Abstract
Synthesis and assembly of the mitochondrial oxidative phosphorylation (OXPHOS) system requires genes located both in the nuclear and mitochondrial genomes, but how gene expression is coordinated between these two compartments is not fully understood. One level of control is through regulated expression mitochondrial ribosomal proteins and other factors required for mitochondrial translation and OXPHOS assembly, which are all products of nuclear genes that are subsequently imported into mitochondria. Interestingly, this cadre of genes in budding yeast has in common a 3′-UTR element that is bound by the Pumilio family protein, Puf3p, and is coordinately regulated under many conditions, including during the yeast metabolic cycle. Multiple functions have been assigned to Puf3p, including promoting mRNA degradation, localizing nucleus-encoded mitochondrial transcripts to the outer mitochondrial membrane, and facilitating mitochondria-cytoskeletal interactions and motility. Here we show that Puf3p has a general repressive effect on mitochondrial OXPHOS abundance, translation, and respiration that does not involve changes in overall mitochondrial biogenesis and largely independent of TORC1-mitochondrial signaling. We also identified the cytoplasmic translation factor Slf1p as yeast metabolic cycle-regulated gene that is repressed by Puf3p at the post-transcriptional level and promotes respiration and extension of yeast chronological life span when over-expressed. Altogether, these results should facilitate future studies on which of the many functions of Puf3p is most relevant for regulating mitochondrial gene expression and the role of nuclear-mitochondrial communication in aging and longevity.
Introduction
The assembly of the mitochondrial oxidative phosphorylation (OXPHOS) system is a remarkably complex process, in part, because expression from two genomes, mtDNA and nuclear DNA, must be coordinated [1]. Furthermore, this process is more intricate given the fact that all of proteins required for replication of mtDNA, as well as transcription and translation of it mRNA products, are encoded by nuclear DNA and imported into the organelle [2], [3]. The budding yeast, Saccharomyces cerevisiae, has been used extensively as a model system to identify and understand signaling pathways that mediate this nuclear-mitochondrial crosstalk [1], [4], [5], [6], [7]. However, even in yeast, these are far from understood and the role of post-transcriptional events as a means of regulating nuclear-encoded mitochondrial gene expression has not been studied to the same extent as transcriptional regulatory circuits.
Budding yeast can respond quickly to changes in nutrient levels by modulating protein expression in both space and time. For example, glucose-repressed genes are regulated by both transcriptional and post-transcriptional mechanisms at the level of mRNA [4], [5]. The nuclear genes that encode mitochondrial ribosome proteins (MRPs) are co-regulated as determined in numerous microarray studies [8]. However, there is lack of lack of obvious transcriptional regulatory elements in the promoters of the genes in this module [9] and it has been postulated that these have been lost during the evolution of glucose-repressed yeasts [10]. Alternatively, 73 of the 74 nucleus-encoded MRP genes possess a UGUANAUA motif in their 3′-UTR that likely directs their co-regulation including during the yeast metabolic cycle [11]. In fact, almost all transcripts encoding for factors involved with mitochondrial protein synthesis or assembly of the mitochondrial OXPHOS system have this Puf3 Element (P3E) [12], [13], which is bound by Puf3p [14] to stimulate the decapping and deadenylation of P3E-containing transcripts [15]. The repression by the PUF family of proteins is conserved and involves the binding of Pop2p, which in turn recruits CCR4-NOT deadenylase machinery as well as decapping factors, Dcp1p and Dhh1p [16].
Early work by Butow and colleagues showed that cytoplasmic ribosomes are attached to the mitochondrial outer membrane [17], strongly suggesting co-translational import of polypeptides into mitochondria [18]. In yeast, many nucleus-derived transcripts encoding mitochondrial proteins are localized to mitochondrial-bound ribosomes via information residing in their 3′-UTR [19]. The mRNA localization phenomenon is apparently conserved, as human MrpS12 mRNA associates with mitochondria in a 3′-UTR dependent fashion [20]. In addition to its role in promoting mRNA decay, Puf3p also is important for mitochondrial transcript localization. That is, mitochondrial association of the majority of P3E transcripts is reduced or abrogated in the absence of Puf3p [18]. The mitochondrial localization of transcripts by Puf3p is likely linked to import of the polypeptide it encodes, as genetic interactions exist between PUF3 and TOM20 [21], a component of the TOM complex responsible for translocation of polypeptides across the outer membrane. Thus, Puf3p appears to be involved in controlling the logistics of OXPHOS assembly and may do so spatially and in response to the yeast metabolic cycle [22]. There is also some indication that the activity of Puf3p in mRNA decay are regulated by the nutrient-sensing TORC1 signaling pathway [12]. Finally, Puf3p physically links the mitochore to the actin cytoskeleton, allowing for bud-directed organellar movement during cell division [23] and is associated with P bodies [24]. With so many assigned functions and locations, how Puf3p operates to control various cellular processes remains unclear, although models have begun to formulate [13], [25]. The goal of this study was to gain additional insight into how Puf3p controls mitochondrial gene expression and respiration and its relationship to the TORC1 pathway, which we have shown represses mitochondrial translation and curtails yeast chronological life span [26], [27].
