Abstract
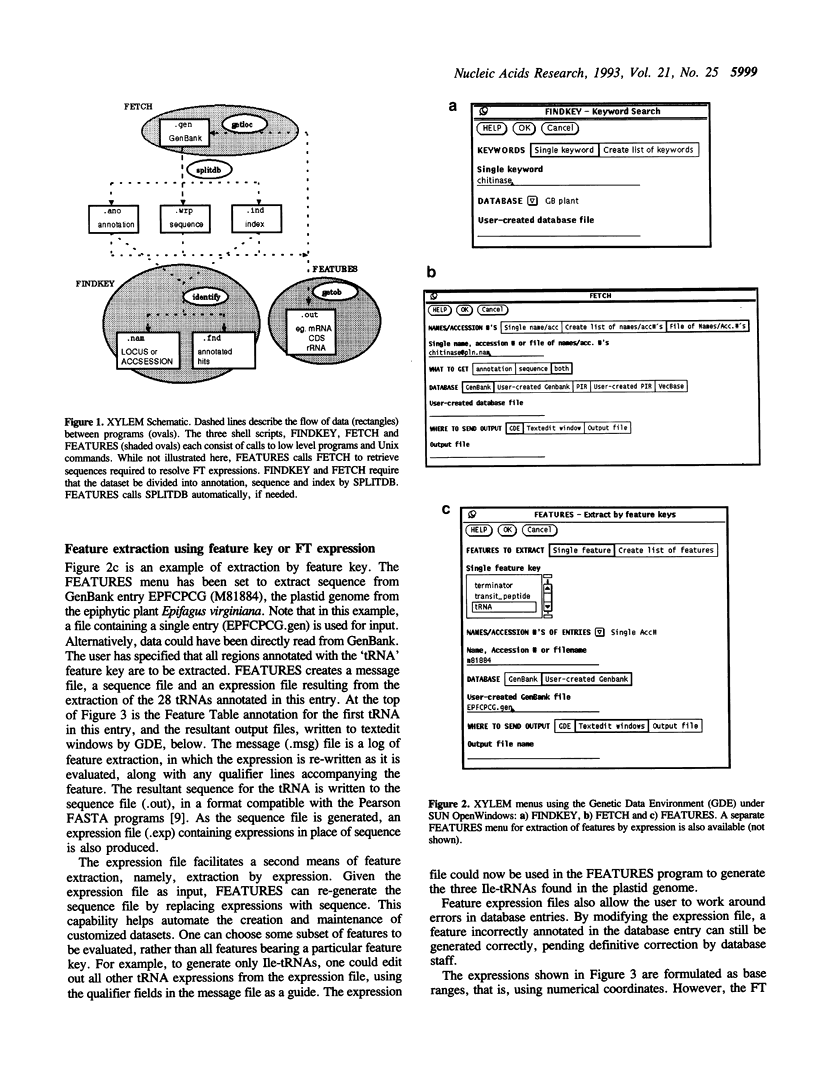
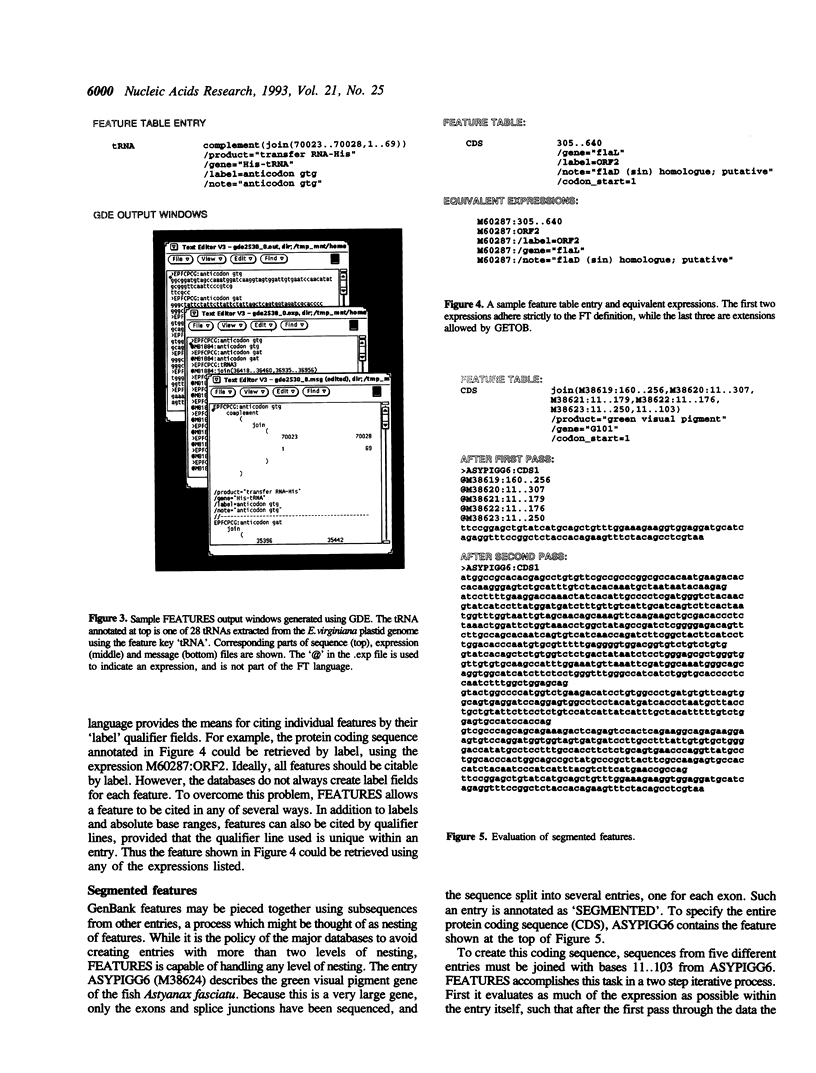
Annotation of features, such as introns, exons and protein coding regions in GenBank/EMBL/DDBJ entries is now standardized through use of the Features Table (FT) language. The essence of the FT language is described by the relation 'expression-->sequence', meaning that each FT expression evaluates to a sequence. For example, the expression M74750:1..50 evaluates to the first 50 bases of the sequence with accession number M74750. Because FT is intrinsic to the database definition, it can serve as a software- and platform-independent lingua franca for sequence manipulation. The XYLEM package makes it possible to create and manipulate sequence datasets using FT expressions. FEATURES is a program that resolves FT expressions into their corresponding sequences. Annotated features can be retrieved either by feature key or by expression. Even unannotated portions of a sequence can be retrieved by user-generated FT expressions. Applications of the FT language include retrieval of subsequences from large sequence entries, generation of chromosome models or artificial DNA constructs, and representation of restriction maps or mutants.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burks C., Cassidy M., Cinkosky M. J., Cumella K. E., Gilna P., Hayden J. E., Keen G. M., Kelley T. A., Kelly M., Kristofferson D. GenBank. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2221–2225. doi: 10.1093/nar/19.suppl.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron G. N. The EMBL data library. Nucleic Acids Res. 1988 Mar 11;16(5):1865–1867. doi: 10.1093/nar/16.