanked by two associative leucine zipper blocks (A) at both ends.132,134 Self-assembly of the leucine zipper domains led to a transient hydrogel network above the critical gelling concentration. However, these hydrogels were reversed to solutions at high pH or high temperature due to denaturation of the leucine zipper domains.131,132 Detailed rheological characterization was performed to investigate the relationship between the equilibrium gel modulus and the concentration, pH as well as ionic strength. The impact of these factors on equilibrium gel modulus was fundamentally attributed to network topology.135 Also, this leucine zipper hydrogel exhibited a sharp pH-dependent transition in viscosity due to the dynamic properties of the network.136 Similar coiled-coil-containing block copolypeptides were presented in recent work by Xu and Kopeček, in which the gelation process was monitored by microrheology, and the sol–gel transition was found reversible on addition or removal of guanidine hydrochloride that denatured the coiled-coil domains.137 These responsive hydrogel systems can be potentially applied for controlled release of DNA and protein delivery.131,132,137
Woolfson and colleagues reported that by incorporating characteristics of two secondary structures, the α-helix and β-hairpin, into one peptide sequence, the resulting peptides were capable of transforming from an α-helix into a β-hairpin upon heating. Following the conformational change, a fibrous gel was formed which was then turned back to a solution at pH 2.124 Recent work from the same group presented an α-helical peptide-based hydrogel system comprised of self-assembled fibres.138 All the peptides studied were based on a coiled-coil heptad repeat motif (abcdefg), and an earlier design for enhancing dimer stability was maintained.139 However, the amino acids at positions b, c and f that are exposed on the surfaces of coiled-coil assemblies were replaced with three alanine residues to promote intermolecular interaction. After complementary peptides were mixed at ice-cold temperature or room temperature, self-supporting physical gels were observed to be stable up to 95 °C. Such a hydrogel system was found to promote both growth and differentiation of neural cells, indicating potential applications for tissue engineering.139
Deming and coworkers studied hydrogelation of polypeptide diblock copolymers that were composed of a hydrophilic charged segment and a hydrophobic segment.140–142 By varying polypeptide sequence, it was found that the charged polyelectrolyte block was responsible for sol–gel transition determination and the equilibrium gel modulus was dependent on hydrophilic to hydrophobic ratio and block length. Factors like conformational differences in the hydrophobic segment and solution ionic strength affected the critical gelling concentration. In addition, these hydrogels were responsive to mechanical shear. After being disrupted under high shear strain (1000%), these gels were capable of recovering 80–90% of initial rigidity in a rapid manner.140–142 The fast recovery was attributed to two possible reasons. First, under high strain, only the interconnections between some gel domains were broken while the rest remained intact, thus enabling faster bulk gel recovery. Second, the hydrophobic segments (polyleucine) were interconnected in a layer of packed α-helices, which required less molecular alignment to reform than hydrogen bonds for β-sheet.143 More shear-thinning and self-healing hydrogels will be discussed in the following section.
(b) Hydrogels based on β-sheet structures
In addition to the coiled coil, β-sheet is another structural motif of peptides ubiquitous in natural proteins such as silk fibroin91 and amyloid fibrils formed as a consequence of neurodegenerative diseases.144 The β-sheet motif is also exploited to design responsive peptidic materials, some of which form amyloid-like fibrils145,146 while others self-assemble and form hydrogel networks that are being studied as injectable hydrogels for tissue engineering and drug delivery.13,147
Messersmith and coworkers designed a 16-amino acid peptide, FEK16, consisting of alternating hydrophobic and hydrophilic residues.148 Above a concentration of 10 mg mL−1, FEK16 peptides formed a β-sheet fibrillar gel in aqueous solution, and gelation was largely accelerated when exposed to monovalent or divalent ions. To initiate hydrogelation, temperature-responsive or light-responsive liposomes were adopted to release CaCl2 at 37 °C or when exposed to near-infrared light.148
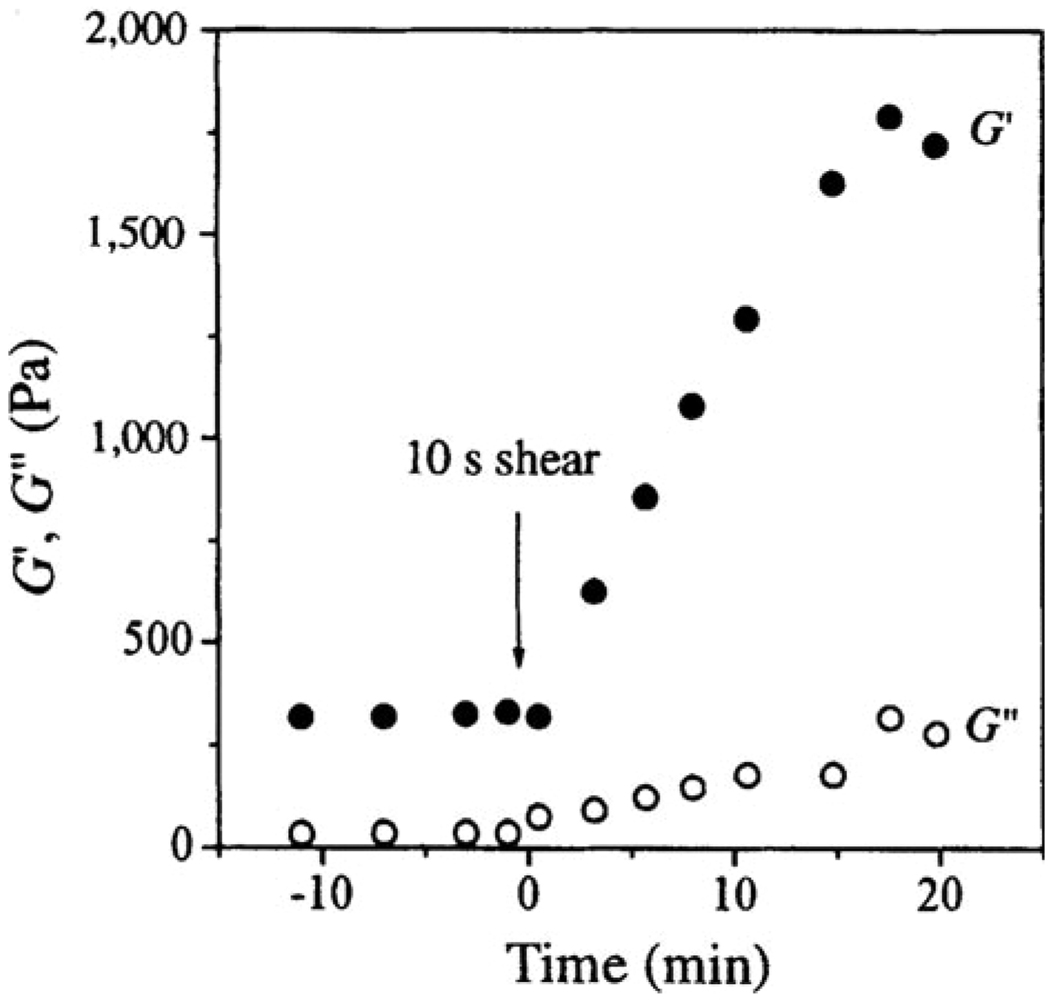
In early work by Aggeli and colleagues, the polypeptide K24 was designed according to the transmembrane domain present in the IsK protein. In 2-chloroethanol, K24 was capable of self-assembling into β-sheet tapes that entangled with each other and formed a hydrogel (~300 Pa) at low peptide concentration. Interestingly, both G′ and G″ displayed an immediate growth in value (Fig. 7) after the hydrogel was sheared at 5/s for ten seconds. The storage modulus settled around 1700 Pa after ten minutes. The authors attributed this unusual rheological behavior to the “annealing of structural defects” induced by shear flow, which remains to be verified.149
Fig. 7.
Increasing shear storage modulus (closed circles) and loss modulus (open circles) after the hydrogel was sheared at 5/s for 10 seconds.149 (Reprinted from A. Aggeli et al., Responsive gels formed by the spontaneous self-assembly of peptides into polymeric β-sheet tapes,149 with permission from Nature Publishing Group).
