Abstract
Background
Non-Typhoidal Salmonella (NTS) is an important cause of invasive bacterial disease and associated with mortality in Africa. However, little is known about the environmental reservoirs and predominant modes of transmission. Our study aimed to study the role of domestic animals in the transmission of NTS to humans in rural area of The Gambia.
Methodology
Human NTS isolates were obtained through an active population-based case-control surveillance study designated to determine the aetiology and epidemiology of enteric infections covering 27,567 Gambian children less than five years of age in the surveillance area. Fourteen children infected with NTS were traced back to their family compounds and anal swabs collected from 210 domestic animals present in their households. Identified NTSs were serotyped and genotyped by multi-locus sequencing typing.
Principal Findings
NTS was identified from 21/210 animal sources in the households of the 14 infected children. Chickens carried NTS more frequently than sheep and goats; 66.6%, 28.6% and 4.8% respectively. The most common NTS serovars were S. Colindale in humans (21.42%) and S. Poona in animals (14.28%). MLST on the 35 NTS revealed four new alleles and 24 sequence types (ST) of which 18 (75%) STs were novel. There was no overlap in serovars or genotypes of NTS recovered from humans or animal sources in the same household.
Conclusion
Our results do not support the hypothesis that humans and animals in close contact in the same household carry genotypically similar Salmonella serovars. These findings form an important baseline for future studies of transmission of NTS in humans and animals in Africa.
Author Summary
Salmonellosis is a neglected tropical disease causing serious dysentery and septicaemia particularly in young infants, elderly and immunocompromised individuals such as HIV patients and associated with substantial mortality in developing countries. Salmonellosis also constitutes a major public health problem as it is considered the most widespread bacterial zoonosis of food origin throughout the world. Many epidemiological data exist from developed countries concerning transmission of Non-Typhoidal Salmonella (NTS) but few are available from developing countries. In addition few studies in sub-Saharan Africa have considered the interface between humans and their environment in relation to animals present in the household and food hygiene. This study describes the prevalence of NTS among fourteen Gambian children and 210 domestic animals living in close proximity (household) to the children in a rural setting in The Gambia. We found that the domestic animals living in the same household as patients carried different NTS serovar and genotypes; indicating that zoonotic transmission does not occur in our setting. This study provides baseline data for future studies of transmission of NTS in rural Africa.
Introduction
Non-Typhoidal Salmonella (NTS) are important causes of invasive bacterial diseases and are associated with substantial mortality. In rural Gambia, NTS was the second most important blood culture isolate after Streptococcus pneumoniae in children with invasive bacterial disease [1]. Similarly, a study in an urban hospital of The Gambia revealed that NTS represented 8.6% of the bacteraemia cases [2]. NTS also cause serious dysentery and septicaemia particularly in young infants [3], [4]. However, little is known about environmental reservoirs and predominant modes of transmission especially in the African context [5], [6]. Various sources including farm animals, pets and reptiles have been potentially implicated in the transmission of NTS between animals and humans but their direct involvement in the transmission has never been demonstrated [7]. The asymptomatic Salmonella carrier state in poultry has serious consequences on food safety and public health due to the risks of food poisoning following consumption of contaminated products. Salmonella enterica serovar Enteritidis can persist in the caecum or ovaries of chickens without triggering any clinical signs. Salmonellosis in young chickens may cause high mortality as a result of severe diarrhoea and dehydration, and include a greater risk of evolving into a carrier state in the surviving animals [8], [9], [10]. Small ruminants, such as sheep and goats, are also potential carriers of Salmonella [11], [12]. In The Gambia, like in most of the African countries, the majority of the population lives in rural areas and depends on agriculture and livestock. In these areas, the most common animals kept in the compound are chickens, sheep and goats. Therefore, farmers and their families often live in close contact with their livestock and even under the same roof and are thus at increased risk of contracting zoonotic infections, particularly children who often play on the ground. Domestic animals may thus play an important role in the maintenance and transmission of NTS to humans at the community level. Investigating the relatedness between human and animal strains of NTS could provide useful information about the epidemiology of these pathogens. Our study seeks to assess the contribution of domestic animals to the transmission of NTS to humans and to study the genetic relatedness between human and animal strains. This study will also provide useful information about the prevalence of NTS in domestic animals in rural areas of The Gambia.
Materials and Methods
Ethics statement
The study protocol and consent form were approved by the Ethics Committees of the Joint Gambian Government/MRC Ethics Committee and the Ethics Committees of the London School of Hygiene and Tropical Medicine, UK and by the Institutional Review Board of the University of Maryland, Baltimore. For any patient eligible for the study written informed consent was obtained prior to their enrollment after the objectives and risks and benefits associated with participation. 95% of eligible subjects agreed to participate. If the participant was illiterate, a witness who was present throughout the consent procedure completed the necessary portions and signed the consent form; the parent/participant marked the consent form (either fingerprint or other notation) cases. If the person was literate, then he/she read and signed the consent form.