Results
Puf3p represses mitochondrial translation, OXPHOS subunit accumulation and respiration without affecting overall mitochondrial biogenesis
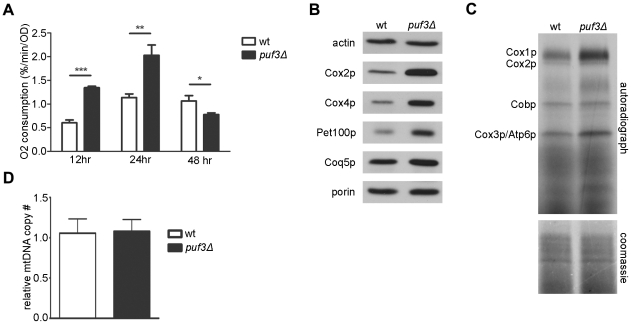
We analyzed multiple mitochondrial parameters in wild-type (DBY2006) and isogenic puf3Δ yeast strains grown in synthetic dextrose medium. Lack of Puf3p increased mitochondrial oxygen consumption during logarithmic growth and at early stationary phase time points, however, this was not sustained (and even decreased slightly) later in stationary phase (Figure 1A). The increase in mitochondrial respiration observed in log-phase growth was accompanied by a corresponding robust increases in the steady-state levels of multiple mitochondrial proteins, including Cox2p and Cox4p, mtDNA-encoded and nucleus-encoded OXPHOS subunits, respectively (Figure 1B) and Pet100p, a nucleus-encoded protein with a Puf3 element in its 3′ UTR. We observed no significant increases in “mitochondrial housekeeping” markers Coq5p and porin compared to the actin loading control, suggesting that overall mitochondrial biogenesis was not increased in puf3Δ cells (Figure 1B). There was also no increase in mtDNA per cell in puf3Δ strains, which is consistent with mitochondrial biogenesis not being altered (Figure 1D). Finally, there was an apparently global up-regulation of mitochondrial translation in puf3Δ strains compared to the wild-type control during log-phase growth (Figure 1C). The up-regulation of mitochondrial translation and OXPHOS subunits per se (i.e. without a corresponding increase in overall mitochondrial biogenesis) is consistent with Puf3p being a negative regulator of the subset of nucleus-encoded genes involved in mitochondrial translation and OXPHOS assembly [23], but not overall organelle biogenesis.
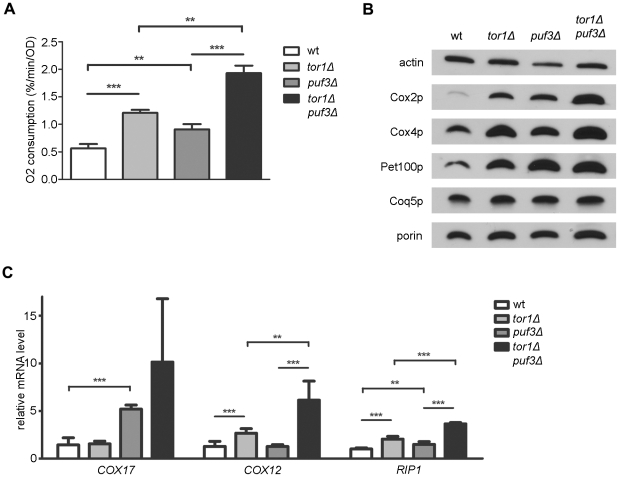
Figure 1. Analysis of puf3Δ Yeast Strains.
Analysis of wild-type (wt) and an isogenic puf3Δ strains are shown. (A) Mitochondrial oxygen consumption assayed at 12 hours (log phase), 24 hours (early stationary phase), and 48 hours post-inoculation (1 day into stationary phase). The bars represent the mean ± SD with p-values as indicated (*<0.05, **<0.01, ***<0.001). Statistical analysis was performed with Prism 5 software, using a Student's t-test (unpaired, two-tailed). (B) Western blot of indicated proteins in log-phase growth (representative of three biological replicates). (C) Autoradiogram of separated mtDNA-encoded proteins labeled with 35S-methionine and 35S -cysteine at logarithmic growth. A section of the same gel stained with coomassie blue to demonstrate loading is shown underneath. (D) mtDNA copy number.
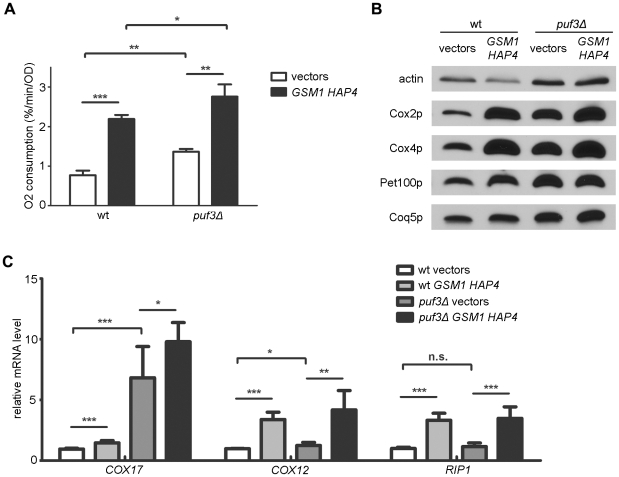
To probe further whether up-regulation of mitochondrial respiration in puf3Δ strains was independent of mitochondrial biogenesis pathways, we over-expressed Hap4p, a known transcriptional regulator of mitochondrial biogenesis, and Gsm1p, another transcription factor implicated in regulating the expression of OXPHOS proteins [28], [29], in wild-type and puf3Δ strains. Simultaneous over-expression of these two transcription factors caused similar increases in respiration in both wild-type and puf3Δ strains (Figure 2A) and, further increases in the already heightened steady-state levels of Cox2p and Cox4p in the puf3Δ strain (Figure 2B). Finally, we assayed the steady-state levels of COX17 transcripts, a canonical Puf3 target [15], and two non-P3E-target transcripts, RIP1 and COX12, by quantitative real-time RT-PCR. As expected, deletion of PUF3 leads to a dramatic increase in COX17 transcripts, but had little or no effect on the non-P3E targets, while over-expression of HAP4 and GSM1 in a wild-type strain led to an increase in COX12 and RIP1 mRNA (Figure 2C). Importantly, Hap4p/Gsm1p over-expression increased all of these transcripts to a similar degree in the puf3Δ strain (Figure 2C), bolstering our conclusion that up-regulation of respiration in the absence of Puf3p is largely independent of pathways that increase mitochondrial biogenesis.
Figure 2. The Regulation of Respiration by Puf3p is Independent of Mitochondrial Biogenesis Pathways.
