Abstract
DNA-dependent activator of IFN regulatory factors (IRF; DAI, also known as ZBP1 or DLM-1) is a cytosolic DNA sensor that initiates IRF3 and NF-κB pathways leading to activation of type I IFNs (IFNα, IFNβ) and other cytokines. In this study, induction of NF-κB is shown to depend on the adaptor receptor-interacting protein kinase (RIP)1, acting via a RIP homotypic interaction motif (RHIM)-dependent interaction with DAI. DAI binds to and colocalizes with endogenous RIP1 at characteristic cytoplasmic granules. Suppression of RIP1 expression by RNAi abrogates NF-κB activation as well as IFNβ induction by immunostimulatory DNA. DAI also interacts with RIP3 and this interaction potentiates DAI-mediated activation of NF-κB, implicating RIP3 in regulating this RHIM-dependent pathway. The role of DAI in activation of NF-κB in response to immunostimulatory DNA appears to be analogous to sensing of dsRNA by TLR3 in that both pathways involve RHIM-dependent signaling that is mediated via RIP1, reinforcing a central role for this adaptor in innate sensing of intracellular microbes.
Host defense against intracellular microbial and viral pathogens involves surveillance for nucleic acids in cellular compartments where they do not naturally exist (1, 2). Sensors that detect such pathogen-associated molecular patterns initiate IFN regulatory factor (IRF)3 3/IRF7 and NF-κB pathways that drive type I IFN and other cytokine responses (3). Sensors for single-stranded RNA and dsRNA are known to contribute to innate immune sensing of viruses (4). With the exception of TLR9 (5), sensors for DNA have not been as thoroughly evaluated. Endocytosed unmethylated bacterial and viral CpG-containing DNA stimulates type I IFN responses via TLR9 (5); however, the existence of TLR9-independent, cytosolic DNA sensing pathways has been supported by the response of a wide variety of cell types to transfected DNA and/or DNA virus infection (6–11). The IFN inducible protein DAI (11, 12), a DNA binding protein previously denoted DLM-1 (12) or ZBP1 (13), is one recently identified DNA sensor (11). The induction of type I IFN (IFNα and IFNβ) by immunostimulatory DNA acting via DAI involves coordinated action of IRF3 and NF-κB transcription factors (11), as occurs with other pathogen sensors (1, 2). Type I IFN mediates a broad host response that restricts virus spread and sculpts the adaptive immune response (14, 15). DAI contains two N-terminal DNA binding domains, Zα and Zβ, plus a C-terminal domain that recruits the TNFR-associated factor family member-associated NF-κB activator (TANK)-binding kinase (TBK), an IκB kinase (IKK) kinase that controls the phosphorylation-dependent activation of IRF3 (11, 16). The mechanism of DAI-mediated NF-κB activation has not been resolved.
Receptor-interacting protein kinase (RIP)1, the founding member of a serine-threonine protein kinase family related to IL-1 receptor-associated kinases, functions as an adaptor in diverse signaling pathways triggered by death receptors, genotoxic stress, and pathogen recognition (17, 18). RIP1 is essential to the induction of NF-κB by TLR3- or TLR4-initiated pathways (19) that act via TIR domain-containing adaptor-inducing IFNβ (TRIF). RIP1 is required for activation of NF-κB and IRF3 (or IRF7) when retinoic acid-inducible gene I-like helicase (RLH) sensors respond to cytosolic RNA (20–22) as well as the induction of NF-κB following death receptor engagement (17, 18, 23). RIP1 carries out this central role as an adaptor through protein-protein interactions mediated by (1) a C-terminal death domain (DD), interacting with other DD adaptors like Fas-associated death domain protein and TNFR-associated death domain protein, (2) a central region that binds TNF-receptor-associated factors, and (3) a RIP homotypic interaction motif (RHIM) that mediates binding to two other RHIM-containing proteins, TRIF and RIP3. RIP3 is a serine-threonine protein kinase closely related to RIP1. TRIF is an adaptor protein acting downstream of TLR3 or TLR4 to control both RIP1-dependent NF-κB activation as well as TBK1-dependent IRF3 activation. RIP3 adaptor regulates RIP1-dependent events by binding to and phosphorylating RIP1 (19). RIP3 suppresses TRIF-dependent NF-κB activation (24) in these two TLR-dependent pathogen sensing pathways. RHIM interactions direct all known TRIF and RIP3 effects on RIP1 adaptor function (17, 18). RIP1 also regulates cell survival signals initiating from pathogen recognition, death receptor engagement or virus infection, contributing to cell fate via apoptosis, necrosis, and autophagy (17, 18, 23, 25, 26). The murine CMV (MCMV) M45 gene product was recently identified as a viral RHIM-containing protein that interacts with RIP1 to block death pathways, including virus-induced premature infected cell death (27).
Previous studies have demonstrated that immunostimulatory DNA activates NF-κB via DAI and this activation promotes the expression of inflammatory cytokines (11); however, the mechanism by which DAI activates NFκB has not been defined. In this study, we identify DAI as a fourth mammalian cellular RHIM protein and the first with pathogen sensor activity. We show that RHIM-dependent signaling dictates immunostimulatory DNA/DAI-mediated NF-κB activation, controlled by interaction with RIP1 and regulated by RIP3.
Materials and Methods
Cell culture and transfections
293T, HeLa, SVEC4-10, and L929 cells were maintained in DMEM containing 4.5 g/ml glucose, 10% FBS (Atlanta Biologicals), 2 mM l-glutamine, 100 U/ml penicillin, and 100 U/ml streptomycin (Invitrogen). Transient transfections were performed using Lipofectamine 2000 according to the manufacturer’s protocol (Invitrogen). A total of 5 µg/ml of Poly(dA-dT)·poly(dT-dA) (Sigma-Aldrich) transfections included a 1:1 (weight to volume ratio (%)) ratio of Lipofectamine 2000 to DNA.
Expression vectors and mutagenesis
The human DAI cDNA (IMAGE clone BC131706) was obtained from Open Biosystems. The open reading frame was PCR amplified and cloned in frame with the N-terminal epitope tags in p3XFLAG-CMV10 (Sigma-Aldrich) or pcDNA3-6myc plasmid (28). Amino acids within the RHIM-like repeat (RLR)-A (aa 206–209), RLR-B (aa 264–267), or RLR-C (aa 333–335) were subjected to site-directed mutagenesis and replaced by four alanines following overlap extension PCR. The RIP3-KD expression vector was generated by deleting the first 51 amino acids of RIP3 in p3XFLAG-RIP3 (28). The pSIREN-RIP1 short hairpin (sh)RNA construct targets nt 1675–1696 of the murine RIP1 open reading frame and was cloned by ligating annealed oligonucleotides (5′GATCCGCCATCTTTGATAACACCACTATTCAAGAGATAGTGGTGTTATCAAAGATGGTTTTTTACGCGTG3′) into the EcoRI/BamHI sites of pSIREN-RetroQ (Clontech). pSIREN-Luc was generated using previously described oligonucleotides (11). The pLKO.1-DAI-62 shRNA construct was obtained from Open Biosystems (RMM3981-97065659) and the pLK0.1-scramble control shRNA vector has been described (29). All other expression plasmids have been described (28).