All animals used in this study were handled by professional veterinary staff in strict accordance with good animal practice as defined by the Gambian Government/ITC code of practice for the care and use of animals for scientific purposes. All animal work were conducted with ethical approval from both the Joint Gambian Government/ MRC Ethics Committee and the Ethics Committees of the London School of Hygiene and Tropical Medicine, UK.
Animal owners gave their written informed consent to examine and take rectal swabs from their animals and gave permission to publish questionnaire results from this study.
Study design and sample collection
Human NTS was obtained through active population-based case-control surveillance between December 2007 and February 2009 which was designed to determine the aetiology and epidemiology of enteric infections in Gambian children less than five years of age as part of the Gates Enteric Multicentre Study (GEMS). The entire surveillance area (figure 1) including all the compounds was mapped under GPS coordinates. This surveillance area represented a total population of 152,393 of which 27,567 are less than five years of age. Children under five years of age who presented with severe diarrhoea (i.e., diarrhoea with dehydration, dysentery, or requiring hospitalization) within 3 days of onset of diarrhea were eligible to participate. For each enrolled child with diarrhoea, one healthy control child without diarrhoea was randomly selected from the community in which the case resided, matched to the case by age, gender, and time of presentation. After providing informed consent from the parent/guardian of each case or control a single, fresh, whole stool specimen was collected from cases and controls and cultured to detect bacteria species (Aeromonas spp., Campylobacter spp, Salmonella Typhi, NTS, Shigella spp, Vibrio spp, diarrheagenic E. coli strains), viral (Rotavirus, Adenovirus, Astrovirus, Norovirus, Sapovirus,) and protozoa (Cryptosporidium spp Entamoeba histolytica, Giardia lamblia).
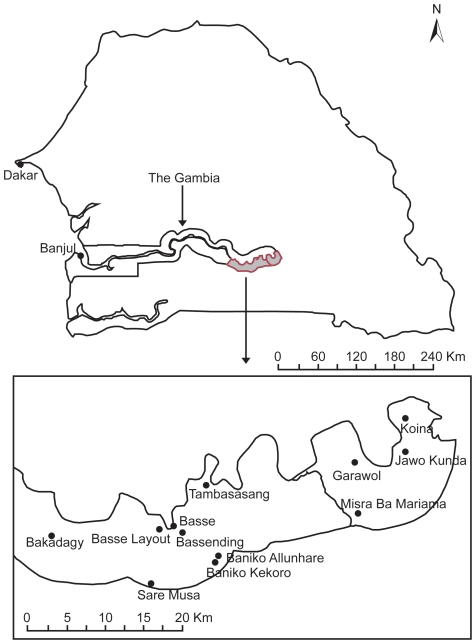
Figure 1. Map of the Gambia showing the locations where Non-Typhoidal Salmonella were recovered in humans from the surveillance area in The Gambia.
During the surveillance period, 495 diarrhoea cases were identified and NTS were isolated from eight patients and six from community healthy controls. The 14 children were enrolled in this study. and traced back to their family compounds and five apparently healthy animals per species (chicken, sheep and goat) residing in the same household as the child were randomly enrolled within a week of isolating NTS from humans.
Anal swabs were collected from 210 household contact animals and 21 NTS strains were isolated from the faeces.
Bacterial analysis
Stool specimens were transported in buffered glycerol saline (BGS) to the laboratory and processed within 6 hours of collection. Stools were plated on Xylose Lactose Desoxycholate (XLD) and MacConkey (MAC) agar and incubated at 36°C for 24 hours. Suspected non lactose fermenter colonies were subjected to biochemical reactions using Analytical Profile Index 20 Enteric (API 20E) according to manufactures' instructions (BioMerieux SA, REF 20 100/20 160). Serotyping was done by slide agglutination using Salmonella polyvalent and monovalent O and H antisera (Diagnostic Pasteur, Paris, France) according to the Kauffmann-White classification scheme [13].
Antimicrobial susceptibility testing
Antimicrobial susceptibility tests were performed on Muller-Hinton agar (Oxoid, USA) using the agar diffusion method with the Bio-Rad discs (Marne-La-Coquette, France) according to the guidelines of the Antibiogram Committee of the French Society for Microbiology (CA-SFM) [14]. Strains were tested with 22 antimicrobial disks (Bio-Rad): amoxicillin (25 mg), amoxicillin (20 mg) plus clavulanic acid (10 mg), ticarcillin (75 mg), cephalotin (30 mg), cefoxitin (30 mg), cefotaxime (30 mg), ceftazidime (30 mg), tobramycin (10 mg), amikacin (30 mg), nalidixic acid (30 mg), pefloxacin (5 mg), norfloxacin (10 mg), trimethoprim (1.2 mg) plus sulfamethoxazole (23.75 mg), tetracycline (30 mg), chloramphenicol (30 mg), gentamicin (10 mg), trimethoprim (300 mg), ciprofloxacin (5 mg), spectinomycin (100 mg), streptomycin (10 mg), sulfonamides (200 mg), and nitrofurantoin (300 mg). Diameters of the inhibition zones were measured with OSIRIS version 3.6xE2.0 (Bio-Rad), and results were interpreted as susceptible, intermediate, or resistant according to the recommendations of the CA-SFM [14]. All these tests were carried out at the MRC microbiology laboratory which is enrolled in the external quality assurance programme of the United Kingdom National External Quality Assessment Scheme [15].