Shown is the analysis of wild-type (wt) and puf3Δ strains with plasmids over-expressing HAP4 and GSM1 (HAP4 GSM1) and corresponding empty vectors (vectors) (A) Mitochondrial oxygen consumption during late-log phase of growth. (B) Western blot analysis (representative of three biological replicates). (C) Steady-state levels of COX17, COX12, and RIP1 transcripts in late log phase of growth. The values indicate the mean +/− SD with p-values indicated as described in the legend of Figure 1.
Over-expression of Puf3p Reduces Mitochondrial Respiration and Steady-state levels of OXPHOS-related Proteins in Late Log-phase Cultures
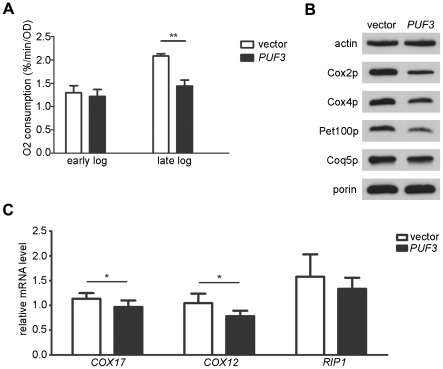
Unlike deletion of PUF3, which increased mitochondrial oxygen consumption and OXPHOS protein accumulation during log-phase growth, over-expression of PUF3 did not significantly repress these parameters until later in log phase (Figure 3A and 3B) when cells have already undergone the diauxic shift and significantly increased mitochondrial biogenesis and respiration. Only a modest decline in the Puf3p-target transcript COX17 was observed under these conditions and the two other mitochondrial transcripts analyzed were either minimally effected or not at all (Figure 3C). These data suggest that the repression of respiration by Puf3p over-expression may involve functions other than mRNA transcript decay.
Figure 3. Analysis in strains that over-express Puf3p.
Shown is a wild-type strain with a plasmid that over-expresses Puf3p (PUF3) or the corresponding empty vector (vector). (A) Mitochondrial oxygen consumption during early and late log phase. (B) Western blot analysis of protein extracts derived from late log cultures (representative of three biological replicates). (C) Steady-state levels of COX17, COX12, and RIP1 transcripts in late log phase. The values indicate the mean +/− SD with p-values indicated as described in the legend of Figure 1.
Over-expression of PUF3 Curtails Life Span Extension in a tor1Δ strain, but is Not a Major Mediator of the Increased Respiration Due to Reduced mTORC1 signaling
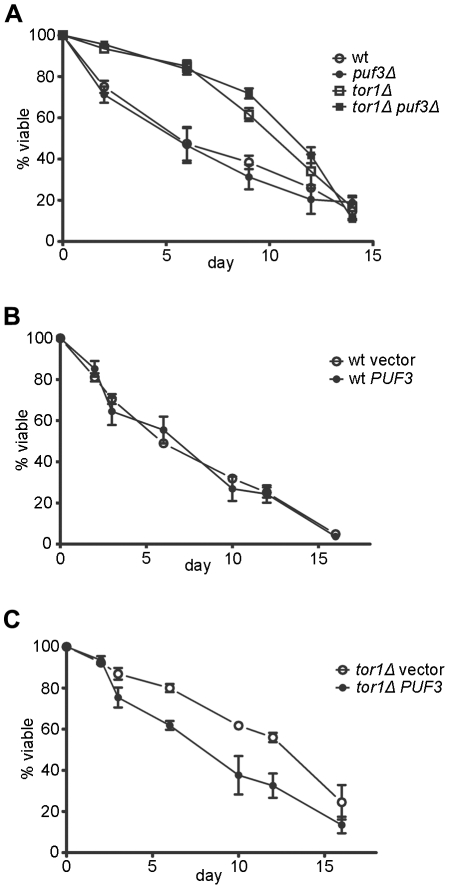
Reactive oxygen species (ROS) are produced during metabolism and are thought to contribute to aging. For example, they are generated by the mitochondrial electron transport chain, placing these organelles at the nexus of oxidative stress and aging according to the mitochondrial theory of aging. We found previously [26], [27] that reduced TORC1 signaling increases the density of OXPHOS complexes in a manner that reduces ROS in stationary phase and extends yeast chronological life span. Given that similar changes in OXPHOS abundance and respiration are observed in a puf3Δ strain (Figure 1), we ascertained whether Puf3p is involved in regulating CLS and/or is a downstream effector of TORC1 effects on mitochondrial function. However, deletion of PUF3 in wild-type or tor1Δ strains did not affect CLS (Figure 4A). Furthermore, no difference in CLS were observed in a wild-type strain that over-expresses Puf3p (Figure 4B). However, over-expression of Puf3p did partially reduce the extended CLS of a tor1Δ strain (Figure 4C).
Figure 4. Chronological Aging in strains that lack or over-express Puf3p.
Shown are chronological life span curves (viability as a function of time in days of stationary phase) of wild-type strain compared to isogenic puf3Δ, tor1Δ, and puf3Δ tor1Δ strains (A), a wild-type strain with an empty vector or a plasmid that over-expresses PUF3 (B), or a tor1Δ strain with an empty vector or a plasmid that over-expresses PUF3 (C). Each point is the mean +/− SD of three biological replicates.
