Abstract
The mosquito Culex quinquefasciatus poses a significant threat to human and veterinary health as a primary vector of West Nile virus (WNV), the filarial worm Wuchereria bancrofti, and an avian malaria parasite. Comparative phylogenomics revealed an expanded canonical C. quinquefasciatus immune gene repertoire compared with those of Aedes aegypti and Anopheles gambiae. Transcriptomic analysis of C. quinquefasciatus genes responsive to WNV, W. bancrofti and non-native bacteria facilitated an unprecedented meta-analysis of 25 vector-pathogen interactions involving arboviruses, filarial worms, bacteria and malaria parasites, revealing common and distinct responses to these pathogen types in three mosquito genera. Our findings provide support for the hypothesis that mosquito-borne pathogens have evolved to evade innate immune responses in three vector mosquito species of major medical importance.
The Southern house mosquito, Culex quinquefasciatus, is a geographically widespread, often abundant mosquito that is an epidemiologically significant vector for an exceptionally diverse array of pathogens including multiple arboviruses, filarial worms and protozoa. C. quinquefasciatus transmits West Nile virus (WNV), St. Louis encephalitis virus and other arboviruses, and acts as the primary vector of the causative agent of lymphatic filariasis, Wuchereria bancrofti, and Plasmodium relictum, an avian malaria parasite. Despite the public health significance of C. quinquefasciatus, knowledge of the insect’s response capacities to this diverse array of pathogens is limited.
Availability of the C. quinquefasciatus genome sequence (1) enabled comparative phylogenomic analyses with Aedes aegypti (2), Anopheles gambiae (3) and Drosophila melanogaster(4) that identified 500 C. quinquefasciatus immunity genes from 39 (sub)families or processes (Table S1). Conservation of C. quinquefasciatus gene family members follows the species phylogeny, showing greatest similarities with A. aegypti. Expansions of C-type lectins (CTLs), fibrinogen-related proteins (FREPs) and serine protease inhibitors (SRPNs) account for much of the 20–30% increase in C. quinquefasciatus immunity gene number compared to A. aegypti (417 genes) and A. gambiae (380 genes) (Figs. S1–S4), This apparent diversification in immune surveillance and immune signal amplification processes seems consistent with selection driven by polluted, microbially complex habitats in which C. quinquefasciatus oviposits and develops (5).
Whole genome microarray analysis revealed dynamic changes in infection response gene (IRG) transcription in WNV-infected mosquitoes (Fig. S5). Significant changes are observed for 22 transcripts in the midgut and 309 in the carcass (i.e., the remainder of the body) at 7 days post-infection (dpi), with the greater number of IRGs in the latter apparently reflecting the diversity of infected cell and tissue types in the carcass. At 14 dpi, more IRGs are modulated in midgut (539) and carcass (490) when WNV infection has spread in midgut cells and has disseminated to the salivary glands (6). Few canonical immunity genes are represented among C. quinquefasciatus WNV IRGs (Fig. S5). Five CTL genes within a C. quinquefasciatus-specific gene expansion (Fig. S3) are up-regulated. Several genes related to the Toll, Imd and JAK-STAT pathways, including Spaetzle, REL1, IAP2 and STAT orthologs, are activated in C. quinquefasciatus and in A. aegypti (7, 8) by WNV and Dengue virus (DENV) infection, respectively, further supporting a key role of these defense systems in controlling viral pathogens.
Although the C. quinquefasciatus genome encodes orthologs for all components of the anti-viral defense RNA interference (RNAi) pathway (9), none of them is transcriptionally modulated significantly during WNV infection. Similarly, RNAi components are not transcriptionally modulated during arbovirus infection in A. aegypti (10), even though RNAi function is key to limiting these infections in mosquitoes (10–12). Apoptosis is evident and C. quinquefasciatus IAP1 is repressed in WNV-infected salivary glands (6, 13). However, no significant changes in transcript abundance for caspases, caspase activators, IAP genes (other than IAP2), or autophagy-related genes are evident in WNV-infected C. quinquefasciatus, even though modulation of apoptosis (14) or autophagy (15) pathway function affects viral infection in flies. The non-responsiveness of these genes appears to reflect the persistent and generally non-cytolytic nature of arbovirus infections in a susceptible vector; overt activation of these responses would counteract virus persistence and transmission.