Multi Locus Sequence Typing (MLST)
MLST was performed on the 35 Salmonella isolates as previously described [3]. The seven genes targeted were aroC, dnaN, hemD, hisD, purE, and thrA. Amplification of all genes was carried out in a 25 µl reaction mixture of the following items: 10xBuffer with 1.5 mm MgCl2 (2.5 µl); 2 mM dNTP'S (0.5 µl); 12.5 mM forward primer (1 µl); 12.5 mM reverse primer (1 µl); 5 U/µl Qiagen Hotstart Taq Polymerase (0.25 µl); Template (cell lysate) (2 µl) and 17.75 µl sterile DNA free water. PCR cycling conditions were as follows: 10 min at 94°C, followed by 32 cycles of 94°C for 1 min, 55°C for 1 min and 72°C for 1 min, and a final extension at 72°C for 5 min. 2 µl aliquots of PCR products were separated on 1% agarose gel electrophoresis, and visualized with ethidium bromide staining and UV illumination, and using a gel documentation system. PCR products were purified using Qiagen kit (Qiagen). Sequencing was done on both strands with BigDye Terminator Cycle Sequencing kit (Applied Biosystems, UK). The labelled fragments were separated by size using 3130xl Genetic Analyser (Applied Biosystems, UK). Sequences were edited and complementary sense antisense fragments were aligned using the Laser Gene DNA star 7.1 software. Finally, the sequences were submitted to the MLST database website [16] and assigned to existing or novel allele or sequence type numbers defined by the database.
Statistical, mapping and cluster analysis
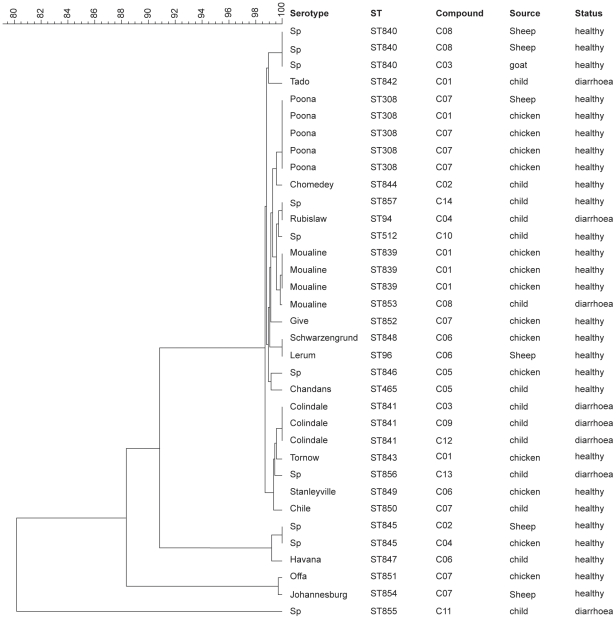
Tests of association were done using Fisher's exact test in Stata 11 (StataCorp. 2009. Stata Statistical Software: Release 11. College Station, TX: StataCorp LP). Stata provides one-sided p-values only for Fisher's exact test unless the table is 2×2 and results with p-values of less than 0.05 for the one-sided test were considered statistically significant. The parameters were grouped for the purpose of the analysis. The secondary diagnosis was categorized into three groups for the Fisher's exact test: group 1, children co-diagnosed with another disease other than malaria; group 2, children co-diagnosed with malaria and group 3, children who were not co-diagnosed with any other disease. The age was also categorized into 2 groups: less than or equal to 18 months (the average age), and more than 18 months. The mapping of the case locations was done using Arc Gis 9.3 software. To perform the cluster analysis of the serovars, MLST data were analysed with Bionumerics software (version 4.0; Applied Maths, Sint-Martens-Latem, Belgium). Analysis using a hierarchic unweighed pair group method (UPGMA) with averaging was used to generate a dendrogram describing the relationship among Salmonella serovars (figure 2).
Figure 2. Dendrogram based on the concatenated sequences of the seven alleles from the 35 NTS.
The scale bar represents percentage (%) of similarity.
Results
Phenotypic diversity of Salmonella serovars in the human and animal population
A diversity of serovars was found in both the human and animal population. None of the compounds showed similar serovars in both humans and animals (table 1). Nevertheless, one serovar namely S. Moualine was simultaneously found in a diarrheic child and in a chicken but in different compounds: C1 in Banico Allunhare and C8 in Koina (table 1, figure 1). MLST revealed a single locus variant at sucA between those 2 strains (table 1). In compounds 9 (C09), 10 (C10), 11 (C11), 12 (C12), 13 (C13) and 14 (C14), no Salmonella was isolated from animals (Table 1). The most prevalent serovars were S. Colindale in the human population (21.42%) and S. Poona in the animal population (14.28%).