Pochan and Schneider et al. previously presented β-hairpin peptides that were designed to fold into β-hairpins by exposure to a desired environmental stimulus and consequently assemble into a highly physically crosslinked hydrogel network.13,150–154 The original twenty-amino acid peptide, MAX1, ((VK)4-VDPPT-(KV)4-NH2) has a tetra-peptide turn group in the middle of the sequence and two neighboring strands of alternating hydrophobic valine residues and hydrophilic lysine residues. When dissolved in solution at pH 7.4 and low ionic strength, MAX1 peptide is in a random coil conformation because the positively charged lysine residues prevent intramolecular folding of the two strands. The addition of salt screens this electrostatic repulsion and allows the peptides to fold into β-hairpins and then, subsequently, self-assemble into a rigid fibrillar gel network. MAX8 peptide (VKVKVKVK-VDPPT-KVEVKVKV-NH2) was designed based on the sequence of MAX1 by replacing the lysine residue at position 15 with a glutamic acid residue.13 This substitution reduced the total positive charge to be screened and enabled swifter folding and self-assembly kinetics. Therefore, at the same peptide concentration and buffer conditions, MAX8 undergoes faster gelation yielding a gel more rigid than MAX1.13,147,155 For both peptides, self-assembly kinetics can be manipulated by varying peptide concentration, ionic strength and temperature.16,147,152,155,156 Correspondingly, gel stiffness and network properties can be optimized for homogeneous cell encapsulation in three-dimensions13 and controlled release of therapeutics.147 It was also validated that both MAX1 and MAX8 hydrogels are cytocompatible13,154,155 and potentially noninflammatory,157 strongly indicating promise as candidates for biomedical applications and biotechnology.
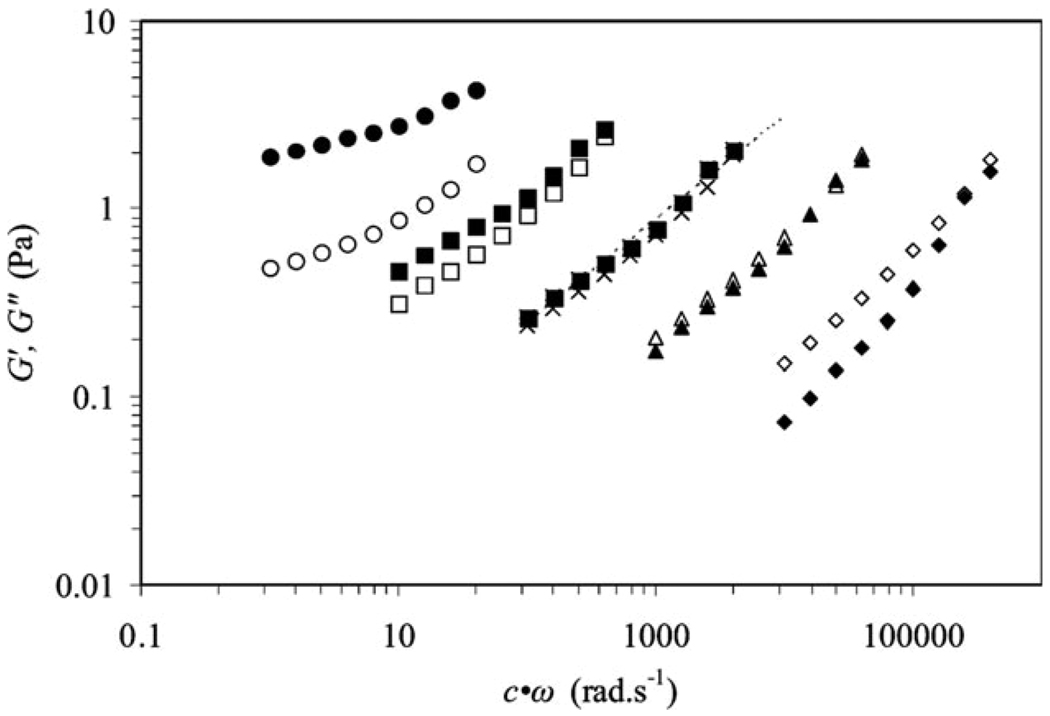
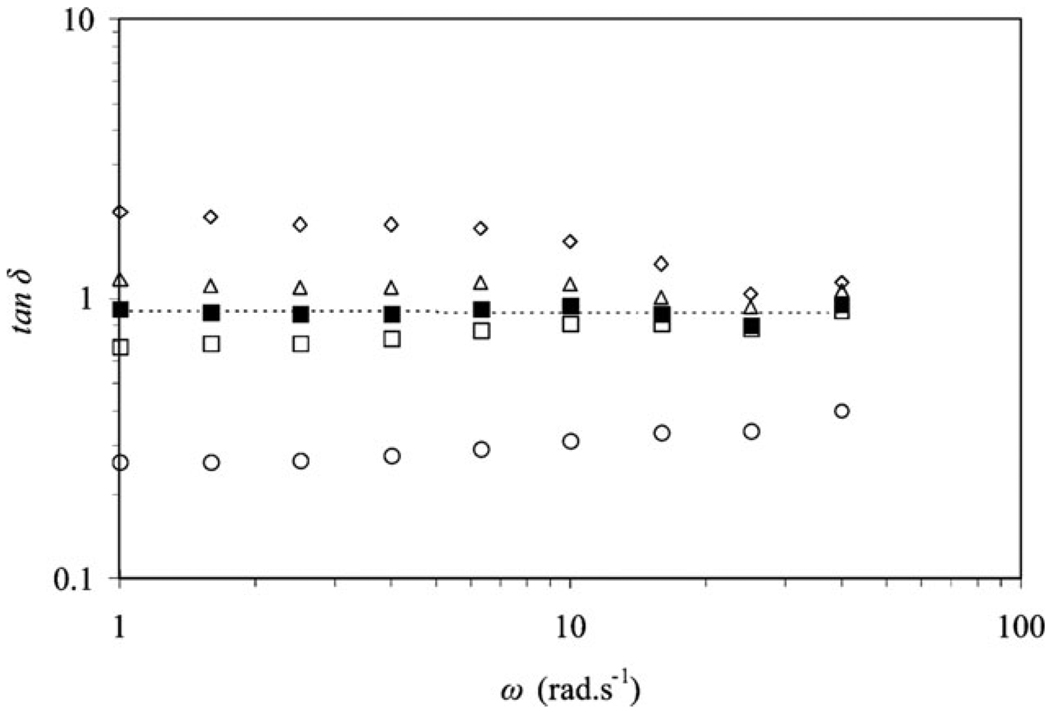
Yucel et al. successfully elucidated mechanisms of early-time hydrogelation for β-hairpin peptides via comprehensive analysis and characterization by rheology, dynamic light scattering (DLS) and cryo-TEM.16 According to Chambon and Winter, the critical gel point corresponds to the timepoint at which the fibril clusters “first span the whole sample volume”,158 corresponding to the moment when G′ and G″ display the same power-law frequency dependence (Fig. 8) and the loss factor, tan δ, remains constant within the frequency range studied (Fig. 9).16 The gelation point in the system Yucel et al. studied was determined to be 56 min after self-assembly was initiated, which was consistent with observations by DLS.16 It should be pointed out that to evaluate the gelation point in the context of Chambon and Winter, it is critical to examine the frequency dependence of G′, G″ and tan δ rather than simply looking for the crossover point of G′ and G″ in a dynamic time sweep. However, for practical reasons of requiring a stiff gel for use as a biomaterial, having a material with G′ > G″ at all frequencies is usually qualitatively sufficient to characterize how a particular gel will behave in an application.
Fig. 8.
Dynamic oscillatory rheology frequency sweep data collected at different time points (γ = 10%): G′ (filled symbols) and G″ (open symbols).16 (Reprinted from T. Yucel et al., Direct observation of early-time hydrogelation in β-hairpin peptide self-assembly,16 with permission from American Chemical Society).
Fig. 9.
Frequency dependence of the loss factor at different time points (γ = 10%).16 (Reprinted from T. Yucel et al., Direct observation of early-time hydrogelation in β-hairpin peptide self-assembly,16 with permission from American Chemical Society).
