Abstract
Genetic analysis of mouse disease models provides a means to investigate how modifying loci cause variation in phenotypic expression. We have shown that polycystic kidney disease (PKD) progression in the juvenile cystic kidney (jck) mutation can be influenced by an epistatic interaction between alleles of different strain backgrounds and we localized one of these loci to chromosome 1. Using a chromosome 1 congenic strain, we improved the genetic analysis and mapped the interacting locus to proximal chromosome 4, with a highly significant lod association of 5.5. Re-analysis of the original F2 cross reveals that in this cohort, while the lod association of the chromosome 4 locus alone is not significant, its effect is apparent when analyzed in combination with the chromosome 1 locus. This result suggests that correlation of paired genotype data with phenotype data will be an effective means to detect epistatic interactions contributing to complex traits, and that these associations can be tested using appropriate congenic lines.
Mouse models of human disorders are potentially powerful tools for understanding heritable contributions to complex genetic traits. The availability of mutations on inbred genetic backgrounds, along with the development of extensive molecular and computational resources for genome-wide analysis, makes these systems ideal for genetic analysis. Of particular interest is the investigation of modifier loci, which influence the variable expression of mutant phenotypes. This has important implications for understanding how traits that are usually benign may, in some cases, be exacerbated and appear as disease phenotypes. Additionally, the characterization of modifying loci for specific disease models may suggest means of therapeutic intervention that can be an alternative to amelioration of the underlying genetic defect.
Polycystic kidney disease (PKD) represents a major cause of human morbidity and mortality. Although PKD1 and PKD2, the genes responsible for the great majority of human autosomal dominant PKD, have both been cloned, the biological basis of this disorder remains elusive. A significant impediment to this has been the difficulty of characterizing polycystin-1, the large (4303 amino acid) membrane glycoprotein that is encoded by PKD1 (European Polycystic Kidney Disease Consortium, 1994; American PKD1 Consortium 1995; International PKD Consortium 1995). The presence of specific protein motifs on the large extracellular domain of polycystin-1 has suggested a role for it in cell–matrix or cell–cell signaling. PKD1 and PKD2 are widely expressed in fetal and adult tissue (Mochizuki et al. 1996; Ward et al. 1996). One hypothesis that reconciles the focal nature of PKD with this widespread expression is the proposal that the disease is recessive, and that renal cysts occur due to loss of heterozygosity as a consequence of somatic mutation in the kidney. Studies of alleles from human cystic epithelium have demonstrated loss of heterozygosity of the wild-type PKD1 allele (Qian et al. 1996; Brasier and Henske 1997). Furthermore, studies of mice carrying targeted mutations in Pkd1 or Pkd2 have also shown that loss of the wild-type allele in a heterozygous mouse will result in cyst development (Wu et al. 1998; Lu et al. 1999).
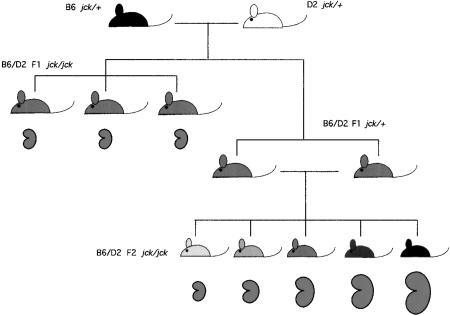
Nonetheless, the exact role of the polycystin genes in maintaining renal tubular integrity is still not known. In this regard, investigation of animal models of PKD may be useful. The juvenile cystic kidney (jck) mutation is a model of autosomal recessive PKD (Atala et al. 1993). We mapped the position of this mutation to chromosome 11 using an intercross of C57BL/6J (B6) carrying jck and DBA/2J (D2) mice (Iakoubova et al. 1995). In the F2 progeny, the size of polycystic kidneys found in age-matched affected mice was markedly variable compared to that found in the parental line (Fig. 1). This suggested that modifying loci affecting severity had been introduced from the D2 background. Initial genetic analysis revealed nonrandom co-segregation with disease phenotype for two loci; one B6 locus on chromosome 1 [polycystic kidney disease modifier 1 (Pkdm1)], and one D2 locus on chromosome 10 [polycystic kidney disease modifier 2 (Pkdm2)]. Quantitative trait locus (QTL) analysis confirmed that inheritance of a B6 locus on chromosome 1 correlated significantly with increased kidney size; this accounted for 74% of the variance in affected F2 progeny and appeared to have its effect as a recessive locus.