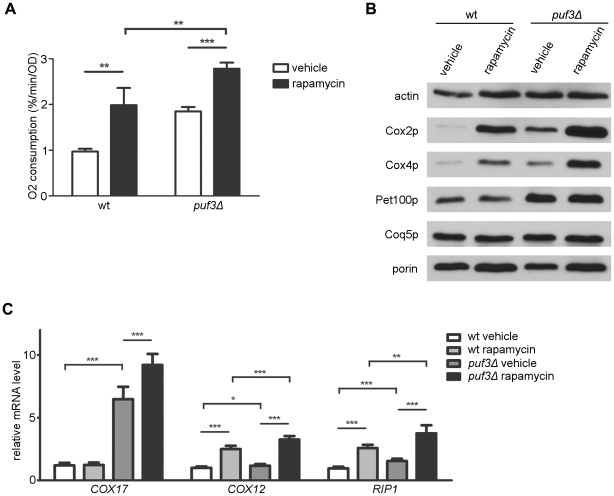
To probe more directly the potential involvement of Puf3p in the regulation of mitochondrial function by TORC1, we compared mitochondrial parameters in tor1Δ and puf3Δ single-mutant strains to those in a tor1Δ puf3Δ double-mutant strain. While both single-mutant strains had increased mitochondrial oxygen consumption and Cox2p and Cox4p levels compared to the isogenic wild-type strains, the double-mutant strains displayed an additive increase in these parameters (Figure 5A and 5B). In contrast to Cox2p and Cox4p, Pet100p (a Puf3p target) did not display an additive increase in the double-mutant strain (Figure 5B). Comparison of the “mitochondrial housekeeping” proteins Coq5p and porin to the loading control actin revealed only modest changes in overall mitochondrial abundance in any of the mutant strains compared to the wild-type control (Figure 5B). Finally, analysis of mitochondrial transcripts in this cadre of strains revealed complex regulation. While loss of Puf3p led to increased levels of the Puf3-target COX17 transcript, this was enhanced further by loss of Tor1p (Figure 5C, left). Similarly, loss of Tor1p resulted in increased amounts of COX12 and RIP1 transcripts, but this too was enhanced further (in an apparently additive manner) by lack of Puf3p (Figure 5C, center and right). Similar results with regard to respiration, steady-state mitochondrial marker protein levels, and target transcripts were observed in wild-type and puf3Δ strains treated with rapamycin (Figure 6). These data indicate that, while there might be some degree of crosstalk/synergism between the Puf3p and TORC1 pathways with regard to specific mitochondrial targets and effects, these pathways operate largely independently with regard to respiration and CLS.
Figure 5. Comparison of mitochondrial parameters in puf3Δ, tor1Δ, and puf3Δ tor1Δ mutant strains.
(A) Mitochondrial oxygen consumption during early log phase. (B) Western blot analysis of protein extracts derived from log phase cultures (representative of three biological replicates). (C) Steady-state levels of COX17, COX12, and RIP1 transcripts in log phase. The values indicate the mean +/− SD with p-values indicated as described in the legend of Figure 1.
Figure 6. Comparison of mitochondrial parameters in wild-type and puf3Δ mutant strains treated with the TORC1 inhibitor rapamycin.
(A) Mitochondrial oxygen consumption of cells treated with rapamycin during early log phase and analyzed during late log phase. (B) Western blot analysis of protein extracts derived from late log phase cultures (representative of three biological replicates). (C) Steady-state levels of COX17, COX12, and RIP1 transcripts in late log phase. The values indicate the mean +/− SD with p-values indicated as described in the legend of Figure 1.
Over-expression of Slf1p Increases Mitochondrial Respiration and Chronological Life Span
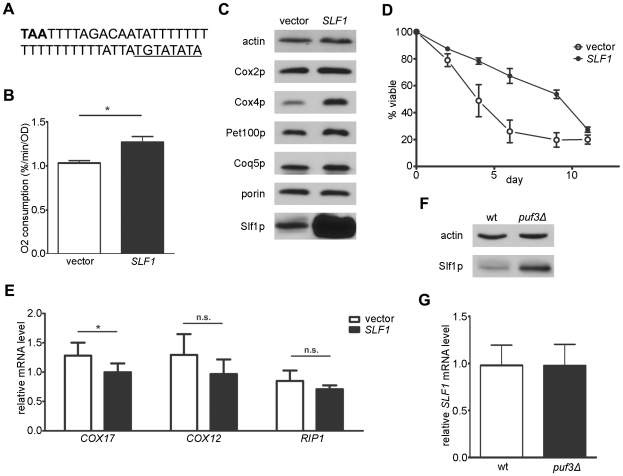
The vast majority of genes in the Puf3 regulon are regulators of mitochondrial translation (e.g. mitochondrial ribosomal proteins and translation factors) or processes immediately downstream (e.g. OXPHOS assembly factors) that control respiration. To find additional genes potentially involved in mitochondrial function, others identified transcripts that were both localized to the mitochondrial periphery and clustered with P3E mRNAs as a function of the yeast metabolic cycle (YMC) [22]. We hypothesized that a subset of the Puf3 regulon is not involved with mitochondrial translation directly, but may nonetheless be important for mitochondrial respiration. To test this hypothesis, we queried the YMC microarray database SCEPTRANS [30] for mRNAs that displayed a YMC pattern of expression, but are not predicted to localize to mitochondria. Of the top 50 transcripts coregulated with COX17 in the YMC, three met this criterion: YOR248W, a dubious ORF; SRL1, a mannoprotein associated with the cell wall [31]; and SLF1, and RNA-binding protein associated with cytoplasmic polysomes [32]. A consensus P3E exists shortly after the stop codon in the SLF1 mRNA (Figure 7A), consistent with it being a target of Puf3p. Thus Slf1p seemed like a likely candidate factor that could potentially regulate respiration and was studied further. While slf1Δ strains did not reveal any obvious growth or respiration phenotype in fermentable or non-fermentable media, over-expression of Slf1p resulted in a modest, but reproducible increase in mitochondrial oxygen consumption (Figure 7B), a pronounced increase in Cox4p, and modest increases in Cox2p and Pet100p (Figure 7C). Over-expression of Slf1p also significantly increased median CLS (Figure 7D) and significantly reduced the amount of the Puf3-target transcript, COX17, while having minimal or no effect on non-puf3-target transcripts (Figure 7E). Finally, Slf1p protein (Figure 7F), but not its transcript (Figure 7G), was up-regulated in a puf3Δ strain. Altogether, these data indicate that Slf1p a P3E-containing gene that encodes a putative regulator of mitochondrial function and CLS and is repressed by Puf3p at the post-transcriptional level.