Immunoblot (IB), immunoprecipitation (IP), immunofluorescence, and luciferase assays
Electrophoretically separated proteins were transferred to membranes that were immunoblotted as previously described (28) using one or more of the following primary Abs: Actin I-19 (Santa Cruz Biotechnology), ZBP1 M-300 (Santa Cruz Biotechnology), RIP1 clone 38 (BD Biosciences), anti-mouse RIP3 (Imgenex), mouse FLAG (M2) peroxidase conjugate (Sigma-Aldrich), c-Myc peroxidase conjugate (Sigma-Aldrich, anti-mouse IgG-HRP (Vector Laboratories), or anti-rabbit IgG-HRP (Vector Laboratories). Protein IP “pull-down” and dual luciferase assay experiments were performed as previously described except 293T cells were harvested at 18 h posttransfection (28). At 14–16 h posttransfection, HeLa grown on coverslips were fixed for 20 min using 4% paraformaldehyde in PBS. Cells were permeabilized for 10 min in 0.2% Triton X-100 (Sigma-Aldrich) in PBS, blocked for 30 min in blocking buffer (PBS containing 1% goat serum), and incubated 2 h at room temperature with rabbit polyclonal FLAG Ab (Sigma-Aldrich) diluted 1/3000 followed by mouse monoclonal RIP1 Ab (BD Biosciences) diluted 1/100 or mouse monoclonal c-Myc 9E10 (Sigma-Aldrich) diluted 1/1000 in blocking buffer. Cells were washed several times with PBS and then incubated for 1 h at room temperature with Alexa Fluor 488 (or 594) goat anti-rabbit Ab and Alexa Fluor 594 (or 488) goat anti-mouse Ab (Invitrogen) diluted 1/1000 in blocking buffer. 4′,6-diamidino-2-phenylindole (Sigma-Aldrich) was used as a DNA counter stain. Following multiple washes with PBS, cells were mounted with Gel Mount (Biomeda). Images were acquired on an AxioCam MRC5 camera attached to a Zeiss Axio Imager. A1 and then processed with AxioVision Release 4.5 software.
Quantitative RT-PCR analysis
Total RNA isolated using the RNeasy Mini Kit (Qiagen) was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) with oligo d(T)8–16 primer (Invitrogen) followed by RNase H (Invitrogen) treatment. Analysis was done using an aliquot of the reverse transcription reaction on the 7500 Fast Real Time PCR System with IFNβ (30) and β-actin (31) primers and SYBR Green Master Mix (Applied Biosystems). Data are presented as relative expression levels normalized to β-actin.
RNA interference
Inhibition of human RIP1 expression in 293T cells used Ontarget Plus RIPK1 (l-00445-00; Dharmacon) siRNA, in comparison to OnTarget Plus siRNA Control. 293T cells seeded on 100-mm plates were transfected with 600 pmol of RNA in 10 µl of Lipofectin 2000 according to the manufacturer’s protocol. After 72 h, cells were seeded into 24-well plates and assayed as described. siRNA knockdown of mouse RIP1 in L929 cells was performed as described (32). In L929 cells, Dharmacon siControl nontargeting siRNA (D-00120-03) was used as a negative control. For retroviral transduction studies, plasmids pSiren-Luc or pSiren-RIP1 together with plasmids LTRVSVG, CMVtat, and JK3 (33) were transfected into 293T cells to produce retroviral particles to infect SVEC4-10 cells. For lentiviral transduction, plasmid pLKO.1-Scramble or pLKO.1-DAI-62 together with plasmids LTRVSVG, CMVtat, and psPAX2 (34) were transfected into 293T cells to produce lentiviral particles to infect SVEC4-10 cells. Transduced cells were selected using 2 µg/ml Puromycin (Invitrogen).
Results
DAI interaction with RIP1 is RHIM-dependent
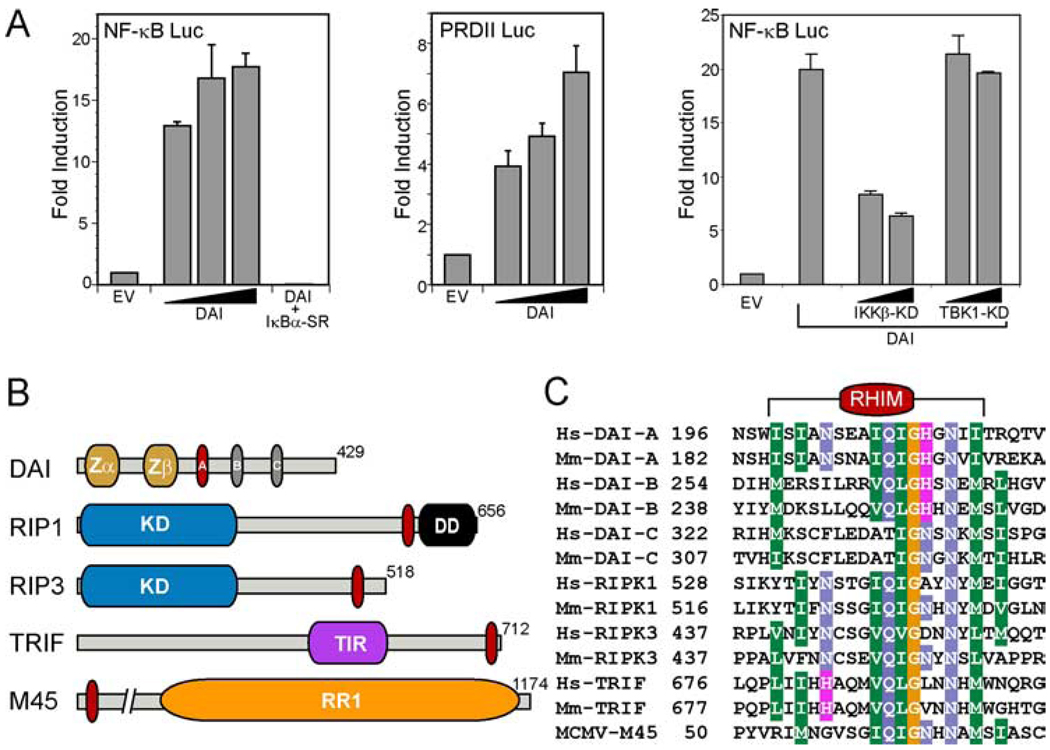
As predicted in the initial reports on DAI (11, 16), when a DAI-expression plasmid was transfected into L929 fibroblasts, DAI-dependent IFNβ expression was induced without additional immunostimulatory DNA treatment. This may have been due to the impact of plasmid DNA as well as the overexpression of the sensor. We used this approach to evaluate the role of TBK1 and IKKβ in DAI-dependent NF-κB activation in 293T cells. Flag-epitope tagged DAI induced NF-κB-dependent luciferase reporter expression 12- to 18-fold over control (Fig. 1A), levels that were also observed when nontagged DAI was transfected into cells (data not shown). Proinflammatory stimuli should promote the accumulation of the RelA NF-κB subunit in the nucleus following the phosphorylation of IκBα by IKKβ (35), an activation that is expected to be blocked by dominant negative IκBα-super repressor (IκBα-SR), which cannot be phosphorylated by IKKβ (36, 37). Coexpression of IκBα-SR and DAI prevented NF-κB activation and established the specificity of this response in DAI-transfected cells. Flag-DAI (or nontagged DAI) also stimulated expression from a second NF-κB-dependent promoter containing the positive regulatory domain II from the IFNβ gene (Fig. 1A and data not shown) (38). DAI is known to interact with TBK1 (11, 16), a kinase related to IKKβ, so we compared NF-κB reporter gene activation by DAI coexpressed with either dominant negative kinase dead (KD) TBK-1 or KD-IKKβ. As expected, KD-IKKβ suppressed reporter gene activity while the dominant negative KD-TBK1 did not (Fig. 1A). Thus, the pathway involved in transfection-dependent DAI-mediated activation of NF-κB appeared to be independent of TBK-dependent signal transduction pathways controlling IRF3 activation (11).
FIGURE 1.
