Abstract
Recently a new strategy using BAC end sequences as sequence-tagged connectors (STCs) was proposed for whole-genome sequencing projects. In this study, we present the construction and detailed characterization of a 15.0 haploid genome equivalent BAC library for the cultivated tomato, Lycopersicon esculentum cv. Heinz 1706. The library contains 129,024 clones with an average insert size of 117.5 kb and a chloroplast content of 1.11%. BAC end sequences from 1490 ends were generated and analyzed as a preliminary evaluation for using this library to develop an STC framework to sequence the tomato genome. A total of 1205 BAC end sequences (80.9%) were obtained, with an average length of 360 high-quality bases, and were searched against the GenBank database. Using a cutoff expectation value of <10−6, and combining the results from BLASTN, BLASTX, and TBLASTX searches, 24.3% of the BAC end sequences were similar to known sequences, of which almost half (48.7%) share sequence similarities to retrotransposons and 7% to known genes. Some of the transposable element sequences were the first reported in tomato, such as sequences similar to maize transposon Activator (Ac) ORF and tobacco pararetrovirus-like sequences. Interestingly, there were no BAC end sequences similar to the highly repeated TGRI and TGRII elements. However, the majority (70.3%) of STCs did not share significant sequence similarities to any sequences in GenBank at either the DNA or predicted protein levels, indicating that a large portion of the tomato genome is still unknown. Our data demonstrate that this BAC library is suitable for developing an STC database to sequence the tomato genome. The advantages of developing an STC framework for whole-genome sequencing of tomato are discussed.
[The BAC end sequences described in this paper have been deposited in the GenBank data library under accession nos. AQ367111–AQ368361.]
Tomato (Lycopersicon esculentum cv. Heinz 1706) is a vegetable crop that ranks second only to potatoes in economic importance (Gould 1992). Tomato is a member of the dicot family Solanaceae, which contains well-known plant species such as potato, tobacco, eggplant, and pepper. Despite differences in genome sizes among Solanaceae members, most, if not all, share the same basic chromosome number of 12 with tomato having the smallest haploid genome size of 953 Mb (Arumuganathan and Earle 1991). Comparative genetic studies revealed that tomato and potato share very conserved colinearity between their genomes (Bonierbale et al. 1988). Tomato has conserved gene repertoires with pepper even though their gene order is different (Tanksley et al. 1988). Among geneticists, tomato is considered an ideal model crop plant. Tomato has an excellent classical morphological map, a high-density molecular map containing >1000 markers (Rick and Yoder 1988; Tanksley et al. 1992; Broun and Tanksley 1996), and a large collection of well-characterized mutants and near-isogenic lines (NILs). In addition, tomato has a relatively small genome size, several large-insert YAC libraries (Martin et al. 1992; Bonnema et al. 1996), a transposon tagging system (Briza et al. 1995), as well as a routine Agrobacterium-mediated transformation system (McCormick et al. 1986). Furthermore, in 1998 several leading researchers in the tomato genomics community began to pursue the discovery and analysis of the majority of genes in tomato using primarily an EST-based approach (S.D. Tanksley, J.J. Giovannoni, G.B. Martin, J.C. Venter, unpubl.; http://www.nsf.gov; http://www.tigr.org). These publicly available resources and enabling technologies will undoubtedly provide an invaluable foundation for plant research, with an ultimate goal of sequencing and understanding the entire tomato genome.
A critical tool for genomic studies in tomato is the availability of deep-coverage large-insert genomic libraries, such as yeast artificial chromosomes (YACs) and bacterial artificial chromosomes (BACs), that can be used for physical mapping, positional cloning, and genome sequencing. The publicly available tomato YAC libraries have served as valuable research tools for the isolation of several agriculturally important genes by positional cloning (e.g., Martin et al. 1993; Alpert and Tanksley 1996). Unfortunately, YAC libraries and clones are cumbersome to construct, screen, and analyze, and DNA inserts are often chimeric (Green et al. 1991), rearranged, or have internal deletions. To date, no comprehensive L. esculentum BAC libraries suitable for whole-genome sequencing and physical mapping have been published. Except for DNA insert-size capacity, BAC libraries have several advantages over YACs in that they can be easily constructed and screened. Moreover, genomic inserts in BACs have been shown to be very stable in Escherichia coli and thus serve as ideal templates in generating whole-genome physical maps by DNA fingerprinting (Marra et al. 1997), developing sequence-tagged connectors (STCs) (Venter et al. 1996; Boysen et al. 1997), and shotgun sequencing (Boysen et al. 1997). These features make the BAC cloning system a popular choice for high-throughput genomics studies.