Figure 1.
Genetic crosses. B6 and D2 jck/+ mice were mated and F1 progeny sacrificed between 6 and 7 weeks of age and scored for the presence of abnormal kidneys. As shown in Fig. 2, these affected kidneys were uniformly small in size. Several F1 jck/+ mice were reserved for intercross analysis and used to generate an F2 population. Affected kidneys from this population showed wide variation in size (Fig. 2).
A highly significant association of recessively inherited B6-related alleles on chromosome 1 with severe disease was unexpected, since the PKD phenotype in the original B6 background is not severe. We proposed that the severe phenotype resulted from a genetic interaction between the B6 locus on chromosome 1 and a D2 gene elsewhere in the genome. To further analyze this, jck was crossed into the D2 background for three generations, testing to ensure that loci spanning chromosome 1 and chromosome 10 carried only D2 alleles. When these mice were intercrossed, variance in kidney size for the affected progeny was markedly less than that seen in the F2 progeny, and not significantly different than that found for the B6 parental mice (Iakoubova et al. 1995; Fig. 2). This observation continues to be valid after serial backcross of the jck mutation into D2 mice for over 10 generations.
Figure 2.
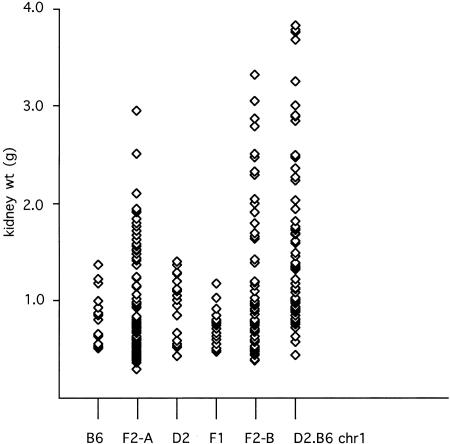
Scattergram of paired-kidney weight distributions. (B6) Distribution in affected inbred C57BL/6J mice (n = 18). (F2-A) Distribution in affected F2 progeny of a cross between C57BL/6J jck/+ and DBA/2 mice (n = 104). (D2) Distribution in affected DBA/2J jck/+ mice (n = 23). (F1) Distribution in affected F1 progeny of a cross between C57BL/6J jck/+ and DBA/2J jck/+ mice (n = 23). (F2-B) Distribution in affected F2 progeny of a cross between (B6 × D2)F1 jck/+ mice (see Fig. 1) (n = 65). (D2.B6 chr1) Distribution in affected F2 progeny of a cross between B6 jck/jck and D2.B6 chr1 congenic mice (n = 67). The data for the B6, F2-A, and D2 cohorts are from Iakoubova et al. (1995).
Thus, the most severe phenotype is found in B6–D2 hybrid mice that retain B6 alleles on mouse chromosome 1. This observation supports the hypothesis that this phenotype requires an interaction between unlinked loci of different strain backgrounds. This result has significant implications for the understanding of the biology of modifying effects, because it suggests that it is not only the genetic backgrounds that are of import, but also their particular combination. Thus, loci which could have potentially profound effects as modifiers, in certain cases may be quiescent in a different genetic background.
In our initial study, the inheritance of a D2 region on distal chromosome 10 was associated with increased severity of kidney disease, which accounted for 12% of the observed variance, with a maximal lod score of 2.1. These results are consistent with linkage; however, we noted it was possible that another undetected D2 locus contributed to the variance in disease severity. However, we analyzed 102 markers in the cohort of severely affected mice and did not detect any additional D2 loci segregating with disease severity (Iakoubova et al. 1995). In this report we have used a congenic strategy to increase the sensitivity of the genetic analysis, and have succeeded in identifying the presumptive interacting locus.