DAI-mediated activation of NF-κB. A, 293T cells were transfected with 100 ng firefly luciferase reporter plasmid carrying five tandem NF-κB sites (left panel; NF-κB-Luc; Stratagene) or carrying two tandem NF-κB sites from the IFNβ promoter (middle panel; positive regulatory domain II-Luc) (28), together with 20, 100, or 500 ng (denoted by triangle) of Flag-DAI and 5 ng of a control noninducible Renilla luciferase expression vector, phRL-TK. IκBα-SR-mediated (400 ng) inhibition of DAI-dependent (100 ng) NF-κB-Luc activation is also shown in the left panel. 293T cells were transfected with 100 or 400 ng of plasmid expressing TBK1-KD or IKKβ-KD (28) together with 100 ng Flag-DAI (right panel). In all experiments, the total DNA transfected was held constant (605 ng) by adding empty vector (EV). Firefly and Renilla luciferase activities were assayed 18 h posttransfection. Firefly luciferase activity was divided by Renilla luciferase activity to normalize for transfection efficiency. Data shown are expressed as the mean relative stimulation in cells receiving DAI compared with the constitutive level present in cells transfected with EV ± SD (n = 3) from one representative experiment of a total of three performed. B, Schematic representation showing domains of RHIM-containing proteins. DAI contains an N-terminal Zα and Zβ domain and three RLR (labeled A, B, and C). The RLR-A motif in DAI is most similar to the RHIM (red oval). RIP1 and RIP3 each have an N-terminal kinase domain (KD), and RIP1 also contains a C-terminal DD. TRIF encodes a TIR domain, and M45 has a region of homology to a ribonucleotide reductase (RR1). C, The RHIM of human (Hs) and mouse (Mm) RIP1, RIP3, and TRIF and the MCMV protein M45 is aligned with each of the RLR motifs of DAI. The alignment was shaded with the BLOSUM62 matrix at a 60% identity threshold. The number adjacent to each sequence in the alignment indicates the starting amino acid within each full-length protein.
To gain insight into potential mechanisms of DAI-dependent NF-κB activation that occur upstream of IKKβ in this setting, we examined the amino acid sequence of DAI for regions of homology to known signal transduction and adaptor protein interaction motifs. We identified three stretches of amino acid common to all mammalian DAI homologues with sequence similarity to the RHIM of RIP1, RIP3, and TRIF (Fig. 1, B and C). Two of these RLR (labeled A and B in DAI schematic; Fig. 1B) exhibited greater similarity to a core motif (I/V)Q(I/L/V)GXXNX(M/L/I) than the third (labeled C in DAI schematic; Fig. 1B). RLR-A appeared most like a RHIM, with two amino acids (I201 and N203) conserved in RIP1 and RIP3. RLR-B also shared a complete core motif with RIP1, RIP3, and TRIF. Similarity was also observed to the RHIM of MCMV protein M45, which interacts with RIP1 to suppress premature infected cell death (27). Of the three, only RLR-A was located within a DAI region (D3) previously implicated in DNA-binding and immunostimulatory DNA-mediated induction of IFNβ expression (11, 16).
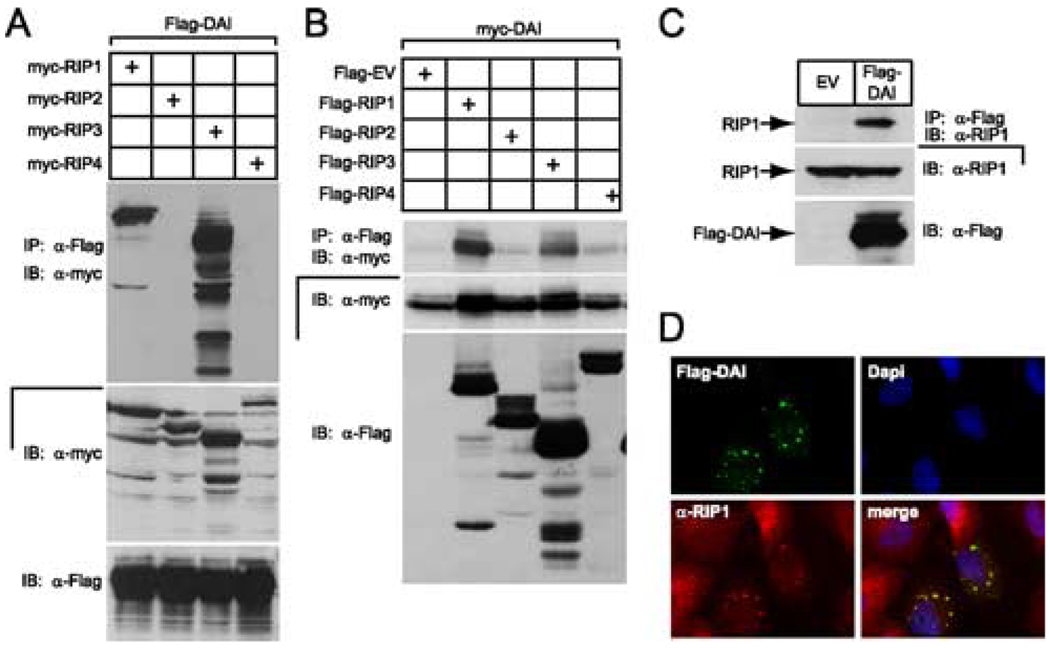
The importance of RHIM-mediated protein-protein interactions in pathways controlled by RIP1, RIP3, and TRIF prompted us to evaluate interactions in IP/IB experiments performed with lysates of 293T cells transfected with Flag epitope tagged DAI and c-Myc epitope tagged RIP kinases 1, 2, 3, and 4 (Fig. 2A). In this overexpression setting, DAI interacted with RHIM-containing members RIP1 and RIP3, but not the others in this family. We detected a similar pattern of interactions when epitope tags were swapped and the experiment was performed with Flag-RIP1 or Flag-RIP3 and c-Myc-DAI (Fig. 2B). In addition to the behavior in dually transfected cells, Flag-DAI immunoprecipitated endogenous RIP1 in 293T cells (Fig. 2C) and colocalized with endogenous RIP1 (Fig. 2D) when immunofluorescence analysis was applied to Flag-DAI-transfected HeLa cells. Endogenous RIP1 was diffusely dispersed throughout the cytoplasm in nontransfected cells; however, RIP1 appeared in a perinuclear, granular pattern colocalized with Flag-DAI in transfected cells (Fig. 2D). This DAI localization pattern has been observed previously (39, 40) and confirmed in recent studies implicating this protein as a DNA sensor (11, 16). We suspect that DAI recruits RIP1 into this cytoplasmic localization pattern as a component of signaling.
FIGURE 2.
DAI interaction with the RHIM-containing RIP kinases, RIP1 and RIP3. A, Autoradiogram of IB and IP of 293T cell extracts following transfection of myc epitope-tagged RIP1, RIP2, RIP3, or RIP4 together with Flag epitope-tagged DAI vectors. B, Autoradiogram of IB and IP of 293T cell extracts following transfection of Flag epitope-tagged RIP1, RIP2, RIP3, and RIP4, or an EV together with myc epitope-tagged DAI vector. Cellular lysates were subjected to anti-Flag IP and analyzed by IB with anti-myc Ab. IB analysis of total cell lysate showed the relative expression of Flag and myc epitope-tagged proteins. C, Demonstration that Flag-DAI binds endogenous RIP1 via IB of 293T cells following transfection with Flag epitope-tagged DAI or an EV. IP was with anti-FLAG M2 beads and IB was with anti-RIP1 Ab. Total cell lysates were examined for the expression of RIP1 and Flag-DAI. D, Immunofluorescent localization of Flag-DAI with endogenous RIP1. HeLa cells were transfected with Flag-tagged DAI and then stained with rabbit anti-Flag Ab and with monoclonal anti-RIP1 Ab for endogenous RIP1. Flag-DAI was detected by an anti-rabbit Alexa-488-conjugated secondary Ab and RIP1 was detected by an anti-mouse Alexa-594-conjugated secondary Ab. Stained cells were examined by epifluorescent microscopy (×1000).