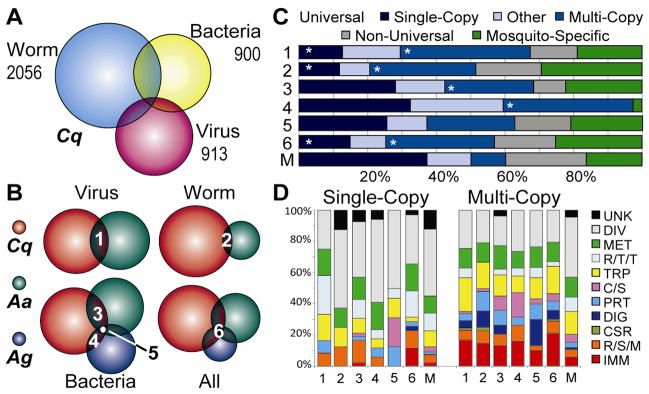
Comparative analysis of ESTs (Tables S3–4) from W. bancrofti-infected C. quinquefasciatus revealed many novel IRGs, presumably because infection with a large metazoan parasite inflicts traumatic injury. Infection with non-native bacteria elicits acute cellular and humoral immune responses in C. quinquefasciatus and other vector mosquitoes (16–18). Approximately 60% of W. bancrofti or bacteria IRGs are of diverse or unknown function (Fig. S6), and only small proportions (4% W. bancrofti and 6% bacteria) are immunity genes. Comparison of C. quinquefasciatus virus, filarial worm, and bacteria IRGs reveals unexpected and extensive overlap (548 genes) between W. bancrofti and bacteria IRGs (Figs. 1A and S6). Overall, 38 C. quinquefasciatus IRGs are common among all three infections (Table S5).
Figure 1.
Infection response genes (IRGs) in the mosquitoes Culex quinquefasciatus (Cq), Aedes aegypti (Aa) and Anopheles gambiae (Ag). (A) Shared and unique infection response genes in C. quinquefasciatus infected with a filarial worm, bacteria, or virus. (B) Proportions of shared and unique IRGs post-infection with viruses (1), filaria (2) or bacteria (3) in C. quinquefasciatus and A. aegypti, in C. quinquefasciatus and A. gambiae (4), and in all three species (5); and common IRGs in C. quinquefasciatus, A. aegypti, and A. gambiae (6). (C) Orthology relationships for IRG sets (Rows 1–6). IRGs with orthologs in at least 20 arthropod species were classified as Universal, as compared to Non-Universal or Mosquito-Specific. Gene copy-number counts distinguish mostly single- and multi-copy orthologous groups. IRG sets 1–6 were compared to 10,083 mosquito OGs (Row M) to identify significantly greater or smaller (asterisks) proportions (Fisher's Exact Tests: p<1e-5). (D) Consensus functional categories of universal single-copy (left) and multi-copy (right) orthologous groups of IRG sets Rows 1–6, and all mosquito groups (Row M). Functional groups are described in SOM, and (24). Each set of IRGs is described in Supplemental Tables S7–12.