RESULTS
The Modifying Effect of the Chromosome 1 B6 Allele Is Recessive
Because the hypothesis that the effect of the chromosome 1 B6-specific modifier is inherited recessively and requires an interaction with a D2 allele has implications for the biological mechanism by which this locus exacerbates PKD, we have investigated this further. It was not initially possible to generate homozygous jck mice in F1 progeny, because the mutation existed on only one background. A congenic line carrying jck on an otherwise D2 genetic background was generated by six generations of serial backcross, at which point the line was statistically likely to be >98% homozygous for D2 alleles in regions unlinked to jck. We tested whether the modifying effects were recessive by crossing this congenic line with B6 jck/+ mice to generate F1 homozygous jck mice, which are heterozygous for B6 and D2 at all loci unlinked to jck (Fig. 1). Remarkably, F1 jck homozygous mice do not show the severe PKD seen in the F2 population; in fact, the F1 state appears to be protective compared to the inbred B6 or D2 backgrounds alone (mean: 0.71 g, variance 0.03 g2, range 0.46–1.16, n = 23; Fig. 2). The small affected kidneys were, in many cases, not distinguished easily from wild-type based on gross examination, and all presumptive affected mice were examined by histology and were tested using markers linked tightly with jck to confirm they were homozygous for the mutation.
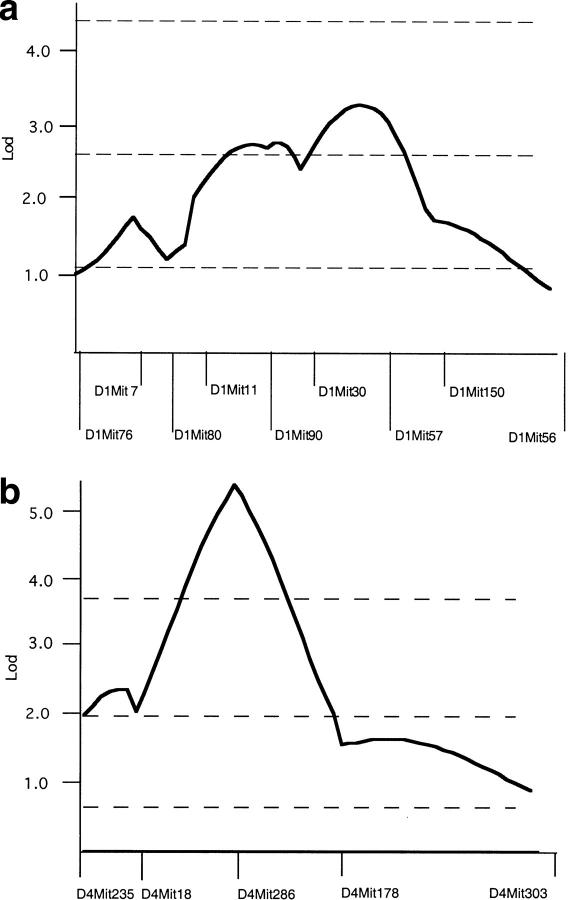
The observation that the F1 mice do not have severe PKD strongly supports the model that the B6 modifier locus is recessive and must be homozygous to have its effect. This result was sufficiently dramatic that the possibility that the modifying loci had been somehow lost in this population was tested by performing an intercross using sibling F1 mice heterozygous for jck. This would generate essentially the same F2 population as was analyzed previously, which should show the same distribution of PKD phenotype (Fig. 1). The F2 mice show a wide variation of PKD severity, confirming that the B6 modifying locus functions as a recessive gene (mean: 2.94 g, variance 0.54 g2, range 0.39-3.33 g, n = 65; Fig. 2). Furthermore, QTL analysis of chromosome 1 again reveals significant linkage of the severe disease phenotype with B6 alleles over a large region of the chromosome, with a maximal lod score of 3.5 (Fig. 3a). This value is likely smaller than that found in the original analysis because this is a smaller data set. QTL analysis of chromosome 10 shows only a suggestive effect, with a lod score of 1.5 (data not shown).