Figure 7. Over-expression of Slf1p increases respiration and CLS, and expression of Sls1p is repressed by Puf3p.
(A) SLF1 DNA sequence depicting the stop codon (bold) and Puf3 element (underlined). (B) Mitochondrial oxygen consumption of wild-type cells carrying an empty vector (YEp351) or YEp351 with SLF1 during early stationary phase. (C) Western blot analysis of protein extracts derived from the same strains in (B) (representative of three biological replicates). (D) CLS analysis of the same strains in (B). (E) Q-RT-PCR analysis COX17, COX12, and RIP1 transcript levels at early stationary phase from the same strains in (B). Analysis of Slf1p levels by western blot (F) and SLF1 mRNA levels by RT-PCR (G) in wild-type and puf3Δ strains in log-phase growth (representative of three biological replicates).
Discussion
The pumilio family (PUF) of proteins in yeast binds distinct sets transcripts and regulates their stability, translation and subcellular localization [25]. In this study, we have examined Puf3p, the member of this family that preferentially binds to nuclear mRNAs that encode proteins involved in mitochondrial translation and OXPHOS assembly [22]. Based on our results, we conclude that 1) Puf3p has a general repressive effect on mitochondrial translation, OXPHOS protein accumulation, and respiration, 2) the Puf3p repressive effect on respiration is mediated by changes in mitochondrial OXPHOS complex density, as opposed to modulating overall mitochondrial biogenesis, 3) that effects of Puf3p on respiration are largely independent of the TORC1 signaling pathway, and 4) that Slf1p is member of the Puf3 regulon that is repressed by Puf3p at the post-transcriptional level and putatively regulates mitochondrial respiration, possibly by influencing cytoplasmic translation of nuclear transcripts that encode mitochondrial proteins. We will also discuss the relevance of effects we observed of over-expression of Puf3p or Slf1p on chronological aging. Altogether, these results should facilitate future studies on which of the many functions of Puf3p is most relevant for regulating mitochondrial gene expression and function.
Our results indicate that, despite its postulated pro-mitochondrial functions of localizing transcripts to the mitochondrial periphery [18] and promoting mitochondrial movement [23], Puf3p is an overall negative regulator of mitochondrial translation and respiration (Figures 1 and 3). This repressive effect on respiration appears to be primarily manifest at the level OXPHOS system abundance as opposed to changes in overall mitochondrial biogenesis as lack of Puf3p increases translation of mtDNA-encoded OXPHOS subunits and steady-state levels of mtDNA-encoded and nuclear-DNA encoded OXPHOS proteins without increasing mitochondrial housekeeping markers or mtDNA (Figure 1). This conclusion is bolstered by similar, but opposite effects observed on respiration and OXPHOS density upon over-expression of Puf3p (Figure 3) and additive effects of over-expression of Hap4p and Gsm1p on these parameters in a puf3Δ strain (Figure 2). These results fully corroborate that the main cadre of transcripts regulated by Puf3p encodes mitochondrial ribosomal proteins and other factors involved in mitochondrial translation and OXPHOS assembly, which does not include genes needed for wholesale organelle biogenesis. At this point we can only speculate on which function of Puf3p is dominating the repressive effects on respiration. At first glance, this may seem contrary to the idea that Puf3p is needed to localize nuclear transcripts to mitochondrial outer membrane for co-translational import of the mitochondrial proteins they encode. However, loss of this function in puf3Δ strains may be compensated for by the overall increase in P3E transcript abundance and/or their release from a translationally repressive condition such as localization to P bodies [24]. That over-expression of Puf3p had little impact on COX17 transcript abundance (Figure 3C) also suggests that functions of Puf3p other than degradation of P3E-target transcripts per se are mediating the repressive effect on respiration under this circumstance.
The TORC1 inhibitor rapamycin causes the collective up-regulation of P3E transcripts in yeast and slows the decay of a reporter transcript bearing a 3′-UTR P3E [12]. This, coupled with the involvement of TORC1 pathway in regulating the deadenylation and decapping of transcripts during the diauxic shift [33], when mitochondrial respiration is up-regulated, suggested that Puf3p may regulated by TORC1 signaling. This hypothesis appeared even more likely when we observed that deletion of PUF3 resulted in up-regulation of OXPHOS density without an increase in overall mitochondrial biogenesis, which is a phenotype we had documented previously in tor1Δ and sch9Δ strains that have reduced TORC1 signaling [27]. Despite these intriguing connections, the simplest interpretation of our results comparing mitochondrial parameters in a tor1Δ puf3Δ double-mutant strain to the respective single-mutant strains (Figure 5) is that Puf3p is not a major downstream effector of TORC1 with regard to repressing mitochondrial respiration. However, our results do indicate some degree of interplay between TORC1 and Puf3p regulation. For example, an epistatic relationship was apparent with regard to the steady-state levels of Pet100p (Figure 5B), whose mRNA has a P3E element, which might indicate that TORC1 and Puf3p regulation may indeed converge on certain P3E-containing transcripts. Furthermore, there were synergistic (more than additive) increases in COX17 and COX12 transcripts in the tor1Δ puf3Δ double-mutant strain (Figure 5C), consistent with crosstalk between the Puf3p and TORC1 pathways. Recently, the mitochondrial protein Fmp48p was found in a global screen to interact with both Tor1p and Puf3p [34], which is intriguing with regard to potential crosstalk between these pathways.