Importantly, both MAX1 and MAX8 hydrogels are highly responsive to mechanical shear. When exposed to a proper shear stress, they shear-thin and flow with very low viscosity.13,152 However, once the stress is removed the gels immediately self-heal into solids and eventually restore their original rigidity with short time relative to the shear rate and duration applied. This unique shear-reversibility indicates the possibility of in vivo delivery by syringe injection of a solid gel construct with a desired therapeutic payload. The power of this strategy is that the well understood material ex vivo has the same properties as the final material in vivo. Therefore, direct correlation of biological properties with material attributes can be made. We are currently both using and studying the hydrogel behavior during and after flow in order to demonstrate that they are excellent candidates for injectable therapeutic delivery vehicles and tissue regeneration substrates. We hope to exactly elucidate the underlying mechanisms that explain the shear-thinning and self-healing behavior.
The decapeptides designed by Yu et al., KVW10 and EVW10, were composed of alternating hydrophobic valines and cationic lysines (for KVW10) or anionic glutamic acids (for EVW10), giving rise to their self-repulsive but intermolecularly attractive nature. Upon mixing these peptides, gelation occurred even at the low peptide concentration of 0.25 wt%.159,160 The resulting decapeptide hydrogel was capable of recovering around 90% of its pre-shear rigidity after being disrupted repeatedly (200% shear strain for 2 min).159,161 By varying peptide sequence, it was found that values of equilibrium gel modulus and yield strain were dependent on hydrophobicity of the apolar residue. Also by varying sequence length, hydrogels based on a shorter sequence were found to be stiffer than those based on a longer sequence.
Recently, Aulisa et al. presented their work on triblock copolypeptides that self-assembled into a hydrogel of β-sheet-rich fibrils under physiological conditions. After these multidomain peptide-based hydrogels experienced 100% shear strain for 60 seconds, they quickly recovered their mechanical rigidity upon removal of shear. It was found that values of plateau gel modulus and ratio of rigidity recovery varied among different peptide sequences.162
(c) Elastin-like polypeptide-based hydrogels
Elastin is a natural protein ubiquitous in elastic tissues like artery, skin and vocal fold.163,164 Elasticity and mechanical reversibility of elastin allow these tissues to extend upon the addition of a stress and recover once the stress is released.164,165 To inherit these characteristics, many elastin-like polypeptides (ELP) were either chemically synthesized or expressed in E. coli with the characteristic pentapeptide repeat unit, valine-proline-glycine-X-glycine (where X is any residue but proline), retained in the primary sequence. The choice of the guest residue determines the critical temperature of inverse phase transition, above which the solubility of ELP in solution decreases and consequent ELP aggregation occurs.166,167 This thermodynamic characteristic was utilized to develop thermal-reversible gelling systems for cartilage repair.168,169 Although ELP coacervates were demonstrated to be cytocompatible,168–170 they were mechanically too weak168 to function like natural connective tissues.171 Therefore, various attempts were taken to fabricate ELP hydrogels in order to enhance their mechanical and rheological properties. The major approach is to design the amino acid sequence of ELP with more covalent cross-link sites incorporated. Cross-linked ELP hydrogels did display improved mechanical properties over ELP coacervates.170,172–175 Among these works, Chilkoti and coworkers performed detailed studies on how temperature, ELP molecular weight, concentration, and lysine content affected the swelling ratio and dynamic shear modulus of ELP hydrogels cross-linked by tris-succinimidyl aminotriacetate.172 It is worth mentioning that these chemical ELP hydrogels were not only cytocompatible but also comparable to natural connective tissue with respect to shear modulus or Young’s modulus.11
A second approach to enhance mechanical properties of ELP hydrogels is to reduce the propensity of phase separation in ELP solutions.11,174–177 These ELPs were triblock copolymer polypeptides, derived from an elastin-mimetic sequence, composed of a central hydrophilic block with a lower critical solution temperature (LCST) well above 37 °C and two hydrophobic end blocks with an LCST below 37 °C. At physiological temperature, the ELP solution underwent gelation instead of phase separation due to the bridging effects of the middle block. Since mechanical and rheological properties were dependent on factors like polypeptide sequence and ionic strength, the range of accessible rheological and mechanical properties were thus expanded.11,174–177
Incorporating residue repeats derived from silk into ELP sequences gives rise to silk–elastin-like polypeptides (SELPs), hydrogels that have been employed for release of drugs178 and DNA.179 Through rational design, SELPs can undergo an irreversible sol–gel transition, the kinetics of which can be accelerated at elevated temperature. Several studies reported in situ physical gelation of SELP solution after it was delivered via syringe injection, indicating potential application of SELP as an injectable gel-forming system.180–183 Since the sequence of SELP is characteristic of both silk and elastin, one can control properties like gelation kinetics, gel rigidity and environmental responsiveness of the resulting SELP-based hydrogels, which is very promising for controlled release of therapeutics.180–183
(d) Hydrogels based on peptide amphiphiles
The fundamental sequence of a designed peptide amphiphile (PA) is a hydrophilic peptide sequence covalently bonded to a hydrophobic aliphatic segment. The PA hybrid motif is being studied for tissue repair and the controlled release of therapeutics184 since PA systems form hydrogel materials and can be further functionalized simply by the use of a biological ligand as a head group of the peptidic segment of an individual PA.185 In addition, amino acids in the peptidic segment and monomers in the alkyl tail can be varied to modify the pathway of self-assembly and the physical features of the final self-assembled structure.186 Varieties of peptide amphiphiles have been designed to function as templates for biomineralization187,188 or as injectable scaffolds for therapeutic delivery189–191 and tissue regeneration.185–192
Due to molecular amphiphilicity, PAs self-assemble into nanofibers185,187–201 or nanobelts.186 As mentioned above, at higher concentration the PA-based nanofibers further assemble into fibrillar hydrogel networks.185,187,192–194,198–201 Stupp and colleagues demonstrated that the sol–gel transition was triggered upon mixing acidic and basic PAs at physiological pH.198 It was also shown that in a PA-based gel network the nanofibers were connected by reversible cross-links offered by cysteine residues.187 With the cell-binding ligand IKVAV as the head group, a resultant PA solution mixed with neural cells was injected in vivo of a rat and a solid gel was formed.185
Gelation behavior and viscoelastic properties of PA-based gels were also studied via dynamic oscillatory measurements. Stendahl et al. performed a detailed investigation on effects of pH, ionic strength and type of metal ions on mechanical stiffness of PA gels.194 The Hartgerink group found that mechanical stiffness of PA-based hydrogels first grew but then decreased with increasing calcium concentration. The authors attributed the decrease of storage modulus to possible phase separation that occurred within the gel.200 They also found that stability and mechanical strength of PA-based hydrogels were dependent on the number and position of glycine residue that can hydrogen bond.201 The PA design motif is certainly a rich area of study that is only beginning to be explored.
(e) Hydrogels based on low-molecular-weight peptidic gelators
Hydrogel systems discussed in the previous sections are all based on polypeptides with at least ten amino acid residues in the sequence. As an alternative, oligopeptides of low molecular weight, such as fluorenylmethoxycarbonyl (Fmoc)-protected amino acids, can self-assemble into supramolecular hydrogels and be studied by rheology. Parallel biological studies indicate that these gels are potentially useful for biomedical applications like tissue regeneration202–204 and drug delivery.205–207 Hydrogelation of these short peptides can either be driven by their responsiveness to pH and temperature,203,205,208–211 or initiated due to the presence of a natural enzyme.206,207,212,213,215–217
There are a few examples of the former gelation mechanism. The Ulijn group reported hydrogelation of Fmoc-functionalized amino acids initiated under physiological conditions. Fmoc– diphenylalanine (Fmoc–F2) molecules were found to self-assemble into fibrous hydrogels that support 3D cell culture of living cells.202,204 Mechanical rigidity of Fmoc–F2 gels were tunable via functionalization of the network with different chemical moieties. The resulting gel networks displayed optimized compatibility with various cells.203 Xu and coworkers also presented a series of Fmoc–dipeptides, of which the sol–gel transition was reversible in response to shift in temperature and pH.209–211 In addition, the same group reported hydrogelation in response to the binding of vancomycin where gel stiffness was observed to increase significantly.214 Mahler et al. demonstrated that diluting an Fmoc–F2 stock solution in water to a proper concentration initiated sol–gel transition, leading to Fmoc–F2 gels that were shear-thinning.215 Recently, Adams et al. reported that gelation of Fmoc–dipeptides was induced by hydrolysis of glucono-δ-lactone. The resulting hydrogels were homogeneous and their mechanical properties were reproducible regardless of the pre-shear history they experienced.216
Several groups have also studied enzyme-induced gelation of low molecular weight peptides. Specifically, the Ulijn group demonstrated that hydrogelation of Fmoc–tyrosine–OH was triggered under physiological conditions by the presence of alkaline phosphatase, the concentration of which determined gelation kinetics as well as final gel stiffness.217 Xu and coworkers studied alkaline phosphate-induced hydrogelation of Fmoc–tyrosine at 37 °C, pH 6.0209 and pH 9.6.210 The final gels were reversed to a solution with kinase.211 Other enzymes like thermolysin,213 β-lactamase218 and MMP-9219 have been used to trigger hydrogelation of various hydrogelators.