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Milosavljevic A. Reconstruction and analysis of human Alu genes. J Mol Evol. 1991 Feb;32(2):105–121. doi: 10.1007/BF02515383. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Pearson W. R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- Pfeiffer F., Gilbert W. A. VecBase, a cloning vector sequence data base. Protein Seq Data Anal. 1988;1(4):269–280. [PubMed] [Google Scholar]
- Read R. L., Davison D., Chappelear J. E., Garavelli J. S. GBPARSE: a parser for the GenBank flat-file format with the new feature table format. Comput Appl Biosci. 1992 Aug;8(4):407–408. doi: 10.1093/bioinformatics/8.4.407. [DOI] [PubMed] [Google Scholar]
- Rudd K. E., Miller W., Ostell J., Benson D. A. Alignment of Escherichia coli K12 DNA sequences to a genomic restriction map. Nucleic Acids Res. 1990 Jan 25;18(2):313–321. doi: 10.1093/nar/18.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T. D., Stormo G. D., Haemer J. S., Gold L. A design for computer nucleic-acid-sequence storage, retrieval, and manipulation. Nucleic Acids Res. 1982 May 11;10(9):3013–3024. doi: 10.1093/nar/10.9.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder J. L., Blattner F. R. Formal description of a DNA oriented computer language. Nucleic Acids Res. 1982 Jan 11;10(1):69–84. doi: 10.1093/nar/10.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidman K. E., George D. G., Barker W. C., Hunt L. T. The protein identification resource (PIR). Nucleic Acids Res. 1988 Mar 11;16(5):1869–1871. doi: 10.1093/nar/16.5.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]