Figure 3.
QTL analysis. (a) MapManager QT was used to analyze chromosome 1 genotype and trait data from affected F2 progeny of a cross between C57BL/6J jck/+ and DBA/2J jck/+ mice. (b) MapManager QT was used to analyze chromosome 4 genotype and trait data from affected F2 progeny of a cross between B6 jck/jck and D2.B6 chr1 congenic mice. For both analyses, a permutation test was used to calculate the threshold levels indicated for suggestive, significant, and highly significant linkage (dotted lines).
The Interacting Modifying Locus Maps to Chromosome 4
These results strongly implicate an interaction between the B6 allele on chromosome 1 and a D2 allele elsewhere in the genome as responsible for severe PKD. Because the B6 allele acts as a recessive locus, however, only one-fourth of affected mice will be informative with respect to the interacting locus. Thus, the identification of the interacting locus may be hindered because most mice will not contribute useful genotype information. The combined analyses of the F2 crosses do not strongly support the chromosome 10 locus as the interacting locus, although it clearly has some contribution to severe PKD, as the sum of the lod scores at D10Mit10 in both B6 × D2 F2 populations is 4.6.
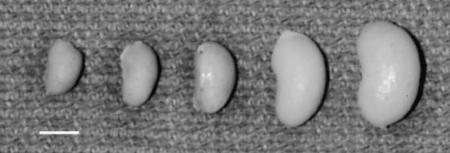
To increase the efficiency of the characterization of the interacting locus, a jck congenic line was generated that was homozygous for B6 alleles from D1Mit7 to D1Mit155 in a D2 background. When this D2.B6 congenic strain is crossed with B6 jck/+ mice, and their progeny intercrossed, the congenic region is fixed as B6, whereas the remainder of the genome is segregating as an F2 population. In this experiment, all affected mice will be informative for the interacting locus and the genetic analysis will be much more efficient. One prediction for progeny of this intercross is that the distribution of PKD severity should be skewed to a more severe disease phenotype, since all of the mice are homozygous in the congenic region, rather than the one-fourth that would be in an F2 population. This is clearly the result, as shown in Figure 2 (mean: 1.53 g, variance: 0.76 g2, range: 0.41-3.8 g, n = 67). In many cases, the range in phenotypic variation was dramatic, even within the same litter, as shown in Figure 4.
Figure 4.
Variation in affected kidney size. A single kidney from each of five affected F2 progeny obtained in one litter of a cross between B6 jck and D2.B6 chr1 congenic mice. Bar, 1 cm.
To identify the interacting locus in this congenic cross, we used a strategy of interval haplotype analysis described in Neuhaus and Beier (1998). This technique allows one to screen for regions most likely to carry a nonrandomly inherited locus using only 2 or 3 markers on a chromosome. This analysis identified two intervals (one on chromosome 2 and one on chromosome 4) as most likely to contain a nonrandomly inherited locus. These regions were analyzed in more detail in all of the affected mice using additional markers, and a locus on proximal chromosome 4 was significantly found to be associated with severity of PKD, with a lod score of 5.5 (Fig. 3b). The MapManager QT program can be constrained to calculate lod associations for dominant, additive (semidominant), and recessive modes of inheritance. For this constrained analysis the additive mode generated the highest lod score of 5.2. The evidence that the mean kidney weight for affected progeny of the (D2.B6 × B6)F2 cohort heterozygous for D4Mit286 (the QTL peak) is intermediate between mice that are either homozygous B6 or homozygous D2 is consistent with the D2 locus having a semidominant effect (Table 1).
Table 1.