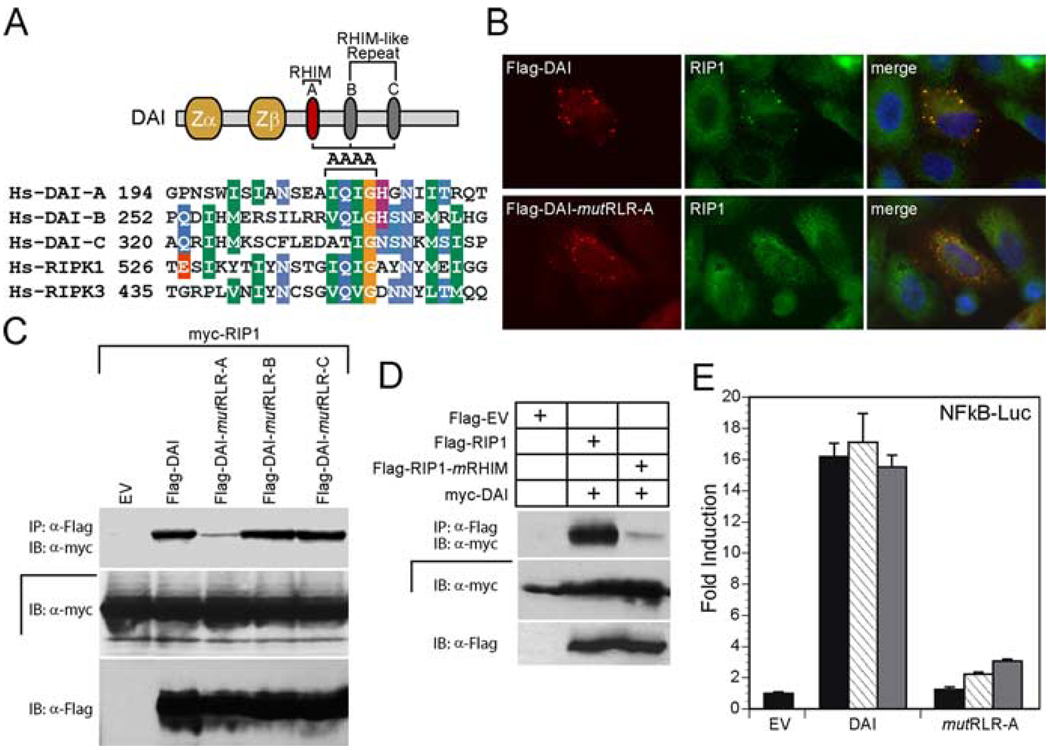
To assess the importance of the putative DAI RLRs in interactions with RIP1 and RIP3, we used alanine substitution mutations in core conserved amino acid (V/I)QIG in RLR-A (DAI-mutRLR-A) or RLR-B (DAI-mutRLR-B) and amino acid QIG in RLR-C (DAI-mutRLR-C) (Fig. 3A). This clustered mutation should disrupt RHIM-dependent protein interactions and signaling based on previous studies (19, 24, 27, 28). To test the interaction of Flag-epitope tagged DAI mutants with RIP1, we transfected 293T cells with plasmids expressing c-Myc-epitope tagged RIP1 and Flag-epitope tagged DAI, and evaluated association by coimmunoprecipitation (Fig. 3C). Only DAI-mutRLR-A showed a reduced interaction with RIP1 in this assay; DAI-mutRLR-B and DAI-mutRLR-C immunoprecipitated RIP1 as efficiently as wild-type DAI. RIP1 and all of the DAI-derived proteins were expressed at similar levels in all samples that were compared. Additionally, DAI RLR-A was found to be necessary for interaction (data not shown) as well as colocalization (Fig. 3B) with endogenous RIP1 protein. To assess the importance of the RIP1 RHIM in this interaction, we evaluated the ability of Flag-epitope tagged RIP1 or RHIM mutant RIP1 (RIP1-mRHIM) to immunoprecipitate c-Myc-epitope tagged DAI (Fig. 3D). RIP1 associated with DAI, but not with RIP1-mRHIM, demonstrating that DAI RLR-A was critical for the RHIM-dependent interaction with RIP1. This data implicates RIP1 as an essential adaptor for DAI-mediated activation and suggests that RHIM-dependent interactions may be necessary for activation via this DNA sensor. Although we relied on overexpression, these data are consistent with the hypothesis that DAI may use RHIM-mediated interaction with the adaptor protein RIP1 to activate the NF-κB signaling pathway.
FIGURE 3.
DAI-mediated, RIP1 RHIM-dependent activation of NF-κB. A, Diagram of RHIM mutations made by alanine substitution of the indicated aa (AAAA above the sequence alignment). B, Immunofluorescence analysis of Flag-DAI and Flag-DAI-mutRLR-A colocalization with endogenous RIP1. HeLa cells were transfected with Flag-tagged DAI or mutRLR-A and then stained with rabbit anti-Flag Ab and with monoclonal anti-RIP1 Ab for endogenous RIP1. Flag-DAI was revealed by an anti-rabbit Alexa-594-conjugated secondary Ab and RIP1 by an anti-mouse Alexa-488-conjugated secondary Ab. Stained cells were examined by microscopy (×1000). C, Autoradiogram following IP and IB to detect RHIM-dependent DAI interaction with RIP1. 293T cells were transfected with c-myc epitope-tagged RIP1 and Flag epitope-tagged DAI, DAI-mutRLR-A, DAI-mutRLR-B, DAI-mutRLR-C, or an EV. IP of cellular lysates was with anti-FLAG M2 beads and IB was with anti-myc Ab. IB analysis of total cell lysate determined the expression of myc-RIP1 and Flag-tagged proteins. D, Autoradiograph following IP and IB analysis to detect DAI-RIP1 interaction. 293T cells were transfected with myc epitope-tagged DAI and Flag epitope-tagged RIP1, RIP1-mRHIM, or an EV. IP of cellular lysates was followed by IB with anti-myc Ab. Anti-Flag Ab revealed the relative expression of epitope-tagged proteins in original lysates. E, Luciferase expression assay in 293T cells transfected with 100 ng of NF-κB-Luc together with 50 ng (black bar), 150 ng (hatched bar), or 450 ng (gray bar) of the indicated Flag-DAI expression vector containing either DAI or mutRLR-A DAI and 5 ng of the Renilla luciferase expression vector. In all samples, the amount of transfected DNA was kept constant with addition of EV plasmid DNA. Luciferase activity was evaluated as described in the legend to Fig. 1.
DAI activates NF-κB via RIP1
To determine whether DAI-dependent NF-κB activation is controlled by RIP1 in a manner analogous to a pathway that has been defined for responses mediated by TNFR, TLR3/TLR4, RLH, and DNA damage (19–23, 41), we performed NF-κB-Luc reporter assays and found that DAI induction of NF-κB-dependent gene expression was highly dependent on RLR-A (Fig. 3E). The small dose-dependent increase in reporter activity was eliminated when we tested DAI with mutated RLR-A and RLR-B, suggesting that the RLR-B motif may contribute to NF-κB activation (data not shown) despite the lack of any independent impact on RIP1 binding (see Fig. 3B). Overall, RLR-A was most critical in mediating RIP1 interaction as well as for biological activity.