Table 1. Salmonella strains isolated from humans and animals in the same compound and their Sequence Types profiles.
| MLST allelic profile | ||||||||||||||
| Strain ID | Compound | Village | Host | Status | Serovar | Antigenic formula | aroC | dnaN | hemD | hisD | purE | sucA | thrA | ST |
| 100661 | C01 | Banico allunhare | child | diarrheic | Tado | 8,20:c:z6:- | 197 | 70 | 153 | 4 | 90 | 182 | 139 | 842 |
| 101 | C01 | chicken | healthy | Moualine | 45:06:00 | 13 | 169 | 8 | 104 | 41 | 23 | 4 | 839 | |
| 102 | C01 | chicken | healthy | Moualine | 45:06:00 | 13 | 169 | 8 | 104 | 41 | 23 | 4 | 839 | |
| 103 | C01 | chicken | healthy | Moualine | 45:6:- | 13 | 169 | 8 | 104 | 41 | 23 | 4 | 839 | |
| 105 | C01 | chicken | healthy | Tornow | 45:gm:- | 156 | 54 | 68 | 49 | 39 | 9 | 14 | 843 | |
| 127 | C01 | chicken | healthy | Poona | 13,22:z:1,6:- | 22 | 104 | 25 | 113 | 12 | 83 | 4 | 308 | |
| 100807 | C02 | Sabi | child | healthy | Chomedey | 8,20:z10:enz15:- | 13 | 11 | 17 | 113 | 59 | 71 | 4 | 844 |
| 133 | C02 | sheep | healthy | sp | OME | 42 | 46 | 152 | 239 | 12 | 197 | 4 | 845 | |
| 102099 | C03 | Bassending | child | diarrheic | Colindale | 6,7:r:1,7:- | 6 | 7 | 8 | 10 | 39 | 10 | 14 | 841 |
| 151 | C03 | goat | healthy | sp | OME | 6 | 157 | 44 | 190 | 159 | 170 | 22 | 840 | |
| 100385 | C04 | Garawol | child | healthy | Rubislaw | 11:r:enx:- | 42 | 46 | 48 | 239 | 12 | 13 | 4 | 94 |
| 145 | C04 | chicken | healthy | sp | OME | 42 | 46 | 152 | 239 | 12 | 197 | 4 | 845 | |
| 100123 | C05 | Basse | child | healthy | Chandans | 11:d:enx:- | 83 | 63 | 49 | 154 | 33 | 58 | 137 | 465 |
| 146 | C05 | chicken | healthy | sp | OME | 37 | 31 | 35 | 14 | 8 | 6 | 4 | 846 | |
| 100225 | C06 | Bakadagy | child | healthy | Havana | 13,23:g,f:- | 106 | 70 | 152 | 132 | 34 | 9 | 168 | 847 |
| 115 | C06 | chicken | healthy | Schwarzengrund | 4,12:d:1,7:- | 43 | 47 | 25 | 49 | 41 | 15 | 3 | 848 | |
| 119 | C06 | sheep | healthy | Lerum | 1,3,19:z:1,7:- | 43 | 47 | 49 | 49 | 41 | 15 | 3 | 96 | |
| 121 | C06 | chicken | healthy | Stanleyville | 4,12:HMC | 17 | 7 | 35 | 9 | 2 | 9 | 14 | 849 | |
| 100400 | C07 | Misra ba mariama | child | Healthy | Chile | 6,7:z:1,2:- | 121 | 7 | 10 | 103 | 2 | 10 | 14 | 850 |
| 109 | C07 | chicken | healthy | Offa | 41:z38:- | 111 | 104 | 17 | 256 | 226 | 197 | 65 | 851 | |
| 110 | C07 | chicken | healthy | Give | 3,10:l,v:z26:- | 42 | 11 | 17 | 42 | 40 | 71 | 102 | 852 | |
| 137 | C07 | chicken | healthy | Poona | 13,22:z:1,6:- | 22 | 104 | 25 | 113 | 12 | 83 | 4 | 308 | |
| 143 | C07 | chicken | healthy | Poona | 13,22:z:1,6: | 22 | 104 | 25 | 113 | 12 | 83 | 4 | 308 | |
| 144 | C07 | chicken | healthy | Poona | 13,22:z:1,6: | 22 | 104 | 25 | 113 | 12 | 83 | 4 | 308 | |
| 107 | C07 | sheep | healthy | Poona | 13,22:z:1,6: | 22 | 104 | 25 | 113 | 12 | 83 | 4 | 308 | |
| 141 | C07 | sheep | healthy | Johannesburg | 40:b:enx:- | 111 | 11 | 17 | 256 | 12 | 197 | 65 | 854 | |
| 100887 | C08 | Koina | child | diarrheic | Moualine | 45:6:- | 13 | 169 | 8 | 104 | 41 | 13 | 4 | 853 |
| 147 | C08 | sheep | healthy | sp | OME | 6 | 157 | 44 | 190 | 159 | 170 | 22 | 840 | |
| 149 | C08 | sheep | healthy | sp | OME | 6 | 157 | 44 | 190 | 159 | 170 | 22 | 840 | |
| 100025 | C09 | Sare musa | child | diarrheic | Colindale | 6,7:r:1,7:- | 6 | 7 | 8 | 10 | 39 | 10 | 14 | 841 |
| 100054 | C10 | Jawo kunda | child | healthy | sp | OME | 13 | 109 | 49 | 13 | 12 | 13 | 4 | 512 |
| 100132 | C11 | Baniko kekoro | child | diarrheic | sp | OME | 61 | 112 | 8 | 10 | 134 | 137 | 4 | 855 |