Kidney Weight by Genotype
| D1Mit30 | D4Mit286 | |||
|---|---|---|---|---|
| BB | BD | DD | ||
| D2.B6 chr1: | BBa | 1.11 ± 0.54 (32) | 1.52 ± 0.85 (44) | 2.21 ± 0.93 (18) |
| F2: | BB | 0.55 ± 0.11 (4) | 1.36 ± 0.53 (14) | 1.81 ± 0.57 (6) |
| F2: | BD/DDb | 0.59 ± 0.35 (9) | 0.67 ± 0.38 (26) | 0.79 ± 0.44 (11) |
The mean kidney weight ± S.D. is shown for various genotypic classes of the paired markers D1Mit30 and D4Mit286 (which represent QTL peaks) in the (D2.B6 chr1 congenic × B6) F2 cohort (labeled D2.B6 chr1) described in this report and in a (B6×D2)F2 population (labeled F2) generated previously (Iakoubova et al. 1995). (BB) homozygous B6; (BD) heterozygous B6/D2; (DD) homozygous D2; (N) sample size.
For D1Mit30, the parental D2.B6 congenic strain is homozygous B6 so this is the only possible genotype class in the (D2.B6 congenic × B6) F2 cohort.
In the (B6×D2)F2 cohort the heterozygous B6/D2 and homozygous D2 classes are merged (BD/DD) because the modifying effect only occurs when this locus is homozygous B6.
To test the hypothesis that the identification of the interacting locus was obscured originally because most mice were not informative for its effect, we reanalyzed progeny from the original F2 intercross. The maximum lod score for loci on chromosome 4 was 1.8, which does not reflect significant linkage. Of interest is that when mean kidney weights are determined for all genotypic classes of the paired markers D1Mit30 and D4Mit286 (which represent QTL peaks), the evidence for the interaction is apparent, as the largest value is found in the class of progeny that is homozygous B6 at D1Mit30 and homozygous D2 at D4Mit286 (Table 1).
DISCUSSION
Our results demonstrate clearly that severe PKD in the jck mutant mouse is associated with recessive inheritance of a B6-derived locus on chromosome 1 that must interact with a D2-derived gene. The statistical effect of this locus will be obscured in an intercross, because the chromosome 1 locus behaves recessively and three-fourths of the mice are therefore not informative for the interaction. To address this, we constructed a D2.B6 jck/+ congenic strain which carries a large interval on chromosome 1 as homozygous B6 and the remainder of the genome as D2. When this mouse is crossed with a B6 jck/+ mouse, the congenic region on chromosome 1 is fixed as B6, while the rest of the genome is segregating as in an F2 population. Thus, all the affected mice should be informative for the interacting D2 locus. Consistent with this hypothesis, we were readily able to localize a D2 locus associated with severe PKD.
Of note is the fact that in our original F2 population, the association of a locus on chromosome 4 is not significant statistically. However, when trait data are stratified with respect to genotype at both the putative chromosome 4 locus and the previously identified chromosome 1 locus, the relationship to disease phenotype is apparent. This observation suggests that a recently described algorithm to test genotype data for pair-wise effects in a genome-wide fashion (G.A. Churchill, unpubl.) would be useful for uncovering genetic interactions similar to what we have demonstrated experimentally in this report.
The determination that a locus on proximal chromosome 4 contributes to severe PKD in the jck mutant mouse is of particular interest because this is the position of the locus Pkdm3 (previously called MOP1), which has been shown to modify the progression of PKD in two different mouse mutations, cpk and pcy (Woo et al. 1997). These mutants have quite distinct phenotypes; cpk causes a rapidly progressive disease that is usually lethal before weaning, while affected pcy mice survive until 25–32 weeks. The jck mutation has an intermediate phenotype between these, with mice first showing histological evidence of cysts at 3-4 weeks of age. Pkdm3 was not previously found to require an interacting locus, which may be due to the fact that the strain combinations tested were different than reported here (D2 vs. Mus castaneus for pcy and B6 vs. M. castaneus for cpk.). However, given the limitations of the resolution of QTL analysis, it is equally likely that the interacting locus described here represents a closely linked but separate modifier from Pkdm3.