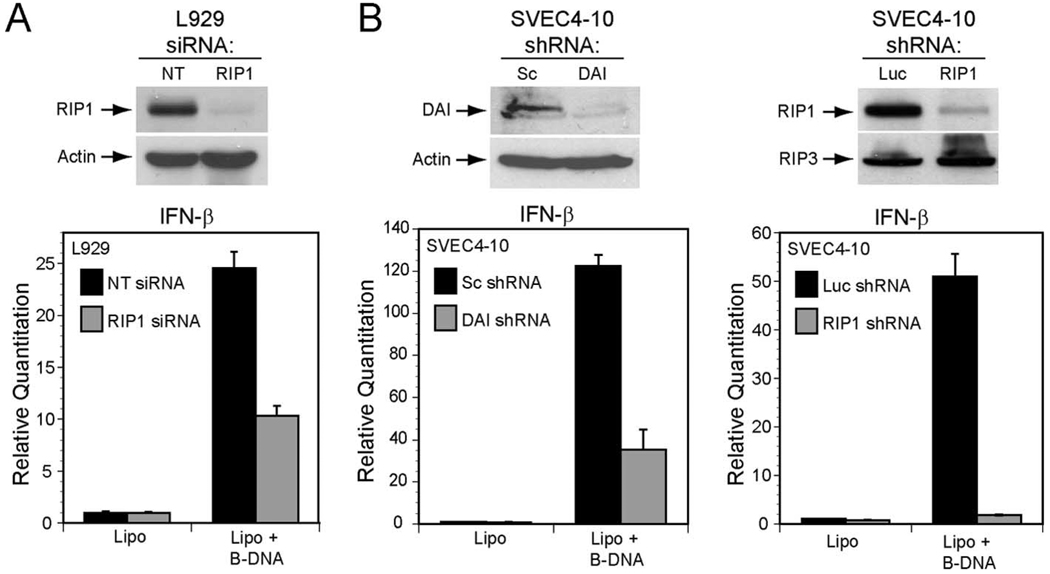
Given evidence that the DNA sensor DAI physically interacts with RIP1 via RLR-A to induce NF-κB, we next tested whether RIP1 was necessary for immunostimulatory dsDNA-induced activation of the innate immune response. Given the fact that DAI has been shown to be necessary for dsDNA-induced IFNβ expression in L929 fibroblast cell line (11), we used RNAi to inhibit RIP1 expression to evaluate the role of natural DAI and RIP1 upon delivery of immunostimulatory poly(dA:dT) DNA delivered by liposome-mediated transfection into L929 cells. Following transfection, IFNβ transcript levels were assessed by quantitative real-time PCR (Fig. 4A). Immunostimulatory DNA-dependent IFNβ expression was suppressed when RIP1 was knocked down in L929 cells line but not in L929 cells transfected with a nontargeting siRNA, consistent with previous reports (11, 16). As discussed in this published work as well as with observations suggesting that DAI is not the only DNA sensor in L929 cells (16), RIP1 siRNA did not completely suppress the response to DNA. Thus, the residual IFNβ induction is likely to be due to a DAI- and RIP1-independent DNA sensor pathway in these cells. To assess immunostimulatory DNA sensing pathways in another cell type, we evaluated murine endothelial cell SVEC4-10 cells stably expressing a scrambled shRNA or a shRNA-targeting DAI. RNAi-mediated knock down of DAI in SVEC4-10 cells suppressed IFNβ induction following delivery of poly(dA:dT) (Fig. 4B) much more dramatically than has been shown in L929 cells (11, 16). Consistent with the impact of RNAi directed at DAI, the SVEC4-10 cellular response to dsDNA was also highly dependent on RIP1. In contrast to control shRNA luciferase expressing cells, RIP1-specific shRNA completely blocked poly(dA:dT)-mediated IFNβ induction. Thus, DAI may be a more critical DNA sensor in endothelial cells than in fibroblasts. In SVEC4-10, DAI-mediated signaling via RIP1 predominates IFNβ activation. This evidence is consistent with RIP1 controlling the cellular NF-κB response that contributes to IFNβ activation following DAI-mediated detection of cytosolic dsDNA. RIP1 may also participate in DAI-independent DNA sensing pathways or affect IRF3/IRF7 activation as has been shown with cytosolic RNA sensors (20–22). Regardless, our evidence clearly demonstrates that RIP1 plays a central role in the cellular response to immunostimulatory cytosolic DNA and suggests that cell type may dictate the relative importance of DAI over other, uncharacterized DNA sensors.
FIGURE 4.
A, RIP1 knockdown attenuates B-DNA-mediated IFNβ gene induction in L929 and SVEC4-10 cells. Top, Autoradiogram of an IB for RIP1 protein in L929 cells 96 h following transfection with nontargeting (NT) siRNA or RIP1-specific siRNA. Actin expression control is shown below. Bottom, IFNβ gene induction in L929 cells assessed by quantitative RT-PCR 9 h after treatment with Lipofectamine 2000 (Lipo) alone or Lipo plus B-DNA (5 µg/ml). B, top left, Autoradiogram of an IB for DAI and actin control in SVEC4–10 cells stably expressing a scramble (Sc) shRNA or a shRNA to DAI. Top right, Autoradiogram of an IB for RIP1 and RIP3 protein in SVEC4–10 cells stably expressing a shRNA to luciferase (Luc) or to RIP1. Bottom, IFNβ gene induction in DAI shRNA (left) and RIP1 shRNA (right) SVEC4–10 cells was assessed by quantative RT-PCR following 5 h of Lipofectamine 2000 (Lipo) or Lipo plus B-DNA (5 µg/ml) stimulation.
DAI and RIP3 synergize to activate NF-κB via RIP1
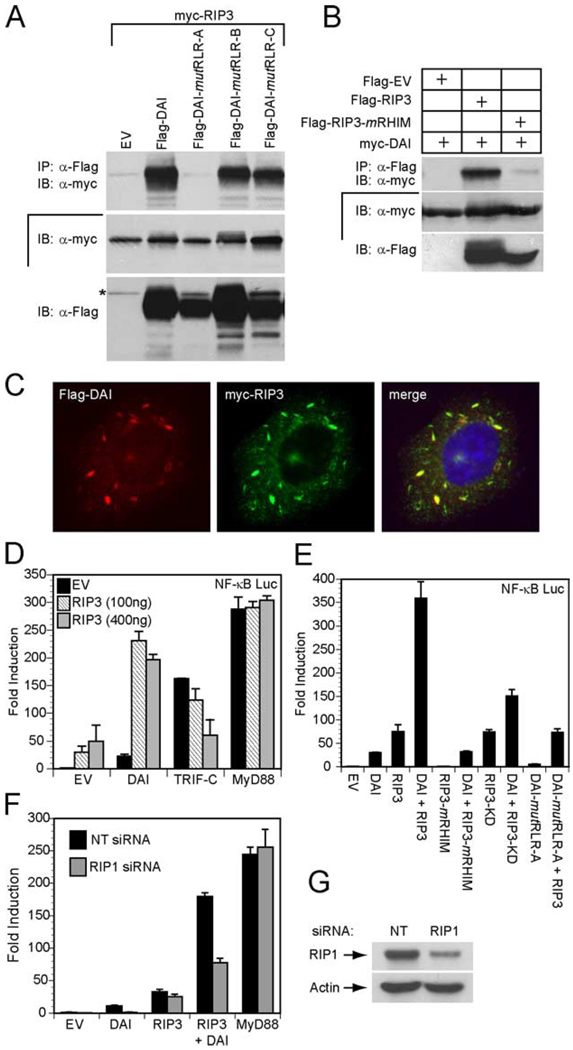
The ability of DAI to interact with RIP3 as well as RIP1 prompted us to investigate RHIM-dependent interaction of DAI with RIP3. Consistent with the RIP1 studies, RLR-A was the only DAI RLR that was required for interaction with RIP3. Thus, based on the behavior of mutRLR-A, RIP1 and RIP3 appear to bind to the same region of DAI (Fig. 5A). Furthermore, the interaction between RIP3 and DAI depended on the RIP3 RHIM (Fig. 5B). Flag-tagged RIP3 containing a mutated RHIM (RIP3-mRHIM) failed to immunoprecipitate c-Myc-DAI. This interaction was further supported by the colocalization of epitope tagged DAI and RIP3 in HeLa cells (Fig. 5C).