| 100288 | C12 | Garawol | child | diarrheic | Colindale | 6,7:r:1,7:- | 6 | 7 | 8 | 10 | 39 | 10 | 14 | 841 |
| 102044 | C13 | Basse layout | child | diarrheic | sp | OME | 70 | 7 | 10 | 10 | 2 | 142 | 14 | 856 |
| 100392 | C14 | Tambasasang | child | diarrheic | sp | OME | 42 | 36 | 48 | 239 | 12 | 13 | 4 | 857 |
The proportion of Salmonella isolated was higher in the chicken population than in other species: 66.6%, 28.6% and 4.8% in chickens, sheep and goats, respectively. In the same compound, a diversity of Salmonella serovars were circulating at animal level especially in chickens; the number varying from 2 to 3 different serovars: in compound 1 (C1), S. Moualine, S. Tornow and S. Poona; in compound 6 (C6), S. Schwarzengrund and S. Stanleyville and in compound 7 (C7), S. Offa, S. Give and S. Poona (table 1).
Epidemiological aspects of salmonellosis in children
The mean age of children enrolled in the study was (to the nearest integer) 18 months and the age varied between 9 and 26 months (table 2). All children who presented with diarrhoea except one were secondarily diagnosed with another disease such as malaria (4 children) or other disease symptoms including fever, or cough (3 children) (table 2). There was a significant association (p-value<0.01) between expressing clinical signs of salmonellosis, i.e. diarrhoea and being co-diagnosed with a secondary disease (table 3). Age was not associated with the expression of clinical signs of salmonellosis (p-value = 0.16).
Table 2. Clinical symptoms present in 14 Gambian children with Non-Typhoidal Salmonella infections.
| ID | age (months) | first diagnosis | second diagnosis |
| 100025 | 26 | diarrhoea | none |
| 100132 | 22 | diarrhoea | malaria |
| 102044 | 26 | diarrhoea | malaria |
| 100288 | 17 | diarrhoea | malaria |
| 100661 | 20 | diarrhoea | cough, fever more than 38°C |
| 100385 | 9 | diarrhoea | malaria |
| 100887 | 22 | diarrhoea | cough with difficult breathing |
| 102099 | 26 | diarrhoea | belly pain, fever more than 38°C |
| 100392 | 10 | Healthy | none |
| 100123 | 20 | Healthy | none |
| 100225 | 9 | Healthy | none |
| 100807 | 21 | Healthy | none |
| 100400 | 9 | Healthy | none |
| 100054 | 10 | Healthy | none |
Table 3. Categorisation of the patient population by diagnostic criteria.
| diagnosis 1 | diagnosis 2 | |||
| other disease symptoms | malaria | none | total | |
| healthy | 0 | 0+ | 6 | 6 |
| diarrhoea | 3 | 4 | 1 | 8 |
| Total | 3 | 4 | 7 | 14 |
Drug susceptibility
All serovars were fully susceptible to all antibiotics tested except one of each the following serovars: Salmonella enteritica serovar Poona, Salmonella enteritica serovar Johannesburg, Salmonella enteritica serovar Chile and Salmonella enteritica serovar Colindale which were resistant to streptomycin.
Genetic diversity of Salmonella serovars using Multilocus Sequence Typing
Four new alleles were discovered: hemD (152), hemD (153), hisD (256) and purE (226) and eighteen novel sequence types (ST) (table 1). Similar serovars exhibited the same allelic profiles, except S. Moualine which had two different allelic profiles for the serovars isolated from humans and animals (table 1).
The seven housekeeping genes were concatenated for all isolates and the UPGMA tree was constructed (figure 2). All Salmonella genotypes had at least 80% similarity and the majority varied between 99% and 100%. Salmonella genotypes causing diarrhoea in children were always clustered with animal genotypes. Both Salmonella enteritica serovar Moualine isolated from a chicken and a child was clustered (figure 2).