Of additional interest is the fact that both the Han/SPRD rat Cy mutation, which causes dominant PKD, and the Wistar rat polycystic kidney (wpk) mutation, which causes recessive PKD, have been localized to a region that has conserved synteny with proximal mouse chromosome 4 (Bihoreau et al. 1997; Nauta et al. 1999).
The low resolution of QTL analysis precludes productive speculation regarding specific genes as candidates for the modifying loci. This is particularly the case because the molecular basis of the jck, cpk, and pcy mutations is not yet known. Additionally, although the causal roles of the Pkd1 and Pkd2 genes in cystogenesis have been confirmed in mouse models (Lu et al. 1997; Wu et al. 1998), the mechanism of these genes remains obscure, and extrapolation regarding pathways cannot be made. As is generally the case for loci causing quantitative effects, a strategy of congenic analysis will be the most appropriate for defining a recombinant interval in which the presumptive modifying locus must reside. We have begun pursuing this for the B6-derived chromosome 1 loci; however, the initial outcome of these studies suggests that there are two linked loci on chromosome 1 contributing to severe PKD, further illustrating the potential complexity of modifying gene analysis (Iakoubova et al. 1999).
Perhaps the most important lesson of this study is to illustrate the complicated nature of modifying effects involving even only few loci. This supports the value of analysis of complex traits in model organisms with defined genetic backgrounds, as it seems unlikely that the loci identified in these studies would be detectable in a population with a highly heterogenous genetic composition. Although the biology of model organisms may be sufficiently divergent such that the loci uncovered are not disease-causing in humans, the expectation that they will provide insight into mechanisms of pathogenesis seems reasonable.
The possibility that the same locus can influence disease progression in three different murine PKD mutations is of considerable significance, as this would suggest that this gene might influence PKD severity irrespective of its cause. As such, this locus could represent a potential target for therapeutic intervention in human PKD. The dominant form of this disorder affects 600,000 people in the United States and 12.5 million worldwide. It is a common cause of renal failure and accounts for 10% of patients on dialysis. There is presently no effective treatment of this disease. Because the symptoms of PKD do not appear until mid-life, even modest therapeutic effects have considerable potential to delay the onset of renal failure and thus extend life span and increase its attendant quality.
METHODS
Mice and Phenotype Characterization
B6 and D2 jck/+ mice are maintained in our mouse colony. D2 jck/+ mice were generated by crossing (B6 x D2)F1 jck/+ mice with D2 wild-type mice and selecting for jck/+ mice carrying only D2 markers on chromosomes 1 and 10. These mice were mated for another five generations with wild-type D2 mice, selecting for jck/+ by genotype analysis of flanking microsatellite markers. For the experiments illustrated in Figure 1, B6 and D2 jck/+ mice were mated and F1 progeny sacrificed between 6-7 weeks of age and scored for the presence of abnormal kidneys. These were removed, weighed, and fixed in Optimal Fix (American Histology Reagent Co.), and tail tissue was reserved for DNA extraction. Kidneys which appeared abnormal but were not obviously polycystic were analyzed by histology and were tested using chromosome 11 markers to confirm they were homozygous for jck. Several F1 jck/+ mice were reserved for intercross analysis and used to generate an F2 population that was analyzed in a similar fashion (Fig. 1). For the congenic analysis, incipient congenic lines were constructed by crossing mice carrying B6 alleles between D1Mit7 and D1Mit155 to D2 mice for 7 generations, selecting for the appropriate chromosome 1 markers by genotype analysis. These mice were then intercrossed and mice homozygous for chromosome 1 markers were identified. These D2.B6 D1Mit7-155 congenic mice were crossed with B6 jck/+ mice, the F1 progeny intercrossed, and the F2 affected progeny analyzed as described above.