Because of recent technological advances and reduced costs in high-throughput genomic DNA sequencing, it may soon become practical and cost effective to sequence the genomes of a number of important crop plants with moderate genome sizes of ∼1000 Mb (e.g., tomato, sorghum, and soybean). The proposed STC strategy in combination with DNA fingerprinting can be used to establish sequence-ready genome frameworks using deep-coverage BAC libraries (Venter et al. 1996; Boysen et al. 1997). Briefly, each genomic insert from a deep-coverage BAC library is sequenced from both ends and fingerprinted to develop an STC and fingerprint databases, respectively. Simultaneously, a genetically anchored “seed” BAC is selected and shotgun sequenced. The complete sequence of the seed BAC is used to screen the STC database to obtain a set of BACs that overlap with the seed BAC based on DNA sequence. The fingerprint database is then consulted to confirm these overlaps and to select a BAC that overlaps minimally with the seed BAC. Incorporating a fingerprinting strategy has proven useful in selecting subsequent BACs for shotgun sequencing in Arabidopsis thaliana (Marra et al. 1999) and to help resolve conflicting data of library hybridization with anchored markers (Mozo et al. 1999). The STC and fingerprinting strategy has been widely adopted and is presently being used for the human, Arabidopsis, and rice genome sequencing projects.
In this study we report the construction and characterization of a comprehensive BAC library for the cultivated tomato L. esculentum cv. Heinz 1706 and analysis of 1205 BAC end sequences as an preliminary evaluation of the STC strategy for establishing a framework to sequence the tomato genome.
RESULTS
Tomato BAC Library Construction
A tomato BAC library was constructed from L. esculentum cv. Heinz 1706 using a HindIII partial digestion of megabase-size DNA embedded in agarose plugs. Several ligation reactions were performed using size-selected DNA fragments from three regions of CHEF gels of 100–150, 150–200, and 200–300 kb. Optimal ligations were obtained from partially restricted DNA in the region of 150–200 kb after a single size selection with a 20-sec constant pulse, 6 V/cm at 14°C for 18 hr. Two ligations yielded a significant number of white clones, averaging 1250 (80.6%) white clones, and 300 (19.4%) blue clones per microliter of ligation mixture as assayed on X-gal/IPTG Luria broth (LB) agar plates. Although the region of the size-selected DNA used for these ligations comigrated between 150 and 200 kb (based on the λ concatemer size markers run side by side on the same gel), the preliminary analysis revealed that the average insert size was between 115 kb and 120 kb. About 100 transformations were performed using the two ligations to produce a total of 129,024 clones. White clones were picked and arrayed into 336 microliter plates with 384 wells using the Genetix Q-Bot.