Discussion
Salmonella serovars are phenotypically diverse, genetically close but remain clonally different between humans and domestic animal sources
The serovar diversity of NTS within the human and animal population was high showing that Salmonella is carried by both humans and animals in the community. The serovars were also widely geographically distributed in our surveillance area located in a typical African rural setting in The Gambia. We showed that several Salmonella serovars were circulating in the chicken population within the same compound or household; such as compounds 1 (C1), 6 (C6) and 7 (C7) respectively in the following 3 villages: Baniko Allunhare, Bagadagy and Misra Ba Mariama (figure 1). The higher proportion of Salmonella serovars in the chicken population compared to the goats and sheep is not surprising as it is known that chicken is the most important reservoir of NTS and thus thought to be the major source of transmission to humans [17]. Salmonella can persist in the chicken cecum or ovaries without triggering clinical signs in the host. Salmonellosis in young chickens may cause high mortality as a result of severe diarrhoea and dehydration, and creates a greater risk of evolving into a carrier state in the animals which survive [8], [9], [10]. The asymptomatic Salmonella carrier state in poultry has serious consequences for food safety and public health due to the risks of food poisoning following consumption of contaminated products. Small ruminants, such as sheep and goats, are also potential carriers of Salmonella [11], [12]. In Ethiopia, a study indicated that Salmonella is common in apparently healthy slaughtered sheep and goats. It also showed the presence of a wide range of Salmonella serovars in sheep and goats, which are of veterinary and public health significance [18]. The high rate of NTS clones circulating in the same compound could lead to mixed infection or carriage within the chicken population. This situation could result in extensive genetic diversity and variability due to frequent intraspecific recombination as it occurs with Helicobacter pylori [19]. This could have as consequence a wider range of clones and thus more difficulties to control NTS infections at animal level.
The diversity of serovars that we observed in this study is different from what we previously reported [3] where Salmonella enterica serovar Enteritidis was the most common serovar (80.6%) followed by Salmonella enterica serovar Typhimurium (8.0%) among NTS isolated from children with pneumonia and/or septicemia patients. It appears that the epidemiology of NTS is changing in The Gambia or the serovars detected might be site or disease specific, i.e. gastroenteritis vs. systemic infections.
MLST provides the best phylogenetic-relationship inference for the Salmonella genus [20]. Therefore, it may be invaluable for determination of the relationship among various Salmonella strains and serovars [21] . As expected, the similarity matrix (figure 2) of the serovars revealed close genetic relationship (>80% and the majority between 99 to 100%) between human and animal serovars, showing the genetic homogeneity of Salmonella. A study done in Senegal has also revealed a high degree of similarity among Salmonella enteritica serovar Brancaster and Salmonella enteritica serovar Enteritidis serovars from poultry and from humans by the use of PFGE techniques [22], but direct evidence of Salmonella transmission from poultry to humans could not be provided.
The high degree of similarity between human and animal serovars supports the theory that Salmonella clones are stable [23]. The genetic tree has also revealed that all lineages contained isolates of mixed origin (human and animal). From the present data, there is therefore no indication of clonal groups or lineages that are adapted to any specific host. These findings support the conclusions of other authors who used the same techniques (MLST) with Salmonella from human and veterinary sources in Denmark [24] or a different technique like Pulsed Field Gel Electrophoresis (PFGE) with isolates from humans and animals, food or the environment in close contact with humans, which was the case in Kenya [25]. Like in the Kenya study [25], we also showed that there was no relatedness between NTS genotypes from humans and those from animals in close contact to humans, this other potential sources of transmission such as environmental or the human-to-human transmission need to be examined.
Expression of clinical signs of salmonellosis is associated with the existence of a secondary disease especially malaria
A statistical significant association (P<0.05) was observed between children expressing clinical signs of salmonellosis (diarrhoea) and co-diagnosis with malaria (table 3). The association between salmonellosis and immunocompromising diseases is well known in the African context. NTS infections are usually associated with opportunistic infections especially in immuno-compromised patients, e.g. HIV-infected adults [26], [27] or with other diseases like malaria or anemia [1], [5], [28], [29]. Children especially those less than 3 years old are the age-group at risk of expressing clinical signs of salmonellosis [1], [26] . The contradiction between our observation and those of most other authors is certainly due to the small sample size and the small range of age groups in our study. All individuals were in fact less than three years old. Malaria has long been suspected to increase the risk of invasive NTS infection and might contribute to the seasonality of NTS disease, although the mechanism underlying the association between malaria and NTS is only partially understood [6].
Animal and human NTS serovars are fully susceptible to commonly used antibiotics in The Gambia
All serovars were susceptible to most commonly used antibiotics for the treatment of clinical infections in The Gambia such as amoxicillin, amoxicillin plus clavulanic acid, trimethoprim plus sulfamethoxazole, tetracycline, streptomycin and chloramphenicol and also to cephalosporins of the third generation which are considered as the drugs of choice for invasive Salmonella infections in humans. This result is in contrast to previous studies done in urban [1], [2] and rural [3] areas in The Gambia where Salmonella strains expressed multi-resistance to several commonly used antibiotics. This could be explained in part by the fact that the NTS isolated from those studies were from invasive cases (pneumonia and sepsis) whereas this study focused on non-invasive cases (diarrhea). In addition, in those studies Salmonella enterica serovar Enteritidis was the most common serovar followed by Salmonella enterica serovar Typhimurium. While these serovars were not detected in this study as a result of temporal trends of childhood NTS infection in The Gambia {Mackenzie, 2010 #33}. There is lack of resistance of serovars to antibiotics in rural areas even to those commonly used in the hospitals to treat bacterial infections. This is due to the fact that in rural areas, NTS infections are not treated because patients are often not conducted to the hospital due to poor access to medical centers, lack of transport facilities and of financial resources. Our findings suggest that these drugs remain suitable for the treatment of salmonellosis in humans and animals. However, we have to interpret these results with caution because the sample size in our study was small.