PCR-Based Genotyping
Genomic DNA was prepared from tail and liver tissue according to standard techniques. PCR primers were purchased from Research Genetics, Inc., and kinase-labeled using 32P-labeled adenosine triphosphates. The PCR fragments were amplified using standard techniques and analyzed on a 6% polyacrylamide denaturing gel.
Interval Haplotype Analysis and Modifier Mapping
To identify the interacting locus we used a strategy of interval haplotype analysis (Neuhaus and Beier 1998). This technique allows one to screen for regions most likely to carry a nonrandomly inherited locus using only two or three markers on a chromosome. From a population of 67 intercross progeny, cohorts of 20 mice with the most and least degree of PKD severity were analyzed separately. All mice with apparent mild disease were tested with chromosome 11 genetic markers to confirm they were homozygous for jck. These cohorts were genotyped using markers 5–10 cM from the ends of each chromosome, with an additional marker added for chromosome 2. A Web-based interface for the program described by Neuhaus and Beier (1998) was used to test for the frequency of nonrecombinant haplotypes in these cohorts (http://aurora.bwh.harvard.edu/cgi-bin/haplotype.pl). This program infers the genotype distribution for markers in the interval, and compares the value of this distribution to the theoretical maximum for the number of mice analyzed. Intervals showing the closest values to the theoretical maximums (and therefore the most potentially skewed distribution of alleles) are candidates for containing the interacting locus. This analysis identified two intervals (one on chromosome 2 and one on chromosome 4) as having an inferred χ2 distribution >75% of the theoretical maximum. This value has been empirically found to be a useful threshold for localizing mutations (Neuhaus and Beier 1998). These regions were analyzed in more detail in all of the affected mice using additional markers.
Statistical Analysis
Kidney weight distributions were analyzed using Excel 5.0. Genotype data were correlated with kidney weight trait data using MapManager QT version b28 (http://mcbio.med.buffalo.edu/mapmgr.html) (Manly and Olson 1999). Permutation tests (Doerge and Churchill 1996) were done in 1-cM steps for 5000 permutations using Map Manager QT. The threshold values of the permutation test, which correspond to suggestive, significant, and highly significant linkage associations, are taken from the guidelines of Lander and Kruglyak (1995) and correspond to the 37th, 95th, and 99.9th percentiles, respectively.
Acknowledgments
We thank Dr. Robert Cardiff for help with histopathological characterization and Dr. Richard Maas as well as the reviewer for helpful comments on the manuscript. This research was supported by a grant from NIDDK (RO1DK45639).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL beier@rascal.med.harvard.edu; FAX (617) 264-5292.
REFERENCES
- American PKD1 Consortium. Analysis of the genomic sequence for the autosomal dominant polycystic kidney disease (PKD1) gene predicts the presence of a leucine-rich repeat. The American PKD1 Consortium (APKD1 Consortium) Hum Mol Genet. 1995;4:575–582. doi: 10.1093/hmg/4.4.575. [DOI] [PubMed] [Google Scholar]
- Atala A, Freeman MR, Mandell J, Beier DR. Juvenile cystic kidneys (jck): A new mouse mutation which causes polycystic kidneys. Kidney Int. 1993;43:1081–1085. doi: 10.1038/ki.1993.151. [DOI] [PubMed] [Google Scholar]
- Bihoreau M, Ceccherini I, Browne J, Kranzlin B, Romeo G, Lathrop G, James M, Gretz N. Location of the first genetic locus, PKDr1, controlling autosomal dominant polycystic kidney disease in Han:SPRD cy/+ rat. Hum Mol Genet. 1997;6:609–613. doi: 10.1093/hmg/6.4.609. [DOI] [PubMed] [Google Scholar]