Insert Size Distribution and Genome Coverage
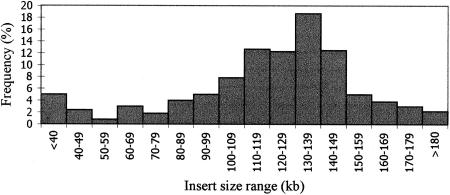
To estimate the genome coverage of the L. esculentum BAC library the average insert size of the clones, the number of nonrecombinant clones, and the number of chloroplast DNA-containing clones were determined. Inserts from 498 random BAC clones were sized by pulsed field gel electorphoresis (PFGE) on 1% agarose CHEF gels. Figure 1 shows an example of the analysis of 27 clones digested with NotI. Inserts of most of the clones did not contain internal NotI sites, which is a typical feature for dicot genomic DNA (Choi et al. 1995; Marek et al. 1997; Danesh et al. 1998; Tomkins et al. 1999). The size distribution of 498 random clones is depicted in Figure 2, which was used to calculate an average insert size of 117.5 kb. Of the clones, 78% have inserts >100 kb; ∼11% contained inserts <70 kb, indicating that there were a significant number of small inserts (<100 kb) trapped in the size-selected fraction used to construct the BAC library. In addition, ∼2.41% of clones did not contain inserts, suggesting the recircularization of vectors with damaged termini. To determine the percentage of BAC clones containing chloroplast DNA in the library, high-density membranes containing the entire library were probed with three chloroplast specific genes from barley (Hordeum vulgare), ndhA, rbcL, and psbA, which are evenly spaced 40 kb apart across the chloroplast genome. The results showed that 1432 positive clones, representing only 1.11% of the entire library, contain chloroplast DNA sequences (data not shown). Of the clones picked robotically, ∼2.11% were later found to be nonrecombinant (blue) by inoculating clones from the first forty 384-well plates of the library onto Q-plates containing X-gal and IPTG and counting the number of blue colonies (325 blue clones/15,360 clones). Taking 1.11% of chloroplast DNA, 2.11% of nonrecombinant clones, and 2.41% of white empty clones into consideration and assuming similar content of mitochondria to the tomato BAC libraries of Hamilton et al. (1999) (0.012%), with an average insert size of 117.5 kb and a haploid genome size of 953 Mb, the library is estimated to contain ∼15.0 haploid genome equivalents.
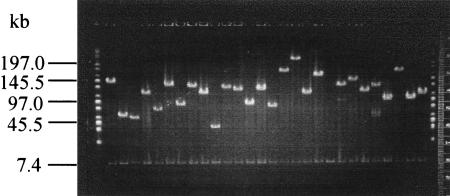
Figure 1.
Sizing of tomato BAC inserts. An ethidium bromide-stained gel of plasmid DNA digested with NotI enzyme to release the tomato genomic inserts from the 7.4-kb pBeloBAC11 vector. Lanes 1 and 30 contain λ mid-range PFGE size markers (New England Biolabs).
Figure 2.
Insert size distribution of the tomato BAC library based on the sizing of 498 random clones by PFGE. Of the clones in the library 78% contained inserts >100 kb.
To further examine the predicted genomic coverage of the BAC library constructed, 19 one- to four-copy restriction fragment length polymorphism (RFLP) markers from tomato chromosome 12 (Table 1) were used to screen 3 of the total 7 filters, which represent a calculated genome coverage of 6.43×. As summarized in Table 1 at least one clone was identified for each marker tested. Considering the approximate copy number of the RFLP markers in the genome (Table 1), an average of 3.33 positive hybridization signals was obtained per probe, which is lower than expected. This result suggests that either our estimation of genome coverage is an overestimate, the library is not entirely random, or our estimation of RFLP copy number is too high.
Table 1.
Screening Results of the Tomato BAC Library with Chromosome 12 RFLP Markers
| RFLP marker | Map position (cM) | Copy no. | No. of hits | Hit address |
|---|---|---|---|---|
| CT211a | 32.5 | 2 | 5 | 42M21, 42N13, 55M10, 70A6, 108O7 |
| TG360 | 40.2 | 1 | 3 | 10C21, 30P19, 75A23 |
| CT99 | 40.2 | 1 | 5 | 29N13, 30P19, 75A23, 92B2, 137L18 |
| TG618 | 41.0 | 1 | 12 | 6P6, 7N7, 39F18, 43A13, 49G18, 76N24, 85O19, 95O7, 97G2, 99B10, 123B1, 138D4 |