IV. Conclusions
Peptide-based hydrogels are a class of biomaterials growing in importance for uses in tissue engineering, drug delivery and microfluidics, and the primary experimental method to explore mechanical properties is rheology. A fundamental understanding of peptidic hydrogel mechanical properties and underlying formation and deformation mechanisms is crucial for determining whether these biomaterials are potentially suitable for biotechnological uses.
In this article, we reviewed rheological properties of peptide and polypeptide-based hydrogels by summarizing bulk mechanical properties, gelation mechanisms, and the behavior of hydrogels during and after flow. Although these rheological results alone can be informative and, with further modeling, yield a rational understanding, rheological studies should always be considered in conjunction with structural characterization data (e.g. microscopy and scattering) in order for better understanding of the observed gel properties. As a matter of fact, techniques combining rheology with microscopy or scattering have been adopted to investigate behavior of shear-thinning gels during and after shear flow.
In the future, studies of hydrogel rheology will not be limited to the characterization of fundamental gel properties. More and more focus will be placed on the interplay of hydrogel mechanical and morphological properties with the behavior of cells and/or tissues in contact with the material. Therefore, rheology will be a critical technique to couple with biological and chemical assays in order to understand any new or established biomaterial, particularly those constructed of peptides.
Acknowledgements
The authors would like to acknowledge funding from the National Institutes of Health through grant 5P20RR017716-07 and the National Institute of Standards and Technology (NIST) through grant number DOC #70NANB7H6178.
Biographies

Congqi Yan
Congqi Yan received her BS in Optical Science and Engineering from Fudan University (Shanghai, China) in 2006. She is currently pursuing her PhD in Materials Science and Engineering under the supervision of Professor Darrin J. Pochan at University of Delaware. Her research focuses on characterizing mechanical properties of hydrogels to judge the feasibility of them for biomedical applications.

Darrin J. Pochan
Darrin Pochan is currently Professor in the Materials Science and Engineering Department as well as the Delaware Biotechnology Institute at the University of Delaware. Since joining the department in 1999 after a PhD in Polymer Science and Engineering at the University of Massachusetts, Amherst, and a National Research Council Postdoctoral fellowship at the National Institute of Standards and Technology in Gaithersburg, MD, he has developed a research program around the construction of new materials and nanostructures via molecular self-assembly mechanisms. Areas of focus are biomaterials and materials for nanotechnology and energy applications through organic/inorganic hybrids. Recent honors for Darrin include an NSF Career Award, the DuPont Young Faculty Award, and the Dillon medal from the American Physical Society. Currently, Darrin also serves as Associate Editor for North America of Soft Matter, a new interdisciplinary journal from the Royal Society of Chemistry in the United Kingdom.
Footnotes
Part of the peptide- and protein-based materials themed issue.
References
- 1.Apostolov AA, Boneva D, Vassileva E, Mark JE, Fakirov S. J. Appl. Polym. Sci. 2000;76:2041. [Google Scholar]
- 2.Rajagopal K, Schneider JP. Curr. Opin. Struct. Biol. 2004;14:480. doi: 10.1016/j.sbi.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Lee KY, Mooney DJ. Chem. Rev. 2001;101:1869. doi: 10.1021/cr000108x. [DOI] [PubMed] [Google Scholar]
- 4.Lutolf MP, Hubbell JA. Nat. Biotechnol. 2005;23:47. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 5.Peppas NA, Bures P, Leobandung W, Ichikawa H. Eur. J. Pharm. Biopharm. 2000;50:27. doi: 10.1016/s0939-6411(00)00090-4. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman AS. Adv. Drug Delivery Rev. 2002;54:3. doi: 10.1016/s0169-409x(01)00239-3. [DOI] [PubMed] [Google Scholar]
- 7.Langer R, Peppas NA. AIChE J. 2003;49:2990. [Google Scholar]
- 8.Beebe DJ, Moore JS, Bauer JM, Yu Q, Liu RH, Devadoss C, Jo BH. Nature. 2000;404:588. doi: 10.1038/35007047. [DOI] [PubMed] [Google Scholar]
- 9.Eddington DT, Beebe DJ. Adv. Drug Delivery Rev. 2004;56:199. doi: 10.1016/j.addr.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 10.Slaughter BV, Khurshid SS, Fisher OZ, Khademhosseini A, Peppas NA. Adv. Mater. 2009;21:3307. doi: 10.1002/adma.200802106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow D, Nunalee ML, Lim DW, Simnick AJ, Chilkoti A. Mater. Sci. Eng., R. 2008;62:125. doi: 10.1016/j.mser.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huebsch N, Mooney DJ. Nature. 2009;462:426. doi: 10.1038/nature08601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haines-Butterick L, Rajagopal K, Branco M, Salick D, Rughani R, Pilarz M, Lamm MS, Pochan DJ, Schneider JP. Proc. Natl. Acad. Sci. U. S. A. 2007;104:7791. doi: 10.1073/pnas.0701980104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell (Cambridge Mass.) 2006;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 15.Gutowska A, Jeong B, Jasionowski M. Anat. Rec. 2001;263:342. doi: 10.1002/ar.1115. [DOI] [PubMed] [Google Scholar]
- 16.Yucel T, Micklitsch CM, Schneider JP, Pochan DJ. Macromolecules. 2008;41:5763. doi: 10.1021/ma702840q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macosko CW. Rheology: principles, measurements and applications. New York, NY: Wiley-VCH, Inc; 1994. [Google Scholar]
- 18.Mezger TG. The rheology handbook: for users of rotational and oscillatory rheometers. 2nd edn. Hannover: Vincentz Network GmbH and Co KG; 2006. [Google Scholar]
- 19.Ross-Murphy SB. Polym. Gels Networks. 1994;2:229. [Google Scholar]
- 20.Kavanagh GM, Ross-Murphy SB. Prog. Polym. Sci. 1998;23:533. [Google Scholar]
- 21.Hiemenz PC, Lodge TP. Polymer Chemistry. 2nd edn. Boca Raton: CRC press; 2007. [Google Scholar]
- 22.Gilsenan PM, Ross-Murphy SB. Int. J. Biol. Macromol. 2001;29:53. doi: 10.1016/s0141-8130(01)00149-0. [DOI] [PubMed] [Google Scholar]
- 23.Kavanagh GM, Clark AH, Ross-Murphy SB. Rheol. Acta. 2002;41:276. [Google Scholar]