We identified Slf1p as a new factor involved, directly or indirectly, in mitochondrial regulation based on its co-regulation in the yeast metabolic cycle with other Puf3 targets (Figure 7). Since this protein is most likely involved in cytoplasmic translation [32], it is one of the minority of Puf3 targets that apparently does not affect mitochondrial translation or OXPHOS assembly in the organelle directly. Over-expression of Slf1p modestly increases respiration (Figure 7B), significantly increases Cox4p levels, and modestly increases Cox2p and Pet100p (Figure 7C). It also decreases COX17 transcripts, but not two other non-P3E targets (Figure 7E). These results indicate that this protein is likely involved in regulating Puf3-target transcripts, but may be somewhat selective. Finally, even though the SLF1 transcript contains a predicted P3E element (Figure 7A), its abundance is not affected by deletion of PUF3 (Figure 7G). However, Slf1p is up-regulated in the puf3Δ strain (Figure 7F). Therefore, Slf1p is a unique member of the Puf3p mitochondrial regulon that is itself regulated by Puf3p at the post-transcriptional level. Slf1p binds RNA, localizes to cytoplasmic polysomes, and influences sensitivity to translational inhibitors [32]. In addition, four out of seven high-throughput physical associations of Slf1 are with factors regulating mRNAs and translation, including eIF4E [35] and Slf1p possesses a putative eIF4E-binding motif (YXXXXLϕ). Thus, it is tempting to speculate that Slf1p promotes translation of a subset of Puf3p targets and/or perhaps other nuclear transcripts that encode mitochondrial proteins to modulate mitochondrial respiration under certain conditions.
We have shown that extension of yeast chronological life span (CLS) by reduced TORC1 signaling requires mitochondrial respiration and involves increased mitochondrial translation and OXPHOS complex density [26], [27]. Even though respiration and OXPHOS complex density vary in response to altered Puf3p levels (Figures 1 and 3), we observed no changes in CLS in either wild-type or tor1Δ strains deleted for PUF3 or in a wild-type strain over-expressing Puf3p (Figure 4A and 4B). However, over-expression of Puf3p reduces CLS in a tor1Δ strain (Figure 4C), but does not eliminate CLS extension completely. The two conclusions we draw from these results are 1) that changes in respiration induced by deletion of Puf3p must be of a different quality than that induced by reduced TORC1 signaling that leads to CLS extension and 2) that increased mitochondrial translation and respiration in a tor1Δ strain is necessary, but not sufficient for full CLS extension (i.e. over-expression of Puf3p provided a direct test of the requirement of these parameters in CLS extension by reduced TORC1 signaling). In this regard it is noteworthy that we recently showed that extension of CLS by TORC1 involves the generation of an adaptive mitochondrial ROS signal during growth (Pan et al. Cell Metabolism, in press). That this signal is not generated even though respiration is up-regulated in puf3Δ strains may explain why they do not have extended CLS (i.e. potentially explaining why increased respiration is necessary, but not sufficient to observe affects on CLS). Finally, over-expression of Slf1p increased median CLS (Figure 7D), which was accompanied by a moderate increase in respiration. Whether these effects of Slf1p are mediated by salient changes in respiration or cytoplasmic translation, which also contribute to aging [36], remains to be determined. It is tempting to speculate that these two processes may conspire to affect CLS, with alterations in cytoplasmic translation regulating the relative rate of translation of nuclear transcripts encoding mitochondrial proteins and hence altering mitochondrial gene expression and respiratory function in ways that influence the quality and quantity of respiration. Dissecting which aspects of increased respiration are beneficial to CLS and how they are regulated remains an important area of future investigation.
Materials and Methods
Yeast Strains
All experiments were performed using the strain background DBY2006 (MATa his3-Δ200 leu2-3,-112 ura3-52 trp1-Δ1 ade2-1) in synthetic dextrose (SD) media with appropriate amino acid supplements. Overnight 5 mL cultures were used to inoculate a 50 mL culture to a final OD600 of 0.1. Where indicated, strains were treated with 80 nM rapamycin (Sigma) or an appropriate volume of vehicle (ethanol) at an OD of 1.0 and assayed after the culture reached an OD of 2.5-3.0. Deletion of the PUF3 ORF was achieved by transformation of DBY2006 with a puf3::kanMx4 amplicon derived using PCR primers flanking the PUF3 ORF and genomic DNA from the Open Biosystems PUF3 knockout strain. To generate single and double knock-outs of TOR1 and PUF3, wild-type and tor1::kanMx4 strains were transformed with a puf3::ura3 PCR amplicon derived using pRS316 as template and primers F (5′-GTT TCT TCT TTA AGC GCC CTG TCC CAT AGT AAC AAC AAC GAG ATT GTA CTG AGA GTG CAC-3′) and R (5′-GTT TCT CAA CAC TGG CTA AAT GTC TAT TTC CTA GGG AGT TCT GTG CGG TAT TTC ACA CCG-3′). In the strains used in Figure 2, the PUF3 locus was replaced with TRP1 using the primers stated above and pRS314 as a template, while the wild-type strain was made congenic by replacing the ade2 locus with pRS314-derived TRP1 using the primers F (5′-GCA AAC AGG CTC AAC ATT AAG ACG GTA ATA CTA GAT GCT GAG ATT GTA CTG AGA GTG CAC-3′) and R (5′-TCT TCT TCT TGC TTT AAT AAA AAC TGT TCC ATT TTC GTT GCT GTG CGG TAT TTC ACA CCG-3′). The PUF3 ORF was amplified with the primers F (5′-GCT CAG CTG CAG TAC GGT AAT TGC GCA CTC TG-3′) and R (5′-GCT CAG GGA TCC CAT GAC AAC AGT ATT CTC AGT CAA A-3′) and cloned into the BamHI/PstI site of the plasmid YEp351. The GSM1 ORF was amplified with the primers F (5′-AAT TAT AAG CTT ATC TCT GAC TGC CGT TTT GC-3′) and R (5′-AAT TAT AAG CTT TTG CCA ATC GGA CAA CAC TA-3′) and cloned into the HindIII site of YEp351. The SLF1 ORF was amplified with the primers F (5′-ATT TAT CCC GGG TGC AGT ATG TTA TTG ATC CAT CG-3′) and R (5′-ATA AAT CCC GGG CTC TGG TGG CAT AAA ACA T-3′) and cloned into the XmaI site of YEp351.