FIGURE 5.
DAI RHIM-dependent interaction with RIP3. A, Autoradiograph following IP and IB to detect mutant DAI interaction with RIP3. 293T cells were transfected with myc epitope-tagged RIP3 and Flag epitope-tagged DAI, DAI-mutRLR-A, DAI-mutRLR-B, DAI-mutRLR-C, or an EV. IP was with anti-Flag M2 beads and IB was with anti-myc Ab. IB analysis of total cell lysate confirmed the expression of myc-RIP3 and Flag-tagged proteins. The asterisk denotes residual signal of myc-RIP3. B, Autoradiograph following IP and IB to detect interaction of DAI and RIP3. 293T cells were transfected with myc epitope-tagged DAI and Flag epitope-tagged RIP3, RIP3-mRHIM, or an EV. IP used anti-Flag Ab and IB used anti-myc Ab. IB analysis of total cell lysate showed the relative expression of epitope-tagged proteins. C, Immunofluorescent localization of Flag-DAI with Myc-RIP3. Transfected HeLa cells were stained with rabbit anti-Flag Ab and with monoclonal 9E10 anti-myc Ab. Flag-DAI (red) was detected by an anti-rabbit Alexa-594-conjugated secondary Ab and Myc-RIP3 (green) was detected by an anti-mouse Alexa-488-conjugated secondary Ab. Stained cells were examined by epifluorescent microscopy (×1000). D, 293T cells were transfected with 100 ng of the pNF-κB luciferase reporter plasmid and 100 ng of Flag-DAI, Flag-TRIF, or Flag-MyD88 together with the indicated amount of Flag-RIP3 and 35 ng of the Renilla luciferase expression vectors. E, 293T cells were transfected with pNF-κB firefly luciferase reporter plasmid and 100 ng of the indicated Flag-DAI construct and/or Flag-RIP3 construct phRL-TK Renilla luciferase expression vector. F, RIP1 siRNA suppresses DAI-induced NF-κB activation. Nontargeting (NT) siRNA or RIP1 siRNA was transfected into 293T cells. At 72 h, cells were transfected with 100 ng of the pNF-κB luciferase reporter plasmid and 100 ng of Flag-DAI and/or Flag-RIP3, or Flag-MyD88 expression vector together with 5 ng of the Renilla luciferase expression vector. D–F, total DNA was held constant by adding EV. Luciferase activity assayed as described in the legend to Fig. 1. G, IB for RIP1 expression is shown using 293T cell extracts 72 h following transfection with the indicated siRNA.
The binding of RIP1 and RIP3 to DAI is reminiscent of the ability of both to interact with the TLR3/TLR4 adaptor protein TRIF through RHIM-dependent interactions (19). RIP3 is not responsible for direct signaling but has been reported to negatively regulate TRIF-mediated, RIP1-dependent activation of NF-κB (19). We evaluated the impact of RIP3 on DAI-dependent activation of NF-κB by coexpressing DAI and RIP3 in the presence of a NF-κB reporter gene (Fig. 5D). DAI or RIP3 alone induced modest (20- to 50-fold) NF-κB-dependent expression, and coexpression induced NF-κB-Luc reporter gene expression over 200-fold. For comparison, control transfections with a truncated constitutively active form of TRIF (TRIF-C) that activates NF-κB in an RHIM-dependent manner, confirmed the previously reported (19) RIP3-dependent suppression of TRIF-dependent NF-κB activation (Fig. 5E). Thus, DAI and RIP3 together activated NF-κB to higher levels, an impact that is opposite the RIP3-dependent down-regulation of NF-κB in TLR3/TLR4 signaling pathways (19). MyD88-induced NF-κB-dependent expression, also an RHIM-independent process, was not affected by coexpression of RIP3 (Fig. 5D). Coexpression of nonfunctional DAI-mutRLR-A and RIP3 induced NF-κB-dependent luciferase activity at levels comparable to RIP3 alone (Fig. 5E), demonstrating the importance of RHIM-dependent interactions with RIP3. Likewise, an intact DAI RLR-A as well as an intact RIP3 RHIM was required for NF-κB activation via RIP1 in this setting (Fig. 5E). Interestingly, maximal activation of NF-κB required RIP3 kinase activity. A kinase-deficient RIP3 (KD-RIP3) activated expression comparable to DAI alone, raising the possibility that RIP3 kinase is responsible for phosphorylation of components within the DAI-induced NF-κB activation pathway, potentially RIP1 or DAI.
To determine whether RIP1 plays a central role in NF-κB reporter gene expression induced by DAI and RIP3, we inhibited RIP1 expression using siRNA (Fig. 5G) and found a reduced activation by DAI as well as by DAI-RIP3 but not by RIP3 alone (Fig. 5F). Thus, endogenous RIP1 mediates the DAI-dependent components of this pathway. The precise pathway of RIP1-independent RIP3-dependent NF-κB activation will require further investigation. As expected, MyD88-dependent activation of NF-κB remained unaffected by inhibition of RIP1 expression. Thus, NF-κB signaling induced by DAI alone or by DAI plus RIP3 proceeds through the RIP1 adaptor.
Discussion
The innate immune system defends the host against microbial invasion by using multiple sensing systems that activate NF-κB and IRF family transcription factors (1–3). RIP1 was initially identified as a crucial adaptor protein in death receptor signaling (42), but accumulating evidence has implicated this adaptor in relaying signals from a number of pathogen recognition receptors, including sensors that detect immunostimulatory nucleic acids (19–22). Following dsRNA stimulation, RIP1 is recruited by TLR3/TRIF to activate NF-κB (19). Stimulation of RLH by cytosolic RNA depends on a complex of RIP1, Cardif/MAVS, and either TNFR-associated death domain protein or Fas-associated death domain protein, activating NF-κB as well as IRF3 (or IRF7) (20–22). In the present study, we reveal for the first time that RIP1 plays a central role in the signal transduction pathways activated by cytosolic DNA and have characterized the steps that RIP1 controls to bridge the DNA sensor DAI to downstream signaling networks.
DAI is likely to respond to cytosolic viral, microbial, or host DNA, stimulating NF-κB activation that drives subsequent proinflammatory and antiviral cytokine production in the cell-intrinsic innate immune response (11, 16). DAI contains three domains that have the sequence elements reminiscent of a RHIM. Only the most N-terminal of these (RLR-A) is critical for RHIM-dependent interactions with RIP1 and RIP3. Although interactions with other RHIM proteins, cellular TRIF and viral M45, remain to be studied, our current evidence strongly implicates DAI in direct induction of NF-κB via RIP1 and downstream IKKβ but independent of TBK-1. The location of RLR-A within the D3 region of DAI is consistent with previous mapping of regions important for induction of IFNβ as well as in DNA recognition (16) and raises the potential that this RHIM is closely associated with DNA binding as well as interaction with RIP1. A more extensive series of amino acid substitution mutants will be needed to fully address these issues. DAI also contains two additional conserved regions, RLR-B and RLR-C, that appear dispensable for interactions with RIP1 or RIP3, although RLR-B may work together with RLR-A for maximal activation of NF-κB. RLR-C maps to the region of DAI that recruits TBK-1 (11) and thus may play a regulatory role influencing IRF3 activation. We are currently evaluating the importance of these additional RHIM-like domains in regulating DAI function.