Our study showed that the use of serotyping data combined with MLST and phylogenetic analysis can provide important information about the epidemiology of NTS in humans and animals. However, our results do not support the hypothesis that humans and animals in close contact in the same household carry genotypically similar Salmonella serovars. Nevertheless these findings have stirred up the problem of the transmission of NTS in African context and suggest that poultry may play an important part in the epidemiology of Salmonella infections of this condition. A better control of malaria may lead to a reduction in the incidence of invasive NTS disease in The Gambia. Multidrug resistance has not yet been a problem in human and animal NTS isolates in this area of the country. Thus, commonly available drugs may still be used for the treatment of NTS infections in rural Gambia. Nevertheless, public authorities must be alert to detect any change in the behavior of Salmonella towards antibiotics, with a view of establishing appropriate control measures for use of these drugs in humans and animals.
Acknowledgments
We acknowledge Kebba Touray for technical assistance with GIS and Mark Achtman for the use of the Salmonella MLST database [30], which is hosted at the University College Cork and funded by the Science Foundation of Ireland (05/FE1/B882).
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the Flemish Interuniversity Council (VLIR-UOS, Brussels, Belgium) and the Medical Research Council, United Kingdom. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Enwere G, Biney E, Cheung YB, Zaman SM, Okoko B, et al. Epidemiologic and clinical characteristics of community-acquired invasive bacterial infections in children aged 2-29 months in The Gambia. Pediatr Infect Dis J. 2006;25:700–705. doi: 10.1097/01.inf.0000226839.30925.a5. [DOI] [PubMed] [Google Scholar]
- 2.Hill PC, Onyeama CO, Ikumapayi UN, Secka O, Ameyaw S, et al. Bacteraemia in patients admitted to an urban hospital in West Africa. BMC Infect Dis. 2007;7:2. doi: 10.1186/1471-2334-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikumapayi UN, Antonio M, Sonne-Hansen J, Biney E, Enwere G, et al. Molecular epidemiology of community-acquired invasive non-typhoidal Salmonella among children aged 2 29 months in rural Gambia and discovery of a new serovar, Salmonella enterica Dingiri. J Med Microbiol. 2007;56:1479–1484. doi: 10.1099/jmm.0.47416-0. [DOI] [PubMed] [Google Scholar]
- 4.Mulholland EK, Ogunlesi O, Adegbola R, Weber M, Sam BE, et al. Aetiology of serious infections in young Gambian infants. Pediatric Infectious Disease Journal. 1999;18(supplement):S35–S41. doi: 10.1097/00006454-199910001-00007. [DOI] [PubMed] [Google Scholar]
- 5.Graham SM, Molyneux EM, Walsh AL, Cheesbrough JS, Molyneux ME, et al. Nontyphoidal Salmonella infections of children in tropical Africa. Pediatr Infect Dis J. 2000;19:1189–1196. doi: 10.1097/00006454-200012000-00016. [DOI] [PubMed] [Google Scholar]
- 6.Morpeth SC, Ramadhani HO, Crump JA. Invasive non-Typhi Salmonella disease in Africa. Clin Infect Dis. 2009;49:606–611. doi: 10.1086/603553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wall PG, Threllfall EJ, Ward LR, Rowe B. Multiresistant Salmonella typhimurium DT104 in cats: a public health risk. Lancet. 1996;348:471. doi: 10.1016/S0140-6736(96)24033-4. [DOI] [PubMed] [Google Scholar]
- 8.Barrow PA, Huggins MB, Lovell M, Simpson JM ( 1987) Observations on the pathogenesis of experimental Salmonella typhimurium infection in chickens. Res Vet Sci. 42:194–199. [PubMed] [Google Scholar]
- 9.Sadeyen JR, Trotereau J, Velge P, Marly J, Beaumont C, et al. Salmonella carrier state in chicken: comparison of expression of immune response genes between susceptible and resistant animals. Microbes and Infection. 2004;6:1278–1286. doi: 10.1016/j.micinf.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Van Immerseel F, De Buck J, De Smet I, Mast J, Haesebrouck F, et al. Dynamics of immune cell infiltration in the caecal lamina propria of chickens after neonatal infection with a Salmonella enteritidis strain. Dev Comp Immunol. 2002;26:355–364. doi: 10.1016/s0145-305x(01)00084-2. [DOI] [PubMed] [Google Scholar]
- 11.Alvseike O, Skjerve E. Prevalence of a Salmonella subspecies diarizonae in Norwegian sheep herds. Prev Vet Med. 2002;52:277–285. doi: 10.1016/s0167-5877(01)00252-5. [DOI] [PubMed] [Google Scholar]
- 12.Uzzau S, Leori GS, Petruzzi V, Watson PR, Schianchi G, et al. Salmonella enterica serovar-host specificity does not correlate with the magnitude of intestinal invasion in sheep. Infect Immun. 2001;69:3092–3099. doi: 10.1128/IAI.69.5.3092-3099.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimont PAD, Weill FX. Antigenic formulae of the Salmonella serovars, 9th ed. Paris: World Health Organization Collaboration Centre for Reference and Research on Salmonella and Institut Pasteur; 2007. [Google Scholar]
- 14.Anomymous Comité de l'Antibiogramme de la Société de Microbiologie. Communiqué 2009. Edition Janvier 2009. 2009. Available from: ( http://www.sfm.asso.fr/publi/general.php?pa=1)
- 15. http://www.ukneqas.org.