Chronological Life Span and Respiration Assays
Chronological life span was assayed as described previously [26]. 100 µl of culture was incubated with 100 µl of 4 mg/ml trypan blue for 5 minutes at 30°, and blue and clear cells were counted with a hemocytometer. Mitochondrial oxygen consumption was tested as described previously [26]. Briefly, 5 mL of yeast cultures were assayed at indicated stages of growth with a Clark-type electrode (model 5300A; YSI Bioanalytical). Addition of sodium azide to cultures ensured oxygen consumption was due to mitochondrial function.
Transcript analysis by quantitative real-time PCR
RNA extraction and precipitation was carried out as described (Bonawitz et al. 2008) and purified further by RNeasy column (Qiagen). Using this RNA, cDNA was synthesized as described [37], using 3.2 µl of 50 µM OligodT23 and 2 µl of 40 µM random hexamer in a 20 µl reaction volume. The diluted cDNA was then mixed with 0.5 µl of 25 µM of each primer and 12.5 µl SYBR green [38] in a total volume of 25 µl and PCR performed using a Biorad C1000 thermocycler/CFX96 RT system used the following program: 1 cycle of 3 min at 95°C, followed by 35 cycles of 30 s at 95°C, 30 s at 53°C, and 30 s at 72°C. Ct values were obtained with Biorad CFX manger software and transcript levels were normalized using the actin control. The primers used were:
Actin RT F 5′-CCCAGGTATTGCCGAAAGAATGC-3′
Actin RT R 5′-GGAAGATGGAGCCAAAGCGG-3′;
Cox17 RT F 5′-CCAGAAAAGGAGGAGCGGGATA-3′
Cox17 RT R 5′-CGAAGCCATAACCCTTCATGCAC-3′;
Cox12 RT F 5′-CCCCAACAAAACCAAACAAAGCA-3′
Cox12 RT R 5′-TCCAAAAGACCTTGCACGGAG-3′;
Rip1 RT F 5′-TGGTCGGTGCTATGGGTCTTT-3′
Rip1 RT R 5′-AACATCGGCAGTAGCGGTCA-3′;
Slf1 RT F 5′-GGAAATTGCCCTTGGAAGCA-3′
Slf1 RT R 5′- GCCCAATTTTCGCGCCTAAT-3′.
Western Immunoblotting
Protein extracts were prepared by TCA precipitation, separated on SDS-PAGE gels and transferred to PVDF (Millipore) membranes as described [39]. Antibodies directed against Cox2p (Invitrogen, 459150), Cox4p (Molecular Probes, A-6432), porin (Invitrogen, 459500), and actin (Chemicon) were diluted 1∶1000, incubated overnight, washed, and probed with goat-anti-mouse secondary diluted 1∶5000. Antibodies versus Coq5p, Pet100p, and Slf1p were obtained from colleagues and utilized as described above but diluted 1∶1000-1∶5000 and probed with donkey-anti-rabbit (Santa Cruz) secondary diluted by 1∶5000.
Mitochondrial Translation Assays
Radiolabeling of mtDNA-encoded proteins was performed as described [40]. Briefly, cells that reached an OD of ∼1.0 were treated with 50 µl of 25 mg/ml cyclohexamide for 5 minutes and pulsed with 50 µCi of EXPRE35S35S Protein Labeling Mix (PerkinElmer) for 15 minutes. Cells were then washed, lysed using glass beads, and the resulting crude mitochondrial extracts were acquired by differential centrifugation. Protein (74 µg) was resolved on Any-kD precast gradient gels (BioRad). Gels were then dried, subjected to autoradiography, followed by rehydration and staining with coomassie blue.
Relative mtDNA copy number
The amount of mtDNA per cell was determined as described previously [38]. Briefly, DNA was isolated using the phenol-chloroform “smash-and-grab” method, followed by incubation in 100 µl TE with 10 µg/mL RNase for 1 hour at 65°. DNA was diluted with water 1∶200-1∶400 and added to a 25 µl reaction mixture consisting of 14 µl of SYBR green and 0.5 µl of 16 µM forward and reverse primer. PCR and subsequent analysis were performed using the conditions described above for real-time PCR. The primers used were:
Cox1 RT F 5′-CTACAGATACAGCATTTCCAAGA-3′
Cox1 RT R 5′-GTGCCTGAATAGATGATAATGGT-3′;
Actin RT F 5′-GTATGTGTAAAGCCGGTTTTG-3′
Actin RT R 5′-CATGATACCTTGGTGTCTTGG-3′.