The identification of C. quinquefasciatus IRGs provided an unprecedented opportunity to undertake a meta-analysis of 25 vector-pathogen interactions in C. quinquefasciatus, A. aegypti, and A. gambiae infected with arboviruses, parasites and bacteria (Fig. 1B, Table S6) within the context of orthologous groups (OGs) that define evolutionarily related genes. A set of 69 arbovirus IRG OGs (representing 93 C. quinquefasciatus and 89 A. aegypti genes) was implicated in C. quinquefasciatus-WNV, A. aegypti-DENV and A. aegypti-Sindbis virus (SINV) responses (Fig. S7, Table S7). A cytochrome P450 DENV IRG from Drosophila (Cyp6a19, FBgn0033979) and mammalian cells (19) is similar to genes that respond significantly in C. quinquefasciatus-WNV (CPIJ004411) infection, and in A. aegypti-SINV and A. aegypti-DENV (AAEL009117) infections, highlighting the potential importance of this molecule as a universal arbovirus IRG. Filarial worm IRGs comprised 41 OGs modulated during C. quinquefasciatus-W. bancrofti infection and infection of A. aegypti with Brugia malayi (Fig. S8, Table S8). The IRGs represented most frequently include serine proteases and cuticle proteins. Changes in the latter may be associated with tissue repair necessitated by parasite invasion, migration and development (20). Increased representation of heat shock protein and cytochrome P450 IRGs appears to reflect stress during the infection response. The most extensive overlap (113 OGs) in bacterial IRGs was observed between Culicine mosquitoes, C. quinquefasciatus and A. aegypti (Table S9). Only 34 OGs and 26 OGs represent IRGs (Fig. S9) in bacteria-infected C. quinquefasciatus and A. gambiae (Table S10), and A. aegypti and A. gambiae, respectively. Among 16 OGs containing bacteria IRGs from all three species, serine proteases, cecropins, myosin light chain, and components of the 26S proteasome are highly represented (Table S11). A meta-analysis of bacteria, filarial worm, virus, and malaria parasite infection datasets from C. quinquefasciatus, A. aegypti and A. gambiae reveals 95 orthologous IRGs that span mosquito species and pathogen types (Figs. 1B and S10, Table S12).
Orthology data (21) were employed to distinguish universal (see Fig. 1) multi- or single-copy OGs from mosquito-specific OGs, revealing that the majority of IRGs have orthologs across Arthropoda (Fig. 1C, Table S13). Universal multi-copy OGs are overrepresented among IRGs for viruses, filaria and bacteria; and universal single-copy OGs are underrepresented among arbovirus and filarial worm IRGs (and among IRGs common to all pathogens in C. quinquefasciatus, A. aegypti and A. gambiae) compared to the complete set of mosquito OGs (Fig 1C). Immunity genes (IMM, Fig. 1D), including CTLs, CLIPs and SRPNs, are generally more prevalent among responsive multi-copy OGs than among responsive single-copy OGs. In fact, no canonical immunity genes are found among arbovirus- or filarial worm-responsive universal single-copy OGs.
The cosmopolitan distribution of C. quinquefasciatus across continents and ecozones generally south of 39° N latitude implies that this species has the plasticity to adapt to diverse habitats, and this plasticity may be enhanced by an expanded immunity gene repertoire. Overrepresentation of universal multi-copy OGs among pathogen IRGs implies that members of expanded gene families have been recruited into pathogen-responsive defense pathways. Arboviral and filarial worm infections constitute susceptible, long-term vector-pathogen interactions in which the pathogen undergoes amplification or develops intracellularly, while acute infections with non-native bacteria trigger systemic immunity and are cleared rapidly (22, 23). Our meta-analysis reveals that arboviral and filarial worm pathogens transmitted by vector mosquitoes modulate very few canonical immunity genes, and fail to affect expression of RNAi and most programmed cell death pathway genes in these vectors. Our results therefore provide strong support for the hypothesis that pathogens that successfully develop in, and are transmitted by, vector mosquitoes have evolved to avoid most immune responses in the three mosquito genera responsible for the vast majority of human morbidity and mortality attributable to insect-transmitted pathogens.