RIP3 is known to bind RIP1 in a RHIM-dependent interaction that suppresses NF-κB activation in response to either TNF-α or the TLR3/TLR4 adaptor protein TRIF (19, 24). DAI also interacts with RIP3 via a RHIM, and this interaction apparently enhances DAI-mediated NF-κB activation. This synergistic NF-κB response required an intact RIP3 kinase domain and was mediated through RIP1, raising the interesting and speculative possibility that RIP3 differentially regulates dsRNA sensing by TLR3 and cytosolic dsDNA sensing by DAI. This would enable a cell to fine-tune the cellular response and differentiate between various immunostimulatory nucleic acids. No phenotype has yet been reported for RIP3-deficient mice (43), but, based on the data we present here, DAI-mediated signaling should be attenuated in some cell types in these animals.
DAI stimulates IFNβ expression through pathways that activate IRF3 and NF-κB much like other sensors, and may play a more important role in certain cell types such as endothelial cells. In addition to adding clarity to the pathway leading to NF-κB activation, our data broadens the role for RIP1 in pathogen recognition. RHIM-dependent pathways have been shown to be critical for sensing Gram-negative bacteria in Drosophila, via the immunodeficiency pathway (44), as well as in mammals, via a MyD88-independent signal transduction pathway that is activated by TLR3 and TLR4. An ortholog of RIP1 has therefore been implicated across a broad evolutionary landscape in host defense to microbes. The importance of RHIM interactions in relaying signals from DAI in response to immunostimulatory DNA is analogous to the role of TLR4 in sensing LPS or TLR3 in sensing dsRNA and signaling via the TRIF adaptor to activate NF-κB and IRF3 (Fig. 6). Other parallels may emerge with regard to the cell type-specific importance of DAI in activation of the type I IFN response. Our data expand the range of RHIM-dependent interactions in innate sensing of intracellular bacterial and viral pathogens, and open new avenues by which pathogens may modulate sensing, suppress cell death, reduce activation of type I IFN, and counteract IFN-induced antiviral activities (45–48).
FIGURE 6.
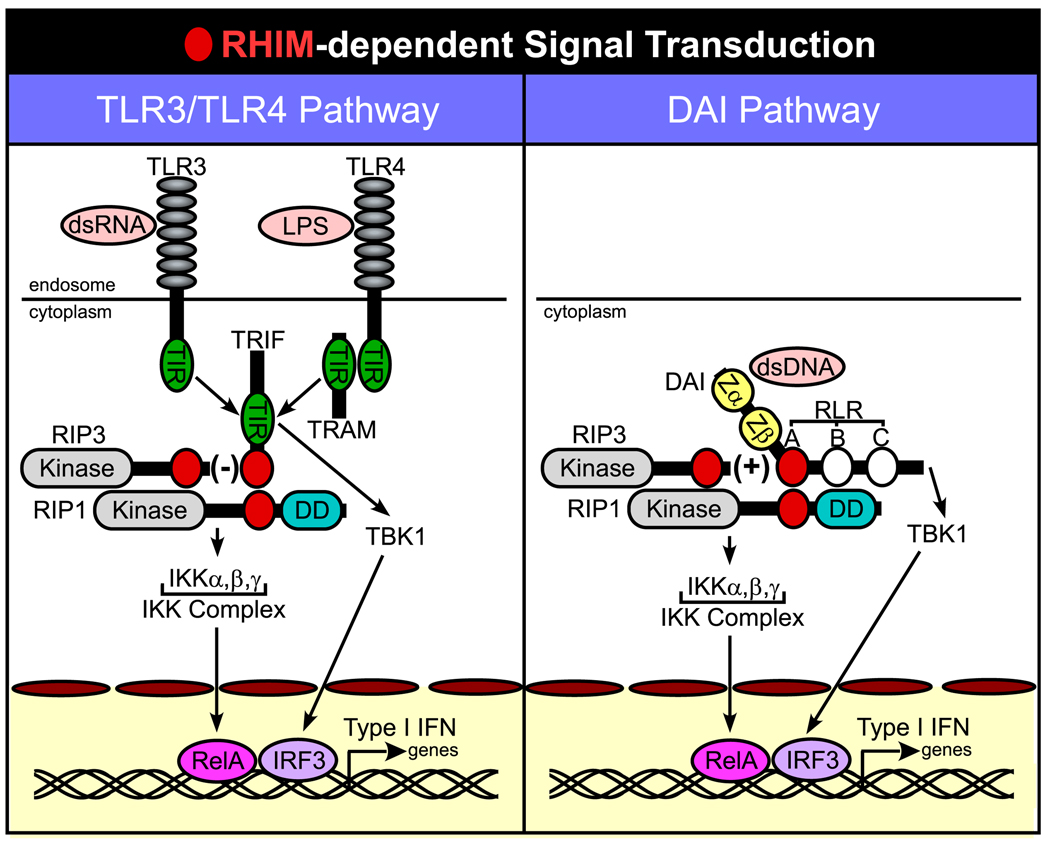
Schematic diagram of RHIM-dependent signaling. Common steps in RHIM-dependent interactions are shown for TLR3/TLR4 signaling via TRIF (left), as summarized in (19), as well as for DAI (right), described in this report. When triggered, DAI and TRIF recruit RIP1 in a pathway that activates NF-κB by release of RelA from the IKK complex. TRIF and DAI also induce TBK1-mediated activation of IRF3. Together, RelA (NF-κB) and IRF3 promote Type I IFN gene expression. RIP3 regulates RIP1-dependent NF-κB activation in opposite ways, depressing RHIM-dependent TRIF-mediated events, but stimulating RHIM-dependent DAI-mediated events. Red circles indicate the RHIM of DAI, TRIF, RIP1, and RIP3. White circles on DAI indicate RLR-B and -C.
Acknowledgments
We acknowledge Carl Davis and Rafi Ahmed for providing reagents and A. L. McCormick and R. Tandon for critical reading of the manuscript.
Footnotes
This work was supported by Public Health Service Grant R01 AI20211 (to E.S.M.) and T32 HL069769 (to J.W.U.) as well as a grant from the Georgia Cancer Coalition.