Acknowledgments
The authors would like to thank Yong Pan for assistance with translation assays, Antoni Barrientos for the HAP4 plasmid, Sandra Wolin for the anti-Slf1p antibody, Robert Poyton for the anti-Pet100p antibody and Cathy Clarke for the anti-Coq5p antibody.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by National Institutes of Health (NIH) grant HL-059655 awarded to GSS. MCL was supported by NIH pre-doctoral Genetics training grant T32 GM007499. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Grivell LA. Nucleo-mitochondrial interactions in mitochondrial gene expression. Crit Rev Biochem Mol Biol. 1995;30:121–164. doi: 10.3109/10409239509085141. [DOI] [PubMed] [Google Scholar]
- 2.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 3.Shutt TE, Shadel GS. A compendium of human mitochondrial gene expression machinery with links to disease. Environ Mol Mutagen. 2010;51:360–379. doi: 10.1002/em.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cereghino GP, Scheffler IE. Genetic analysis of glucose regulation in saccharomyces cerevisiae: control of transcription versus mRNA turnover. EMBO J. 1996;15:363–374. [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 6.Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 7.Forsburg SL, Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- 8.Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, et al. Revealing modular organization in the yeast transcriptional network. Nat Genet. 2002;31:370–377. doi: 10.1038/ng941. [DOI] [PubMed] [Google Scholar]
- 9.Lai LC, Kosorukoff AL, Burke PV, Kwast KE. Metabolic-state-dependent remodeling of the transcriptome in response to anoxia and subsequent reoxygenation in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:1468–1489. doi: 10.1128/EC.00107-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ihmels J, Bergmann S, Gerami-Nejad M, Yanai I, McClellan M, et al. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- 11.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 12.Foat BC, Houshmandi SS, Olivas WM, Bussemaker HJ. Profiling condition-specific, genome-wide regulation of mRNA stability in yeast. Proc Natl Acad Sci U S A. 2005;102:17675–17680. doi: 10.1073/pnas.0503803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devaux F, Lelandais G, Garcia M, Goussard S, Jacq C. Posttranscriptional control of mitochondrial biogenesis: spatio-temporal regulation of the protein import process. FEBS Lett. 2010;584:4273–4279. doi: 10.1016/j.febslet.2010.09.030. [DOI] [PubMed] [Google Scholar]
- 14.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olivas W, Parker R. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 2000;19:6602–6611. doi: 10.1093/emboj/19.23.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 17.Kellems RE, Allison VF, Butow RA. Cytoplasmic type 80S ribosomes associated with yeast mitochondria. IV. Attachment of ribosomes to the outer membrane of isolated mitochondria. J Cell Biol. 1975;65:1–14. doi: 10.1083/jcb.65.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saint-Georges Y, Garcia M, Delaveau T, Jourdren L, Le Crom S, et al. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS One. 2008;3:e2293. doi: 10.1371/journal.pone.0002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zarnack K, Feldbrugge M. mRNA trafficking in fungi. Mol Genet Genomics. 2007;278:347–359. doi: 10.1007/s00438-007-0271-8. [DOI] [PubMed] [Google Scholar]
- 20.Russo A, Russo G, Cuccurese M, Garbi C, Pietropaolo C. The 3′-untranslated region directs ribosomal protein-encoding mRNAs to specific cytoplasmic regions. Biochim Biophys Acta. 2006;1763:833–843. doi: 10.1016/j.bbamcr.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Eliyahu E, Pnueli L, Melamed D, Scherrer T, Gerber AP, et al. Tom20 mediates localization of mRNAs to mitochondria in a translation-dependent manner. Mol Cell Biol. 2010;30:284–294. doi: 10.1128/MCB.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lelandais G, Saint-Georges Y, Geneix C, Al-Shikhley L, Dujardin G, et al. Spatio-temporal dynamics of yeast mitochondrial biogenesis: transcriptional and post-transcriptional mRNA oscillatory modules. PLoS Comput Biol. 2009;5:e1000409. doi: 10.1371/journal.pcbi.1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Rodriguez LJ, Gay AC, Pon LA. Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J Cell Biol. 2007;176:197–207. doi: 10.1083/jcb.200606054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SI, Dudley AM, Drubin D, Silver PA, Krogan NJ, et al. Learning a prior on regulatory potential from eQTL data. PLoS Genet. 2009;5:e1000358. doi: 10.1371/journal.pgen.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quenault T, Lithgow T, Traven A. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 2011;21:104–112. doi: 10.1016/j.tcb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Y, Shadel GS. Extension of chronological life span by reduced TOR signaling requires down-regulation of Sch9p and involves increased mitochondrial OXPHOS complex density. Aging (Albany NY) 2009;1:131–145. doi: 10.18632/aging.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng Y, He T, Wu Y, Vanka P, Yang G, et al. Computationally analyzing the possible biological function of YJL103C—an ORF potentially involved in the regulation of energy process in yeast. Int J Mol Med. 2005;15:123–127. [PubMed] [Google Scholar]
- 29.Ho SW, Jona G, Chen CT, Johnston M, Snyder M. Linking DNA-binding proteins to their recognition sequences by using protein microarrays. Proc Natl Acad Sci U S A. 2006;103:9940–9945. doi: 10.1073/pnas.0509185103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudlicki A, Rowicka M, Otwinowski Z. SCEPTRANS: an online tool for analyzing periodic transcription in yeast. Bioinformatics. 2007;23:1559–1561. doi: 10.1093/bioinformatics/btm126. [DOI] [PubMed] [Google Scholar]
- 31.Hagen I, Ecker M, Lagorce A, Francois JM, Sestak S, et al. Sed1p and Srl1p are required to compensate for cell wall instability in Saccharomyces cerevisiae mutants defective in multiple GPI-anchored mannoproteins. Mol Microbiol. 2004;52:1413–1425. doi: 10.1111/j.1365-2958.2004.04064.x. [DOI] [PubMed] [Google Scholar]
- 32.Sobel SG, Wolin SL. Two yeast La motif-containing proteins are RNA-binding proteins that associate with polyribosomes. Mol Biol Cell. 1999;10:3849–3862. doi: 10.1091/mbc.10.11.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Albig AR, Decker CJ. The target of rapamycin signaling pathway regulates mRNA turnover in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2001;12:3428–3438. doi: 10.1091/mbc.12.11.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mehta R, Chandler-Brown D, Ramos FJ, Shamieh LS, Kaeberlein M. Regulation of mRNA translation as a conserved mechanism of longevity control. Adv Exp Med Biol. 2010;694:14–29. doi: 10.1007/978-1-4419-7002-2_2. [DOI] [PubMed] [Google Scholar]
- 37.Cotney J, Wang Z, Shadel GS. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 2007;35:4042–4054. doi: 10.1093/nar/gkm424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lebedeva MA, Shadel GS. Cell cycle- and ribonucleotide reductase-driven changes in mtDNA copy number influence mtDNA Inheritance without compromising mitochondrial gene expression. Cell Cycle. 2007;6:2048–2057. doi: 10.4161/cc.6.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodeheffer MS, Boone BE, Bryan AC, Shadel GS. Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase. J Biol Chem. 2001;276:8616–8622. doi: 10.1074/jbc.M009901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodeheffer MS, Shadel GS. Multiple interactions involving the amino-terminal domain of yeast mtRNA polymerase determine the efficiency of mitochondrial protein synthesis. J Biol Chem. 2003;278:18695–18701. doi: 10.1074/jbc.M301399200. [DOI] [PMC free article] [PubMed] [Google Scholar]