- 24.Winer JP, Oake S, Janmey PA. PLoS One. 2009;4:e6382. doi: 10.1371/journal.pone.0006382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reinhart-King CA, Dembo M, Hammer DA. Langmuir. 2003;19:1573. [Google Scholar]
- 26.Cicuta P, Donald AM. Soft Matter. 2007;3:1449. doi: 10.1039/b706004c. [DOI] [PubMed] [Google Scholar]
- 27.Waigh TA. Rep. Prog. Phys. 2005;68:685. [Google Scholar]
- 28.Asghar A, Henrickson RL. Adv. Food Res. 1982;28:231. doi: 10.1016/s0065-2628(08)60113-5. [DOI] [PubMed] [Google Scholar]
- 29.Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen J, Lu H, Richmond J, Kaplan DL. Biomaterials. 2003;24:401. doi: 10.1016/s0142-9612(02)00353-8. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto M, Tabata Y, Ikada Y. J. Bioact. Compat. Polym. 1999;14:474. [Google Scholar]
- 31.Young S, Wong M, Tabata Y, Mikos AG. J. Controlled Release. 2005;109:256. doi: 10.1016/j.jconrel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 32.Nijenhuis KT. Polym. Bull. (Berlin) 2007;58:27. [Google Scholar]
- 33.Clark AH, Ross-Murphy SB. Adv. Polym. Sci. 1987;83:57. [Google Scholar]
- 34.Ross-Murphy SB. Polymer. 1992;33:2622. [Google Scholar]
- 35.Balian G, Bowes JH. In: The Science and Technology of Gelatin. Ward AG, Courts A, editors. New York: Academic Press; 1977. [Google Scholar]
- 36.Veis A. The Macromolecular Chemistry of Gelatin. New York: Academic Press; 1964. [Google Scholar]
- 37.Ramshaw JAM, Brodsky B. In: Industrial Proteins in Perspective; Progress in Biotechnology. Aalbersberg WY, Hamer RJ, Jasperse P, De Jong HHJ, De Kruijf CG, Walstra P, De Wolf FA, editors. Amsterdam, Boston: Elsevier; 2003. [Google Scholar]
- 38.Gilsenan PM, Ross-Murphy SB. Food Hydrocolloids. 2000;14:191. [Google Scholar]
- 39.Gilsenan PM, Ross-Murphy SB. J. Rheol. (N. Y.) 2000;44:871. [Google Scholar]
- 40.Foegeding EA. Food Biophys. 2006;1:41. [Google Scholar]
- 41.Clark AH, Lee-Tuffnell CD. In: Functional Properties of Food Macromolecules. Mitchell JR, Ledward DA, editors. Essex: Elsevier; 1986. [Google Scholar]
- 42.Langton M, Hermansson AM. Food Hydrocolloids. 1992;5:523. [Google Scholar]
- 43.Stading M, Hermansson AM. Food Hydrocolloids. 1991;5:339. [Google Scholar]
- 44.Langton M, Hermansson AM. Food Hydrocolloids. 1996;10:179. [Google Scholar]
- 45.Tombs MP. In: Proteins as Human Food. Lawrie RA, editor. Connecticut: AVI Publishing; 1970. [Google Scholar]
- 46.Clark AH, Tuffnell CD. Int. J. Pept. Protein Res. 1980;16:339. doi: 10.1111/j.1399-3011.1980.tb02595.x. [DOI] [PubMed] [Google Scholar]
- 47.Clark AH, Judge FJ, Richards JB, Stubbs JM, Suggett A. Int. J. Pept. Protein Res. 1981;17:380. doi: 10.1111/j.1399-3011.1981.tb02005.x. [DOI] [PubMed] [Google Scholar]
- 48.Clark AH, Saunderson DHP, Suggett A. Int. J. Pept. Protein Res. 1981;17:353. doi: 10.1111/j.1399-3011.1981.tb02002.x. [DOI] [PubMed] [Google Scholar]
- 49.Richardson RK, Ross-Murphy SB. Br. Polym. J. 1981;13:11. [Google Scholar]
- 50.Heertje I, van Kleef FSM. Food Microstruct. 1986;5:91. [Google Scholar]
- 51.Doi E. Trends Food Sci. Technol. 1993;4:1. [Google Scholar]
- 52.Durand D, Gimel JC, Nicolai T. Physica A (Amsterdam) 2002;304:253. [Google Scholar]
- 53.Gosal WS, Clark AH, Pudney PDA, Ross-Murphy SB. Langmuir. 2002;18:7174. [Google Scholar]
- 54.Veerman C, Ruis H, Sagis LMC, van der Linden E. Biomacromolecules. 2002;3:869. doi: 10.1021/bm025533+. [DOI] [PubMed] [Google Scholar]
- 55.Sagis LM, Veerman C, van der Linden E. Langmuir. 2004;20:924. doi: 10.1021/la035390s. [DOI] [PubMed] [Google Scholar]
- 56.Clark AH. In: Functional Properties of Food Macromolecules. 2nd edn. Hill SE, Ledward DA, Mitchell JR, editors. Maryland: Aspen; 1998. [Google Scholar]
- 57.Gosal WS, Ross-Murphy SB. Curr. Opin. Colloid Interface Sci. 2000;5:188. [Google Scholar]
- 58.Totosaus A, Montejano JG, Salazar JA, Guerrero I. Int. J. Food Sci. Technol. 2002;37:589. [Google Scholar]
- 59.Foegeding EA. In: Food Colloids: Interactions, Microstructure and Processing. Dickinson E, editor. Cambridge: The Royal Chemical Society; 2005. [Google Scholar]
- 60.Tobitani A, Ross-Murphy SB. Macromolecules. 1997;30:4855. [Google Scholar]
- 61.Kavanagh GM, Clark AH, Ross-Murphy SB. Langmuir. 2000;16:9584. doi: 10.1016/s0141-8130(00)00144-6. [DOI] [PubMed] [Google Scholar]
- 62.Kavanagh GM, Clark AH, Ross-Murphy SB. Int. J. Biol. Macromol. 2000;28:41. doi: 10.1016/s0141-8130(00)00144-6. [DOI] [PubMed] [Google Scholar]
- 63.Kavanagh GM, Clark AH, Ross-Murphy SB. Food Hydrocolloids. 2001;15:383. [Google Scholar]
- 64.Gosal WS, Clark AH, Ross-Murphy SB. Biomacromolecules. 2004;5:2408. doi: 10.1021/bm049659d. [DOI] [PubMed] [Google Scholar]
- 65.Gosal WS, Clark AH, Ross-Murphy SB. Biomacromolecules. 2004;5:2420. doi: 10.1021/bm049660c. [DOI] [PubMed] [Google Scholar]
- 66.Gosal WS, Clark AH, Ross-Murphy SB. Biomacromolecules. 2004;5:2430. doi: 10.1021/bm0496615. [DOI] [PubMed] [Google Scholar]
- 67.Veerman C, de Schiffart G, Sagis LMC, van der Linden E. Int. J. Biol. Macromol. 2003;33:121. doi: 10.1016/s0141-8130(03)00076-x. [DOI] [PubMed] [Google Scholar]
- 68.Veerman C, Sagis LMC, Heck J, van der Linden E. Int. J. Biol. Macromol. 2003;31:139. doi: 10.1016/s0141-8130(02)00074-0. [DOI] [PubMed] [Google Scholar]
- 69.Pouzot M, Durand D, Nicolai T. Macromolecules. 2004;37:8703. [Google Scholar]
- 70.Pouzot M, Nicolai T, Durand D, Benyahia L. Macromolecules. 2004;37:614. [Google Scholar]
- 71.Pouzot M, Benyahai L, Nicolai T. J. Rheol. (N. Y.) 2004;48:1123. [Google Scholar]
- 72.Mehalebi S, Nicolai T, Durand D. Soft Matter. 2008;4:893. doi: 10.1039/b718640a. [DOI] [PubMed] [Google Scholar]
- 73.Mehalebi S, Nicolai T, Durand D. Int. J. Biol. Macromol. 2008;43:129. doi: 10.1016/j.ijbiomac.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 74.Yan H, Saiani A, Gough JE, Miller AF. Biomacromolecules. 2006;7:2776. doi: 10.1021/bm0605560. [DOI] [PubMed] [Google Scholar]
- 75.Yan H, Frielinghaus H, Nykanen A, Ruokolainen J, Saiani A, Miller AF. Soft Matter. 2008;4:1313. doi: 10.1039/b716966c. [DOI] [PubMed] [Google Scholar]
- 76.Yan H, Nykanen A, Ruokolainen J, Farrar D, Saiani A, Miller AF. Faraday Discuss. 2008;139:71. doi: 10.1039/b717748h. [DOI] [PubMed] [Google Scholar]
- 77.Perez-Rigueiro J, Viney C, Llorca J, Elices M. J. Appl. Polym. Sci. 2000;75:1270. [Google Scholar]