Supplementary Material
Acknowledgments
This work was supported by Iowa Agriculture and Home Economics Experiment Station, Ames, IA, USA, to L.C.B.; funding from the Foundation for the National Institutes of Health, Grand Challenges in Global Health Initiative to C.L.C.; NIH grant P20 RR017686 to K.M.; NIH grant R21 AI067642 to R.J.C.; CDC grant T01CCT622892 to S.H.; NIAID/NIH/DHHS contract number HHSN266200400039C to University of Notre Dame (VectorBase); NIAID/NIH/DHHS contract number HHSN266200400001C to the Broad Institute; Swiss National Science Foundation grant 3100A0 to E.M.Z.; NIH grant R01 AI59492 to A.S.R.; NIH grants R01 AI19769 and R01 AI67698 to B.M.C; NIH grants R01 AI061576, R01 AI059492 and R01 AI078997 and Johns Hopkins Malaria Research Institute Core Facilities at the Johns Hopkins Malaria Research Institute to G.D.; and the DeLuca Professorship from Boston College to M.A.T.M. R.M.W. was supported by the Giorgi-Cavlieri Foundation, Swiss National Science Foundation grant 3100A0 to E.M.Z., and a Wellcome Trust Ph.D. fellowship. J.L.R. was supported by NIH individual NRSA training grant F31 AI080161-01A1 and by the American Society for Microbiology Robert D. Watkins Graduate Research Fellowship. S.H.P.P.K. was supported by a Wellcome Trust Fellowship. D.L.V. was supported by NIH grant T32 A107536. The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (accession #: GSE23045).
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References and Notes
- 1.Arensburger P, et al. Science. 2010 submitted. [Google Scholar]
- 2.Waterhouse R, et al. Science. 2007;316:1738. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christophides GK, et al. Science. 2002;298:159. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- 4.Sackton TB, et al. Nat Genet. 2007;39:1461. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- 5.Silverly R. Mosquitoes of Indiana. Indiana State Board of Health; Muncie, IN: 1972. pp. 92–93. [Google Scholar]
- 6.Girard YA, Popov V, Wen J, Han V, Higgs S. J Med Entomol. 2005;42:429. doi: 10.1093/jmedent/42.3.429. [DOI] [PubMed] [Google Scholar]
- 7.Souza-Neto JA, Sim S, Dimopoulos G. Proc Natl Acad Sci U S A. 2009;106:17841. doi: 10.1073/pnas.0905006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xi Z, Ramirez JL, Dimopoulos G. PLoS Pathog. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell CL, Black WC, Hess AM, Foy BD. BMC Genomics. 2008;9:425. doi: 10.1186/1471-2164-9-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell CL, et al. BMC Microbiol. 2008;8:47. doi: 10.1186/1471-2180-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez-Vargas I, et al. PLoS Pathog. 2009;5:e1000299. doi: 10.1371/journal.ppat.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keene KM, et al. Proc Natl Acad Sci USA. 2004;101:17240. doi: 10.1073/pnas.0406983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girard Y, et al. J Med Entomol. 2010;47:421. doi: 10.1603/me09249. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Blair CD, Olson KE, Clem RJ. J Gen Virol. 2008;89:2651. doi: 10.1099/vir.0.2008/005314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Immunity. 2009;30:588. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hillyer JF, Schmidt SL, Christensen BM. J Parasitol. 2003;89:62. doi: 10.1645/0022-3395(2003)089[0062:RPAMOB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 17.Bartholomay LC, Farid HA, Ramzy R, Christensen BM. Mol Biochem Parasitol. 2003;130:43. doi: 10.1016/s0166-6851(03)00143-9. [DOI] [PubMed] [Google Scholar]
- 18.Dimopoulos G, et al. Proc Natl Acad Sci U S A. 2002;99:8814. doi: 10.1073/pnas.092274999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sessions OM, et al. Nature. 2009;458:1047. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erickson SM, et al. PLoS Negl Trop Dis. 2009;3:e529. doi: 10.1371/journal.pntd.0000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kriventseva EV, Rahman N, Espinosa O, Zdobnov EM. Nucleic Acids Res. 2008;36:D271. doi: 10.1093/nar/gkm845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartholomay LC, Fuchs JF, Vizioli J, Lowenberger C, Christensen BM. Insect Mol Biol. 2004;13:125. doi: 10.1111/j.0962-1075.2004.00467.x. [DOI] [PubMed] [Google Scholar]
- 23.Hillyer JF, Schmidt SL, Christensen BM. Microbes and Infection. 2004;6:448. doi: 10.1016/j.micinf.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Nene V, et al. Science. 2007;316:1718. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.