- Brasier JL, Henske EP. Loss of the polycystic kidney disease (PKD1) region of chromosome 16p13 in renal cyst cells supports a loss-of-function model for cyst pathogenesis. J Clin Invest. 1997;99:194–199. doi: 10.1172/JCI119147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Polycystic Kidney Disease Consortium. The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. Cell. 1994;77:881–894. doi: 10.1016/0092-8674(94)90137-6. [DOI] [PubMed] [Google Scholar]
- Iakoubova OA, Dushkin H, Beier DR. Localization of a murine recessive polycystic kidney disease mutation and modifying loci which affect disease severity. Genomics. 1995;26:107–114. doi: 10.1016/0888-7543(95)80088-4. [DOI] [PubMed] [Google Scholar]
- Iakoubova, O.A., H. Dushkin, L. Pacella, and D.R. Beier. 1999. Genetic analysis of modifying loci on mouse chromosome 1 affecting severity in a model of recessive polycystic kidney disease. Physiol. Genom. (in press). [DOI] [PubMed]
- International PKD Consortium. Polycystic kidney disease: The complete structure of the PKD1 gene and its protein. Cell. 1995;81:289–298. doi: 10.1016/0092-8674(95)90339-9. [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: Guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lu W, Peissel B, Babakhanlou H, Pavlova A, Geng L, Fan X, Larson C, Brent G, Zhou J. Perinatal lethality with kidney and pancreas defects in mice with a targetted Pkd1 mutation. Nat Genet. 1997;17:179–181. doi: 10.1038/ng1097-179. [DOI] [PubMed] [Google Scholar]
- Lu W, Fan X, Basora N, Babakhanlou H, Law T, Rifai N, Harris P, Perez-Atayde A, Rennke H, Zhou J. Late onset of renal and hepatic cysts in Pkd1-targeted heterozygotes. Nat Genet. 1999;21:160–161. doi: 10.1038/5944. [DOI] [PubMed] [Google Scholar]
- Manly, K.F. and J.M. Olson. 1999. Overview of QTL mapping software and introduction to Map Manager. Mamm. Genome (in press). [DOI] [PubMed]
- Mochizuki T, Wu G, Hayashi T, Xenophontos SL, Veldhuisen B, Saris JJ, Reynolds DM, Cai Y, Gabow PA, Pierides A, et al. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science. 1996;272:1339–1342. doi: 10.1126/science.272.5266.1339. [DOI] [PubMed] [Google Scholar]
- Nauta J, Goedbloed M, van Herk HJ, Wright CJ, Guay-Woodford LM. The rat wpk mutation, a new model of autosomal recessive polycystic kidney disease (ARPKD) maps to chromosome 5 (abstract) J Am Soc Nephrol. 1999;10:422A. [Google Scholar]
- Neuhaus I, Beier D. Efficient localization of mutations by interval haplotype analysis. Mamm Genome. 1998;9:150–154. doi: 10.1007/s003359900706. [DOI] [PubMed] [Google Scholar]
- Qian F, Watnick TJ, Onuchic LF, Germino GG. The molecular basis of focal cyst formation in human autosomal dominant polycystic kidney disease type I. Cell. 1996;87:979–987. doi: 10.1016/s0092-8674(00)81793-6. [DOI] [PubMed] [Google Scholar]
- Ward CJ, Turley H, Ong AC, Comley M, Biddolph S, Chetty R, Ratcliffe PJ, Gattner K, Harris PC. Polycystin, the polycystic kidney disease 1 protein, is expressed by epithelial cells in fetal, adult, and polycystic kidney. Proc Natl Acad Sci. 1996;93:1524–1528. doi: 10.1073/pnas.93.4.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo DD, Nguyen DK, Khatibi N, Olsen P. Genetic identification of two major modifier loci of polycystic kidney disease progression in pcy mice. J Clin Inv. 1997;100:1934–1940. doi: 10.1172/JCI119724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, D'Agati V, Cai Y, Markowitz G, Park JH, Reynolds DM, Maeda Y, Le TC, Hou H, Jr, Kucherlapati R, et al. Somatic inactivation of Pkd2 results in polycystic kidney disease. Cell. 1998;93:177–188. doi: 10.1016/s0092-8674(00)81570-6. [DOI] [PubMed] [Google Scholar]