- 78.Cunniff PM, Fossey SA, Auerbach MA, Song JW, Kaplan DL, Adams WW, Eby RK, Mahoney D, Vezie DL. Polym. Adv. Technol. 1994;5:401. [Google Scholar]
- 79.Pins G, Christiansen D, Patel R, Silver F. Biophys. J. 1997;73:2164. doi: 10.1016/S0006-3495(97)78247-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meinel L, Hofmann S, Karageorgiou V, Kirker-Head C, McCool J, Gronowicz G, Zichner L, Langer R, Vunjak-Novakovic G, Kaplan DL. Biomaterials. 2005;26:147. doi: 10.1016/j.biomaterials.2004.02.047. [DOI] [PubMed] [Google Scholar]
- 81.Dal Pra I, Freddi G, Minic J, Chiarini A, Armato U. Biomaterials. 2005;26:1987. doi: 10.1016/j.biomaterials.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 82.Arai T, Freddi G, Innocenti R, Tsukada M. J. Appl. Polym. Sci. 2004;91:2383. [Google Scholar]
- 83.Motta A, Migliaresi C, Faccioni F, Torricelli P, Fini M, Giardino R. J. Biomater. Sci., Polym. Ed. 2004;15:851–864. doi: 10.1163/1568562041271075. [DOI] [PubMed] [Google Scholar]
- 84.Fini M, Motta A, Torricelli P, Giavaresi G, Nicoli Aldini N, Tschon M, Giardino R, Migliaresi C. Biomaterials. 2005;26:3527. doi: 10.1016/j.biomaterials.2004.09.040. [DOI] [PubMed] [Google Scholar]
- 85.Aoki H, Tomita N, Morita Y, Hattori K, Harada Y, Sonobe M, Wakitani S, Tamada Y. Biomed. Mater. Eng. 2003;13:309. [PubMed] [Google Scholar]
- 86.Gil ES, Spontak RJ, Hudson SM. Macromol. Biosci. 2005;5:702. doi: 10.1002/mabi.200500076. [DOI] [PubMed] [Google Scholar]
- 87.Gil ES, Frankowski DJ, Spontak RJ, Hudson SM. Biomacromolecules. 2005;6:3079. doi: 10.1021/bm050396c. [DOI] [PubMed] [Google Scholar]
- 88.Vepari C, Kaplan DL. Prog. Polym. Sci. 2007;32:991. doi: 10.1016/j.progpolymsci.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hardy JG, Romer LM, Scheibel TR. Polymer. 2008;49:4309. [Google Scholar]
- 90.Inoue S, Tanaka K, Arisaka F, Kimura S, Ohtomo K, Mizuno S. J. Biol. Chem. 2000;275:40517. doi: 10.1074/jbc.M006897200. [DOI] [PubMed] [Google Scholar]
- 91.Matsumoto A, Chen J, Collette AL, Kim UJ, Altman GH, Cebe P, Kaplan DL. J. Phys. Chem. B. 2006;110:21630. doi: 10.1021/jp056350v. [DOI] [PubMed] [Google Scholar]
- 92.Jin HJ, Kaplan DL. Nature. 2003;424:1057. doi: 10.1038/nature01809. [DOI] [PubMed] [Google Scholar]
- 93.Horan RL, Antle K, Collette AL, Wang Y, Huang J, Moreau JE, Volloch V, Kaplan DL, Altman GH. Biomaterials. 2005;26:3385. doi: 10.1016/j.biomaterials.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 94.Kim UJ, Park J, Kim HJ, Wada M, Kaplan DL. Biomaterials. 2005;26:2775. doi: 10.1016/j.biomaterials.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 95.Nazarov R, Jin HJ, Kaplan DL. Biomacromolecules. 2004;5:718. doi: 10.1021/bm034327e. [DOI] [PubMed] [Google Scholar]
- 96.Kim UJ, Park J, Li C, Jin HJ, Valluzzi R, Kaplan DL. Biomacromolecules. 2004;5:786. doi: 10.1021/bm0345460. [DOI] [PubMed] [Google Scholar]
- 97.Ayub Z, Arai M, Hirabayashi K. Biosci., Biotechnol., Biochem. 1993;57:1910. [Google Scholar]
- 98.Wang H, Zhang Y, Shao H, Hu X. Int. J. Biol. Macromol. 2005;36:66. doi: 10.1016/j.ijbiomac.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 99.Kang GD, Nahm JH, Park JS, Moon JY, Cho CS, Yeo JH. Macromol. Rapid Commun. 2000;21:788. [Google Scholar]
- 100.Yoo MK, Kweon HY, Lee KG, Lee HC, Cho CS. Int. J. Biol. Macromol. 2004;34:263. doi: 10.1016/j.ijbiomac.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 101.Wang XQ, Kluge JA, Leisk GG, Kaplan DL. Biomaterials. 2008;29:1054. doi: 10.1016/j.biomaterials.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yucel T, Cebe P, Kaplan DL. Biophys. J. 2009;97:2044. doi: 10.1016/j.bpj.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vollrath F, Knight DP, Hu XW. Proc. R. Soc. Ser. B. 1998;265:817–820. [Google Scholar]
- 104.Huemmerich D, Scheibel T, Vollrath F, Cohen S, Gat U, Ittah S. Curr. Biol. 2004;14:2070. doi: 10.1016/j.cub.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 105.Slotta U, Hess S, Spiess K, Stromer T, Serpell L, Scheibel T. Macromol. Biosci. 2007;7:183–188. doi: 10.1002/mabi.200600201. [DOI] [PubMed] [Google Scholar]
- 106.Rammensee S, Huemmerich D, Hermanson KD, Scheibel T, Bausch AR. Appl. Phys. A: Mater. Sci. Process. 2006;82:261. [Google Scholar]
- 107.Zisch AH, Lutolf MP, Hubbell JA. Cardiovasc. Pathol. 2003;12:295. doi: 10.1016/s1054-8807(03)00089-9. [DOI] [PubMed] [Google Scholar]
- 108.Janmey PA, Winer JP, Weisel JW. J. R. Soc. Interface. 2009;6:1. doi: 10.1098/rsif.2008.0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hubbell JA. Curr. Opin. Biotechnol. 2003;14:551. doi: 10.1016/j.copbio.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 110.Fukada E, Kaibara M. Thromb. Res. 1976;8 suppl. II:49. doi: 10.1016/0049-3848(76)90047-5. [DOI] [PubMed] [Google Scholar]
- 111.Kaibara M, Date M. Biorheology. 1985;22:197. doi: 10.3233/bir-1985-22304. [DOI] [PubMed] [Google Scholar]
- 112.Kaibara M, Fukada E. Biochim. Biophys. Acta. 1977;499:352. doi: 10.1016/0304-4165(77)90066-6. [DOI] [PubMed] [Google Scholar]
- 113.Kaibara M, Fukada E. Biorheology. 1980;17:255. [PubMed] [Google Scholar]
- 114.Kaibara M, Fukada E, Sakaoku K. Biorheology. 1981;18:22. doi: 10.3233/bir-1981-18104. [DOI] [PubMed] [Google Scholar]
- 115.Fukada E, Kaibara M. Biorheology. 1973;10:129. doi: 10.3233/bir-1973-10207. [DOI] [PubMed] [Google Scholar]
- 116.Kaibara M, Fukada E. Polym. Eng. Rev. 1983;3:383. [Google Scholar]
- 117.Kaibara M, Fukada E. Biorheology. 1970;6:329. doi: 10.3233/bir-1970-6407. [DOI] [PubMed] [Google Scholar]
- 118.Kaibara M, Fukada E. Biorheology. 1969;6:73. doi: 10.3233/bir-1969-6202. [DOI] [PubMed] [Google Scholar]
- 119.Ryan EA, Mockros LF, Weisel JW, Lorand L. Biophys. J. 1999;77:2813. doi: 10.1016/S0006-3495(99)77113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ryan EA, Mockros LF, Stern AM, Lorand L. Biophys. J. 1999;77:2827–2836. doi: 10.1016/S0006-3495(99)77114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Urech L, Bittermann AG, Hubbell JA, Hall H. Biomaterials. 2005;26:1369. doi: 10.1016/j.biomaterials.2004.04.045. [DOI] [PubMed] [Google Scholar]
- 122.Shah JV, Janmey PA. Rheol. Acta. 1997;36:262. [Google Scholar]
- 123.Storm C, Pastore JJ, MacKintosh FC. Nature. 2005;435:191. doi: 10.1038/nature03521. [DOI] [PubMed] [Google Scholar]
- 124.Ciani B, Hutchinson EG, Sessions RB, Woolfson DN. J. Biol. Chem. 2002;277:10150. doi: 10.1074/jbc.M107663200. [DOI] [PubMed] [Google Scholar]