| CT223b | 41.8 | 2 | 5 | 7L6, 43A13, 108K6, 112F4, 138D4 |
| CD4 | 42.6 | 2 | 9 | 12A8, 15I17, 33A22, 47K14, 47K22, 106G5, 113E22, 113G20, 115C2 |
| CD22 | 42.6 | 1 | 9 | 13P13, 56M9, 79N7, 84L2, 92C18, 108F16, 112I1, 118G20, 143J10 |
| TG387 | 42.6 | 2 | 11 | 6P3, 42C5, 45G17, 62D10, 64N20, 65G5, 77E17, 82L18, 86H6, 58N11, 108N15 |
| CT91 | 42.6 | 4 | 5 | 3I4, 7L20, 11P15, 38P8, 77I7 |
| CT134 | 42.6 | 1 | 8 | 6C20, 19A3, 20B12, 63A2, 70F13, 94B8, 97C12, 135E3 |
| CT187 | 42.6 | 2–3 | 4 | 17G9, 62E19, 74J17, 132L22 |
| TG381 | 42.6 | 1 | 12 | 4H1, 4M13, 14N13, 16D3, 19J5, 37H17, 41O14, 46K15, 58K8, 60J21, 85M4, 118K10 |
| TG394 | 47.1 | 1 | 5 | 22D20, 25M21, 40O3, 4205, 60N12 |
| TG367 | 47.1 | 1 | 2 | 10B24, 116P8 |
| TG111 | 47.1 | 1 | 1 | 128E10 |
| CT184 | 53.8 | 2 | 5 | 9D7, 20J11, 24F2, 88M15, 131O1 |
| CT106 | 53.8 | 3 | 1 | 70J8 |
| CT239 | 67.8 | 1–2 | 1 | 142G13 |
| TG296 | 69.8 | 2 | 6 | 23P17, 36O4, 54K18, 68P24, 84N24, 95O4 |
BAC End Sequencing
To examine the feasibility of using a STC strategy to establish a framework to sequence the tomato genome, we sequenced and analyzed the ends of the first 745 clones of the BAC library. A total of 1490 sequencing reactions were performed resulting in a set of 1205 high-quality sequences (a success rate of 80.9%). High-quality sequences were defined as those having >75 high-quality bases other than vector and E. coli sequences. The average length of the end sequence, after removal of the vector sequence using CROSS-MATCH software, was 360 bases with a standard deviation of 142 bases.
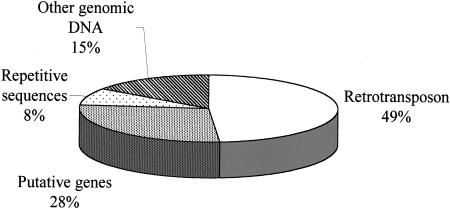
To determine the sequence composition of the BAC end sequences, they were searched against Genbank using BLASTN, BLASTX, and TBLASTX (Altschul et al. 1990). A probability cutoff value (E value) of at least 10−6 was used to assign putative identities to the STCs. The BLASTN search resulted in 194 sequences with E < 10−6, whereas BLASTX and TBLASTX resulted in 90 and 37 sequences, respectively. Fifteen STCs were found to contain chloroplast DNA making up 1.2% of the total high-quality BESs. This value is very close to the estimation made by hybridizing the library with probes from the chloroplast genome (1.11%, as described above). For the remaining 306 STCs sharing significant sequence similarities in Genbank, sequences similar to retrotransposons constitute the major component of each category. A total of 149 out of 306 STCs (48.7%) were found to be similar to various retrotransposons. The BLASTN search resulted in 78 out of 194 STCs that were retrotransposon-like sequences including both copia-like (51 STCs) and gypsy-like (27 STCs) retrotransposons. However, it appears that the sequences of gypsy-like retrotransposons are more conserved than copia-like retrotransposons as 51 out of 78 STCs similar to retrotransposons picked up by BLASTN search turned out to be gypsy-like retrotransposons. It is interesting that some of the previously determined repetitive sequences were found to be part of the retrotransposons. For example, 20 STCs were similar to a tomato random amplified polymorphic DNA sequence. A BLASTX search of this sequence (GenBank accession no. AJ223850) showed that it is to some extent similar to a Vicia faba retrotransposon (accession no. AB007467). The BLASTX database search resulted in 58 out of 59 sequences that are similar to the protein domains of copia-like retrotransposons, suggesting that tomato copia-like retrotransposons are more degenerated on the DNA level but still very conserved at the protein level. We were able to find 12 STCs that are similar to retrotransposon-like sequences in 37 STCs obtained from TBLASTX with E < 10−6. Four STCs were similar to a tobacco pararetrovirus sequences and another three STCs were similar to maize transposon Activator (Ac) open reading frame.