Abbreviations used in this paper: IRF, IFN regulatory factor; TANK, TNFR-associated factor family member-associated NF-κB activator; TBK, TANK-binding kinase; IKK, IκB kinase; TRIF, TIR domain-containing adaptor-inducing IFNβ; RLH, retinoic acid-inducible gene I-like helicase; DD, death domain; RHIM, RIP homotypic interaction motif; RIP, receptor-interacting protein kinase; MCMV, murine CMV; RLR, RHIM-like repeat; shRNA, short hairpin RNA; IκBα-SR, IκBα-super repressor; KD, kinase dead; EV, empty vector; IB, immunoblot.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Takaoka A, Taniguchi T. Cytosolic DNA recognition for triggering innate immune responses. Adv. Drug Delivery Rev. 2007;60:847–857. doi: 10.1016/j.addr.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Ishii KJ, Akira S. Innate immune recognition of, and regulation by, DNA. Trends Immunol. 2006;27:525–532. doi: 10.1016/j.it.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Kawai T, Akira S. Innate immune recognition of viral infection. Nat. Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 4.Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Kumagai Y, Takeuchi O, Akira S. TLR9 as a key receptor for the recognition of DNA. Adv. Drug Delivery Rev. 2008;60:795–804. doi: 10.1016/j.addr.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat. Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 7.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, Akira S. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 9.Abate DA, Watanabe S, Mocarski ES. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J. Virol. 2004;78:10995–11006. doi: 10.1128/JVI.78.20.10995-11006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paladino P, Cummings DT, Noyce RS, Mossman KL. The IFN-independent response to virus particle entry provides a first line of antiviral defense that is independent of TLRs and retinoic acid-inducible gene I. J. Immunol. 2006;177:8008–8016. doi: 10.4049/jimmunol.177.11.8008. [DOI] [PubMed] [Google Scholar]
- 11.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 12.Fu Y, Comella N, Tognazzi K, Brown LF, Dvorak HF, Kocher O. Cloning of DLM-1, a novel gene that is up-regulated in activated macrophages, using RNA differential display. Gene. 1999;240:157–163. doi: 10.1016/s0378-1119(99)00419-9. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz T, Behlke J, Lowenhaupt K, Heinemann U, Rich A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 2001;8:761–765. doi: 10.1038/nsb0901-761. [DOI] [PubMed] [Google Scholar]
- 14.Honda K, Takaoka A, Taniguchi T. Type I interferon gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, Choi MK, Ban T, Yanai H, Negishi H, Lu Y, Tamura T, Takaoka A, Nishikura K, Taniguchi T. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc. Natl. Acad. Sci. USA. 2008;105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Festjens N, Vanden Berghe T, Cornelis S, Vandenabeele P. RIP1, a kinase on the crossroads of a cell’s decision to live or die. Cell Death Differ. 2007;14:400–410. doi: 10.1038/sj.cdd.4402085. [DOI] [PubMed] [Google Scholar]
- 18.Meylan E, Tschopp J. The RIP kinases: crucial integrators of cellular stress. Trends Biochem. Sci. 2005;30:151–159. doi: 10.1016/j.tibs.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Meylan E, Burns K, Hofmann K, Blancheteau V, Martinon F, Kelliher M, Tschopp J. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-κB activation. Nat. Immunol. 2004;5:503–507. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- 20.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 21.Balachandran S, Thomas E, Barber GN. A FADD-dependent innate immune mechanism in mammalian cells. Nature. 2004;432:401–405. doi: 10.1038/nature03124. [DOI] [PubMed] [Google Scholar]
- 22.Michallet MC, Meylan E, Ermolaeva MA, Vazquez J, Rebsamen M, Curran J, Poeck H, Bscheider M, Hartmann G, Konig M, et al. TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity. 2008;28:651–661. doi: 10.1016/j.immuni.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P. The death domain kinase RIP mediates the TNF-induced NF-κB signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- 24.Sun X, Yin J, Starovasnik MA, Fairbrother WJ, Dixit VM. Identification of a novel homotypic interaction motif required for the phosphorylation of receptor-interacting protein (RIP) by RIP3. J. Biol. Chem. 2002;277:9505–9511. doi: 10.1074/jbc.M109488200. [DOI] [PubMed] [Google Scholar]
- 25.Vivarelli MS, McDonald D, Miller M, Cusson N, Kelliher M, Geha RS. RIP links TLR4 to Akt and is essential for cell survival in response to LPS stimulation. J. Exp. Med. 2004;200:399–404. doi: 10.1084/jem.20040446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Upton JW, Kaiser WJ, Mocarski ES. Cytomegalovirus M45 cell death suppression requires RHIM-dependent interaction with receptor-interacting protein 1 (RIP1) J. Biol. Chem. 2008;283:16966–16970. doi: 10.1074/jbc.C800051200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaiser WJ, Offermann MK. Apoptosis induced by the toll-like receptor adaptor TRIF is dependent on its receptor interacting protein homotypic interaction motif. J. Immunol. 2005;174:4942–4952. doi: 10.4049/jimmunol.174.8.4942. [DOI] [PubMed] [Google Scholar]
- 29.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 30.O’Donnell SM, Hansberger MW, Connolly JL, Chappell JD, Watson MJ, Pierce JM, Wetzel JD, Han W, Barton ES, Forrest JC, et al. Organ-specific roles for transcription factor NF-κB in reovirus-induced apoptosis and disease. J. Clin. Invest. 2005;115:2341–2350. doi: 10.1172/JCI22428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hida S, Ogasawara K, Sato K, Abe M, Takayanagi H, Yokochi T, Sato T, Hirose S, Shirai T, Taki S, Taniguchi T. CD8+ T cell-mediated skin disease in mice lacking IRF-2, the transcriptional attenuator of interferon-α/β signaling. Immunity. 2000;13:643–655. doi: 10.1016/s1074-7613(00)00064-9. [DOI] [PubMed] [Google Scholar]
- 32.Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 33.Bartz SR, Vodicka MA. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods. 1997;12:337–342. doi: 10.1006/meth.1997.0487. [DOI] [PubMed] [Google Scholar]
- 34.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 35.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 36.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of IκB-α proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 37.Traenckner EB, Pahl HL, Henkel T, Schmidt KN, Wilk S, Baeuerle PA. Phosphorylation of human IκB-α on serines 32 and 36 controls IκB-α proteolysis and NF-κB activation in response to diverse stimuli. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lenardo MJ, Fan CM, Maniatis T, Baltimore D. The involvement of NF-κB in β-interferon gene regulation reveals its role as widely inducible mediator of signal transduction. Cell. 1989;57:287–294. doi: 10.1016/0092-8674(89)90966-5. [DOI] [PubMed] [Google Scholar]
- 39.Pham HT, Park MY, Kim KK, Kim YG, Ahn JH. Intracellular localization of human ZBP1: differential regulation by the Z-DNA binding domain, Zα, in splice variants. Biochem. Biophys. Res. Commun. 2006;348:145–152. doi: 10.1016/j.bbrc.2006.07.061. [DOI] [PubMed] [Google Scholar]
- 40.Deigendesch N, Koch-Nolte F, Rothenburg S. ZBP1 subcellular localization and association with stress granules is controlled by its Z-DNA binding domains. Nucleic Acids Res. 2006;34:5007–5020. doi: 10.1093/nar/gkl575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janssens S, Tinel A, Lippens S, Tschopp J. PIDD mediates NF-κB activation in response to DNA damage. Cell. 2005;123:1079–1092. doi: 10.1016/j.cell.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 42.Stanger BZ, Leder P, Lee TH, Kim E, Seed B. RIP: a novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell. 1995;81:513–523. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- 43.Newton K, Sun X, Dixit VM. Kinase RIP3 is dispensable for normal NF-κBs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and Toll-like receptors 2 and 4. Mol. Cell. Biol. 2004;24:1464–1469. doi: 10.1128/MCB.24.4.1464-1469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, Oshima Y, Peach C, Erturk-Hasdemir D, Goldman WE, Oh BH, et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat. Immunol. 2006;7:715–723. doi: 10.1038/ni1356. [DOI] [PubMed] [Google Scholar]
- 45.Finlay BB, McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 46.Schroder M, Bowie AG. An arms race: innate antiviral responses and counteracting viral strategies. Biochem. Soc. Trans. 2007;35:1512–1514. doi: 10.1042/BST0351512. [DOI] [PubMed] [Google Scholar]
- 47.McLean JE, Ruck A, Shirazian A, Pooyaei-Mehr F, Zakeri ZF. Viral manipulation of cell death. Curr. Pharm. Des. 2008;14:198–220. doi: 10.2174/138161208783413329. [DOI] [PubMed] [Google Scholar]
- 48.Roy CR, Mocarski ES. Pathogen subversion of cell-intrinsic innate immunity. Nat. Immunol. 2007;8:1179–1187. doi: 10.1038/ni1528. [DOI] [PubMed] [Google Scholar]