- 125.Klok HA. Macromolecules. 2009;42:7990. [Google Scholar]
- 126.Wang C, Stewart RJ, Kopeček J. Nature. 1999;397:417. doi: 10.1038/17092. [DOI] [PubMed] [Google Scholar]
- 127.Wang C, Kopeček J, Stewart RJ. Biomacromolecules. 2001;2:912. doi: 10.1021/bm0155322. [DOI] [PubMed] [Google Scholar]
- 128.Yang JY, Xu CY, Kopečková P, Kopeček J. Macromol. Biosci. 2006;6:201. doi: 10.1002/mabi.200500208. [DOI] [PubMed] [Google Scholar]
- 129.Yang J, Xu C, Wang C, Kopeček J. Biomacromolecules. 2006;7:1187. doi: 10.1021/bm051002k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wu K, Yang J, Konák C, Kopečková P, Kopeček J. Macromol. Chem. Phys. 2008;209:467. [Google Scholar]
- 131.Petka WA, Harden JL, McGrath KP, Wirtz D, Tirrell DA. Science. 1998;281:389. doi: 10.1126/science.281.5375.389. [DOI] [PubMed] [Google Scholar]
- 132.Shen W, Zhang KC, Kornfield JA, Tirrell DA. Nat. Mater. 2006;5:153. doi: 10.1038/nmat1573. [DOI] [PubMed] [Google Scholar]
- 133.Jing P, Rudra JS, Herr AB, Collier JH. Biomacromolecules. 2008;9:2438. doi: 10.1021/bm800459v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shen W, Lammertink RGH, Sakata JK, Kornfield JA, Tirrell DA. Macromolecules. 2005;38:3909. [Google Scholar]
- 135.Shen W, Kornfield JA, Tirrell DA. Soft Matter. 2007;3:99. doi: 10.1039/b610986a. [DOI] [PubMed] [Google Scholar]
- 136.Shen W, Kornfield JA, Tirrell DA. Macromolecules. 2007;40:689. [Google Scholar]
- 137.Xu C, Kopeček J. Pharm. Res. 2008;25:674. doi: 10.1007/s11095-007-9343-z. [DOI] [PubMed] [Google Scholar]
- 138.Banwell EF, Abelardo ES, Adams DJ, Birchall MA, Corrigan A, Donald AM, Kirkland M, Serpell LC, Butler MF, Woolfson DN. Nat. Mater. 2009;8:596. doi: 10.1038/nmat2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pandya MJ, Spooner GM, Sunde M, Thorpe JR, Rodger A, Woolfson DN. Biochemistry. 2000;39:8728. doi: 10.1021/bi000246g. [DOI] [PubMed] [Google Scholar]
- 140.Nowak AP, Breedveld V, Pakstis L, Ozbas B, Pine DJ, Pochan D, Deming TJ. Nature. 2002;417:424. doi: 10.1038/417424a. [DOI] [PubMed] [Google Scholar]
- 141.Pochan DJ, Pakstis L, Ozbas B, Nowak AP, Deming TJ. Macromolecules. 2002;35:5358. [Google Scholar]
- 142.Breedveld V, Nowak AP, Sato J, Deming TJ, Pine DJ. Macromolecules. 2004;37:3943. [Google Scholar]
- 143.Deming TJ. Soft Matter. 2005;1:28. doi: 10.1039/b500307e. [DOI] [PubMed] [Google Scholar]
- 144.Kelly JW. Curr. Opin. Struct. Biol. 1996;6:11. doi: 10.1016/s0959-440x(96)80089-3. [DOI] [PubMed] [Google Scholar]
- 145.Lamm MS, Rajagopal K, Schneider JP, Pochan DJ. J. Am. Chem. Soc. 2005;127:16692. doi: 10.1021/ja054721f. [DOI] [PubMed] [Google Scholar]
- 146.Koga T, Matsuoka M, Higashi N. J. Am. Chem. Soc. 2005;127:17596. doi: 10.1021/ja0558387. [DOI] [PubMed] [Google Scholar]
- 147.Branco MC, Pochan DJ, Wagner NJ, Schneider JP. Biomaterials. 2009;30:1339. doi: 10.1016/j.biomaterials.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Collier JH, Hu BH, Ruberti JW, Zhang J, Shum P, Thompson DH, Messersmith PB. J. Am. Chem. Soc. 2001;123:9463. doi: 10.1021/ja011535a. [DOI] [PubMed] [Google Scholar]
- 149.Aggeli A, Bell M, Boden N, Keen JN, Knowles PF, McLeish TCB, Radford SE. Nature. 1997;386:259. doi: 10.1038/386259a0. [DOI] [PubMed] [Google Scholar]
- 150.Schneider JP, Pochan DJ, Ozbas B, Rajagopal K, Pakstis L, Kretsinger J. J. Am. Chem. Soc. 2002;124:15030. doi: 10.1021/ja027993g. [DOI] [PubMed] [Google Scholar]
- 151.Pochan DJ, Schneider JP, Kretsinger J, Ozbas B, Rajagopal K, Haines L. J. Am. Chem. Soc. 2003;125:11802. doi: 10.1021/ja0353154. [DOI] [PubMed] [Google Scholar]
- 152.Ozbas B, Krestsinger J, Rajagopal K, Schneider JP, Pochan DJ. Macromolecules. 2004;37:7331. [Google Scholar]
- 153.Haines LA, Rajagopal K, Ozbas B, Salick DA, Pochan DJ, Schneider JP. J. Am. Chem. Soc. 2005;127:17025. doi: 10.1021/ja054719o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kretsinger JK, Haines LA, Ozbas B, Pochan DJ, Schneider JP. Biomaterials. 2005;26:5177. doi: 10.1016/j.biomaterials.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 155.Hule RA, Nagarkar RP, Altunbas A, Ramay HR, Branco MC, Schneider JP, Pochan DJ. Faraday Discuss. 2008;139:251. doi: 10.1039/b717616c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Veerman C, Rajagopal K, Palla CS, Pochan DJ, Schneider JP, Furst EM. Macromolecules. 2006;39:6608. [Google Scholar]
- 157.Haines-Butterick LA, Salick DA, Pochan DJ, Schneider JP. Biomaterials. 2008;29:4164. doi: 10.1016/j.biomaterials.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chambon F, Winter HH. Polym. Bull. (Berlin) 1985;13:499. [Google Scholar]
- 159.Ramachandran S, Tseng Y, Yu YB. Biomacromolecules. 2005;6:1316. doi: 10.1021/bm049284w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ramachandran S, Flynn P, Tseng Y, Yu YB. Chem. Mater. 2005;17:6583. [Google Scholar]
- 161.Ramachandran S, Trewhella J, Tseng Y, Yu YB. Chem. Mater. 2006;18:6157. [Google Scholar]
- 162.Aulisa L, Dong H, Hartgerink JD. Biomacromolecules. 2009;10:2694. doi: 10.1021/bm900634x. [DOI] [PubMed] [Google Scholar]
- 163.Cliff WJ. Blood vessels (Biological structure and function) Cambridge, New York: Cambridge University Press; 1976. [Google Scholar]
- 164.Jia X, Kiick K. Macromol. Biosci. 2009;9:140. doi: 10.1002/mabi.200800284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P, editors. Molecular Biology of the Cell. 4th edn. New York: Garland Science; 2002. [Google Scholar]
- 166.Urry DA. J. Phys. Chem. B. 1997;101:11007. [Google Scholar]
- 167.Meyer DE, Chilkoti A. Biomacromolecules. 2004;5:846. doi: 10.1021/bm034215n. [DOI] [PubMed] [Google Scholar]
- 168.Betre H, Setton LA, Meyer DE, Chilkoti A. Biomacromolecules. 2002;3:910. doi: 10.1021/bm0255037. [DOI] [PubMed] [Google Scholar]
- 169.Betre H, Ong SR, Guilak F, Chilkoti A, Fermor B, Setton LA. Biomaterials. 2006;27:91. doi: 10.1016/j.biomaterials.2005.05.071. [DOI] [PubMed] [Google Scholar]
- 170.McHale MK, Setton LA, Chilkoti A. Tissue Eng. 2005;11:1768. doi: 10.1089/ten.2005.11.1768. [DOI] [PubMed] [Google Scholar]
- 171.Setton LA, Mow VC, Howell DS. J. Orthop. Res. 1995;13:473. doi: 10.1002/jor.1100130402. [DOI] [PubMed] [Google Scholar]
- 172.Trabbic-Carlson K, Setton LA, Chilkoti A. Biomacromolecules. 2003;4:572. doi: 10.1021/bm025671z. [DOI] [PubMed] [Google Scholar]