- 16.Aanensen DM, Spratt BG. The multilocus sequence typing network: mlst.net. Nucleic Acids Res. 2005;33:W728–733. doi: 10.1093/nar/gki415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braden CR. Salmonella enterica serotype Enteritidis and eggs: a national epidemic in the United States. Clin Infect Dis. 2006;43:512–517. doi: 10.1086/505973. [DOI] [PubMed] [Google Scholar]
- 18.Woldemariam E, Molla B, Alemayehu D, Muck A. Prevalence and distribution of Salmonella in apparently healthy slaughtered sheep and goats in Debre Zeit, Ethiopia. Small Ruminants Research. 2005;58:19–24. [Google Scholar]
- 19.Suerbaum S, Achtman M. Helicobacter pylori: recombination, population structure and human migrations. Int J Med Microbiol. 2004;294:133–139. doi: 10.1016/j.ijmm.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Cooper JE, Feil EJ. Multilocus sequence typing--what is resolved? . Trends Microbiol. 2004;12:373–377. doi: 10.1016/j.tim.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Kotetishvili M, Stine OC, Kreger A, Morris JG, Jr, Sulakvelidze A. Multilocus sequence typing for characterization of clinical and environmental salmonella strains. J Clin Microbiol. 2002;40:1626–1635. doi: 10.1128/JCM.40.5.1626-1635.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardinale E, Perrier Gros-Claude JD, Tall F, Gueye EF, Salvat G. Risk factors for contamination of ready-to-eat street-vended poultry dishes in Dakar, Senegal. Int J Food Microbiol. 2005;103:157–165. doi: 10.1016/j.ijfoodmicro.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 23.Spratt BG. Exploring the concept of clonality in bacteria. Methods Mol Biol. 2004;266:323–352. doi: 10.1385/1-59259-763-7:323. [DOI] [PubMed] [Google Scholar]
- 24.Torpdahl M, N. SM, Sandvang D, Baggesen DL. Genotypic characterization of Salmonella by multilocus sequence typing, pulsed-field gel electrophoresis and amplified fragment length polymorphism. J Microbiol Methods. 2005;63:173–184. doi: 10.1016/j.mimet.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Kariuki S, Revathi G, Gakuya F, Yamo V, Muyodi J, et al. Lack of clonal relationship between non-typhi Salmonella strain types from humans and those isolated from animals living in close contact. FEMS Immunol Med Microbiol. 2002;33:165–171. doi: 10.1111/j.1574-695X.2002.tb00587.x. [DOI] [PubMed] [Google Scholar]
- 26.Gordon MA, Graham SM, Amanda L, Walsh AL, Wilson L, et al. Epidemics of Invasive Salmonella enterica Serovar Enteritidis and S. enterica Serovar Typhimurium Infection Associated with Multidrug Resistance among Adults and Children in Malawi. Clinical Infectious Diseases. 2008;46:963–969. doi: 10.1086/529146. [DOI] [PubMed] [Google Scholar]
- 27.Sow AI, Seydi M, Thiaw CT, Ndour CT, Soumare MF, et al. Les salmonelloses au centre hospitalier universitaire de Fann à Dakar: aspects bactériologiques. Méd Mal Infect. 2000;30:657–660. [Google Scholar]
- 28.Berkley JA, Lowe BS, Mwangi I, Williams T, Bauni E, et al. Bacteremia among children admitted to a rural hospital in Kenya. N Engl J Med. 2005;352:39–47. doi: 10.1056/NEJMoa040275. [DOI] [PubMed] [Google Scholar]
- 29.Mandomando I, Macete E, Sigaúque B, Morais L, Quinto L, et al. Invasive non-typhoidal Salmonella in Mozambican children. Tropical Medicine and International Health. 2009;14:1467–1147. doi: 10.1111/j.1365-3156.2009.02399.x. [DOI] [PubMed] [Google Scholar]
- 30. http://mlst.ucc.ie/mlst/dbs/Senterica.