The number of putative genes detected by BLASTN, BLASTX, and TBLASTX is 85. This represents only one-quarter of those STCs with known sequence matches in GenBank (27.8%) and 7% of the high-quality tomato sequences. Some of the high-scoring sequences that share significant similarity with genes are summarized in Table 2.
Table 2.
Example of Similarity Search Results Using BLASTN, BLASTX, and TBLASTX
| Accession no. | Clone ID | E-value | Organism | Putative identity |
|---|---|---|---|---|
| BLASTN | ||||
| AQ367695 | toxb0002J16f | 5e-90 | tomato | 17S–25S ribosomal DNA spacer |
| AQ367792 | toxb0001C22f | 3e-70 | tomato | rbcS3A gene for ribulose 1,5-bisphosphate carboxylae/oxygenase small subunit |
| AQ368209 | toxb0001N20f | 5e-69 | tomato | 1-aminocyclopropane-1-carboxylate synthase (ACC4) gene |
| AQ368316 | toxb0002I05f | 6e-68 | tomato | gene for cell wall degrading polygalacturonase |
| AQ367218 | toxb0002C04f | 2e-55 | tomato | cytosolic Cu,Zn superoxide dismutase (Sod) gene and dehydroquinate |
| AQ368051 | toxb0001P17r | 1e-44 | tomato | LAT59 gene 5′–flanking region, expressed during pollen maturation |
| BLASTX | ||||
| AQ367638 | toxb0002D20f | 3.7-60 | L. pimpinellifolium | (AJ002236) Hcr9-9E |
| AQ367206 | toxb0002A12r | 3.7e-50 | tomato | (U65391) PRF |
| AQ367838 | toxb0001I12f | 1.9e-18 | tomato | (X95269) LRR protein |
| AQ368350 | toxb0002M01r | 8.1e-18 | tomato | probable lipid transfer protein precursor |
| TBLASTX | ||||
| AQ367838 | toxb0001I12f | 3e-17 | Capsicum annuum | (AF082727) leaf mRNA cDNA clone I23 similar to leucine-rich-repeat protein |
| AQ367265 | toxb0002G06r | 3e-27 | A. thaliana | ATTS0294 Gif cDNA clone YAP201T7 5′ similar to tomato protein P59 precursor |
| AQ367696 | toxb0002J16r | 4e-84 | Medicago truncatula | 00559 MtRHE cDNA 5′ similar to 26S rRNA |
| AQ367633 | toxb0002D14r | 8e-16 | Saccharum sp. | EST149 sugarcane leaf roll cDNA clone D64-rev similar to 5′ end |
Twenty-five STCs were similar to repetitive sequences from various plants, including 21 STCs that are similar to a tomato microsatellite repeat, a tomato sequence containing a GATA microsatellite, and a high-copy repeat (GenBank accession no. X90770).
Finally, 41 (8%) sequences are associated with genes and have E < 10−6 but are neither part of gene coding sequences nor can be classified into any categories described above.
The results described above for the 306 STCs sharing significant similarity with sequences deposited in GenBank are summarized in Figure 3 , and the complete BLAST search statistics for the 1490 ends can be viewed in detail at the Clemson University Genomics Institute (CUGI) web site (http://www.genome.clemson.edu).
Figure 3.
The distribution of 306 STCs originated from tomato genomic DNA based on putative identification with an E < 10−6.
DISCUSSION
The study of tomato has a rich breeding history over the centuries and recently has fostered pioneering research in genomics including the development of high-density molecular genetic maps (Tanksley et al. 1992), comparative molecular genetic maps between closely related species (e.g., potato, pepper, and tobacco; Bonierbale et al. 1988; Tanksley et al. 1988), plant transformation (McCormick et al. 1986), the molecular dissection of quantitative inheritance (Paterson et al. 1988), and the positional cloning of disease resistance (Martin et al. 1993) and fruit-ripening genes (Wilkinson et al. 1995). Although the soon-to-be sequenced A. thaliana (120-Mb genome) is touted as the model flowering plant to study plant biology, tomato continues to play a pivotal and leading role as a model crop plant. Because of this foundation and popularity and a haploid genome size of 953 Mb—by far the smallest among the Solanacae family (e.g., potato, pepper, and tobacco)—it can be argued that tomato is an ideal candidate for generation of a complete genome sequence.