- 173.Annabi N, Mithieux SM, Boughton EA, Ruys AJ, Weiss AS, Dehghani F. Biomaterials. 2009;30:4550. doi: 10.1016/j.biomaterials.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 174.Lee J, Macosko CW, Urry DW. Macromolecules. 2001;34:5968. [Google Scholar]
- 175.Lim DW, Nettles DL, Setton LA, Chilkoti A. Biomacromolecules. 2007;8:1463. doi: 10.1021/bm061059m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wright ER, McMillan RA, Cooper A. Adv. Funct. Mater. 2002;12:149. [Google Scholar]
- 177.Nagapudi K, Brinkman WT, Thomas BS, Park JO, Srinivasarao M, Wright E, Conticello VP, Chaikof EL. Biomaterials. 2005;26:4695. doi: 10.1016/j.biomaterials.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 178.Dinerman AA, Cappello J, Ghandehari H, Hoag SW. J. Controlled Release. 2002;82:277. doi: 10.1016/s0168-3659(02)00134-7. [DOI] [PubMed] [Google Scholar]
- 179.Megeed Z, Haider M, Li D, O’Malley BW, Jr, Cappello J, Ghandehari H. J. Controlled Release. 2004;94:433. doi: 10.1016/j.jconrel.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 180.Megeed Z, Cappello J, Ghandehari H. Pharm. Res. 2002;19:954. doi: 10.1023/a:1016406120288. [DOI] [PubMed] [Google Scholar]
- 181.Hatefi A, Cappello J, Ghandehari H. Pharm. Res. 2007;24:773. doi: 10.1007/s11095-006-9200-5. [DOI] [PubMed] [Google Scholar]
- 182.Boyd LM, Carter AJ. Eur. Spine J. 2006;15:S414. doi: 10.1007/s00586-006-0172-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cappello J, Crissman JW, Crissman M, Ferrari FA, Textor G, Wallis O, Whitledge JR, Zhou X, Burman D, Aukerman L, Stedronsky ER. J. Controlled Release. 1998;53:105. doi: 10.1016/s0168-3659(97)00243-5. [DOI] [PubMed] [Google Scholar]
- 184.Jun HW, Paramonov SE, Hartgerink JD. Soft Matter. 2006;2:177. doi: 10.1039/b516805h. [DOI] [PubMed] [Google Scholar]
- 185.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303:1352. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 186.Cui H, Muraoka T, Cheetham AG. Nano Lett. 2009;9:945. doi: 10.1021/nl802813f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 188.Yuwono VM, Hartgerink JD. Langmuir. 2007;23:5033. doi: 10.1021/la0629835. [DOI] [PubMed] [Google Scholar]
- 189.Rajangam K, Arnold MS, Rocco MA, Stupp SI. Biomaterials. 2008;29:3298. doi: 10.1016/j.biomaterials.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Rajangam K, Behanna HA, Hui MJ, Han X, Hulvat JF, Lomasney JW, Stupp SI. Nano Lett. 2006;6:2086. doi: 10.1021/nl0613555. [DOI] [PubMed] [Google Scholar]
- 191.Webber MJ, Tongers J, Renault MA, Roncalli JG, Losordo DW, Stupp SI. Acta Biomater. 2010;6:3. doi: 10.1016/j.actbio.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tysseling-Mattiace VM, Sahni V, Niece KL, Birch D, Czeisler C, Fehlings MG, Stupp SI, Kessler JA. J. Neurosci. 2008;28:3814. doi: 10.1523/JNEUROSCI.0143-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Beniash E, Hartgerink JD, Storrie H, Stendahl JC, Stupp SI. Acta Biomater. 2005;1:387. doi: 10.1016/j.actbio.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 194.Stendahl JC, Rao MS, Guler MO, Stupp SI. Adv. Funct. Mater. 2006;16:499. [Google Scholar]
- 195.Bull SR, Guler MO, Bras RE, Meade TJ, Stupp SI. Nano Lett. 2005;5:1. doi: 10.1021/nl0484898. [DOI] [PubMed] [Google Scholar]
- 196.Behanna HA, Donners J, Gordon AC, Stupp SI. J. Am. Chem. Soc. 2005;127:1193. doi: 10.1021/ja044863u. [DOI] [PubMed] [Google Scholar]
- 197.Guler MO, Stupp SI. J. Am. Chem. Soc. 2007;129:12082. doi: 10.1021/ja075044n. [DOI] [PubMed] [Google Scholar]
- 198.Niece KL, Hartgerink JD, Donners J, Stupp SI. J. Am. Chem. Soc. 2003;125:7146. doi: 10.1021/ja028215r. [DOI] [PubMed] [Google Scholar]
- 199.Hartgerink JD, Beniash E, Stupp SI. Proc. Natl. Acad. Sci. U. S. A. 2002;99:5133. doi: 10.1073/pnas.072699999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Jun HW, Yuwono V, Paramonov SE, Hartgerink JD. Adv. Mater. 2005;17:2612. [Google Scholar]
- 201.Paramonov SE, Jun HW, Hartgerink JD. J. Am. Chem. Soc. 2006;128:7291. doi: 10.1021/ja060573x. [DOI] [PubMed] [Google Scholar]
- 202.Zhou M, Smith AM, Das AK, Hodson NW, Collins RF, Ulijn RV, Gough JE. Biomaterials. 2009;30:2523. doi: 10.1016/j.biomaterials.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 203.Jayawarna V, Richardson SM, Hirst AR, Hodson NW, Saiani A, Gough JE, Ulijn RV. Acta Biomater. 2009;5:934. doi: 10.1016/j.actbio.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 204.Jayawarna V, Ali M, Jowitt TA, Miller AF, Saiani A, Gough JE, Ulijn RV. Adv. Mater. 2006;18:611. [Google Scholar]
- 205.Yang ZM, Xu KM, Wang L, Gu HW, Wei H, Zhang MJ, Xu B. Chem. Commun. 2005:4414. doi: 10.1039/b507314f. [DOI] [PubMed] [Google Scholar]
- 206.Yang ZM, Liang GL, Ma ML, Gao Y, Xu B. Small. 2007;3:558. doi: 10.1002/smll.200700015. [DOI] [PubMed] [Google Scholar]
- 207.Sutton S, Campbell NL, Cooper AI, Kirkland M, Frith WJ, Adams DJ. Langmuir. 2009;25:10285. doi: 10.1021/la9011058. [DOI] [PubMed] [Google Scholar]
- 208.Smith AM, Williams RJ, Tang C, Coppo P, Collins RF, Turner ML, Saiani A, Ulijn RV. Adv. Mater. 2008;20:37. [Google Scholar]
- 209.Yang Z, Xu B. Chem. Commun. 2004:2424. doi: 10.1039/b408897b. [DOI] [PubMed] [Google Scholar]
- 210.Yang Z, Gu H, Fu D, Gao P, Lam JK, Xu B. Adv. Mater. 2004;16:1440. [Google Scholar]
- 211.Yang Z, Liang G, Wang L, Xu B. J. Am. Chem. Soc. 2006;128:3038. doi: 10.1021/ja057412y. [DOI] [PubMed] [Google Scholar]
- 212.Zhang Y, Gu H, Yang Z, Xu B. J. Am. Chem. Soc. 2003;125:13680. doi: 10.1021/ja036817k. [DOI] [PubMed] [Google Scholar]
- 213.Toledano S, Williams RJ, Jayawarna V, Ulijn RV. J. Am. Chem. Soc. 2006;128:1070. doi: 10.1021/ja056549l. [DOI] [PubMed] [Google Scholar]
- 214.Zhang Y, Yang Z, Yuan F, Gu H, Gao P, Xu B. J. Am. Chem. Soc. 2004;126:15028. doi: 10.1021/ja044401g. [DOI] [PubMed] [Google Scholar]
- 215.Mahler A, Rechter M, Cohen S, Reches M, Gazit E. Adv. Mater. 2006;18:1365. [Google Scholar]
- 216.Adams DJ, Butler MF, Frith WJ, Kirkland M, Mullen L, Sanderson P. Soft Matter. 2009;5:1856. [Google Scholar]
- 217.Thornton K, Smith AM, Merry CLR, Ulijn RV. Biochem. Soc. Trans. 2009;37:660. doi: 10.1042/BST0370660. [DOI] [PubMed] [Google Scholar]
- 218.Yang ZM, Ho PL, Liang GL. J. Am. Chem. Soc. 2007;129:266. doi: 10.1021/ja0675604. [DOI] [PubMed] [Google Scholar]
- 219.Yang ZM, Ma ML, Xu B. Soft Matter. 2009;5:2546. [Google Scholar]