As a prelude to establish a STC framework of the tomato genome we constructed and characterized a 15.0 haploid genome equivalent BAC library and sequenced and analyzed the insert ends of the first 745 clones in the library.
The BAC library was constructed from the cultivated tomato L. esculentum cv. Heinz 1706 and contains 129,024 clones with an average insert size of 117.5 kb with a low chloroplast DNA contamination (1.11%). Heinz 1706 was chosen because it has been used as the recurrent parent for generating many near-isogenic lines (Philouze 1991) and is the L. esculentum parent of an interspecific cross that is being used to map the jointless-2 locus (Zhang et al. 1999) for positional cloning in our laboratory. In comparison to the previously described tomato YAC libraries (Martin et al. 1992; Bonnema et al. 1996) and BAC libraries (Hamilton et al. 1999), the genomic coverage of this BAC library far exceeds both tomato YAC and BAC libraries (5.5×, Martin et al. 1992; Bonnema et al. 1996; 4.6×, Hamilton et al. 1999; 3×). It should be noted that the genome coverage estimates for all of these libraries, including the library described here, were calculated similarly. This result makes the Heinz 1706 library the deepest coverage large-insert tomato library available to date. Although YAC libraries usually have larger average insert sizes (130 kb, Martin et al. 1992; 250 kb, Bonnema et al. 1996), the depth of the BAC library can compensate for its relatively smaller average insert size. Analysis of chloroplast DNA content showed that high-molecular-weight DNAs prepared from tomato nuclei contained less chloroplast DNA than those derived from protoplasts, such as has been shown for other BAC libraries using similar nuclei extraction methods (Choi et al. 1995; Zhang et al. 1995; Hamilton et al. 1999; M. Budiman and R. Wing, unpubl.). For example, BAC and YAC libraries produced from megabase-size DNA from rice (Nakamura et al. 1997), sorghum (Woo et al. 1994), and tomato (Martin et al. 1992) protoplasts had chloroplast DNA contamination of 7%, 14%, and 10%, respectively. Thus, the development of a 15-fold BAC library for tomato should facilitate physical mapping, positional cloning of many agronomically important genes and genomic regions, and whole-genome sequencing.
The primary purpose of the STC strategy is to generate frameworks for genome assembly and sequencing by providing informative DNA sequence for the precise selection of minimally overlapping BAC clones for subsequent sequencing substrates. This procedure was performed previously through end-sequence walking or PCR-based screening using sequence-tagged sites (STSs) on cosmid clones (Venter et al. 1996). The STC approach and derivatives incorporating BAC fingerprinting (Boysen et al. 1997) are now practiced widely and are being used to assemble the genomes of human, mouse, Drosophila, Arabidopsis, and rice. In our effort to develop a similar STC database for tomato, BAC insert ends from 745 clones were sequenced as a preliminary study for the feasibility of using this tomato library for whole-genome sequencing. Among 1205 high-quality BAC end sequences, 94.6% are from tomato nuclear DNA. Analysis of these STCs using the statistically significant E ≤ 10−6 revealed that 26.6% of the STCs have significant matches in GenBank, of which nearly half were transposable elements and other repetitive sequence. STCs with putative functions or similar to ESTs with unknown functions make up 24.3% of all of the STCs that have a match in GenBank. Of the total STCs analyzed 70.3% appear to represent previously uncharacterized or unique sequences. This fraction demonstrates that a significant portion of the tomato genome remains unknown. The major repetitive sequences are retrotransposons and may be distributed throughout the genome. Surprisingly, we did not identify any BAC end sequences sharing significant similarities with the well-characterized and highly repetitive sequences TGRI and TGRII, which comprise ∼1.85% of the tomato genome. This result is likely due to the fact that both TGRI and TGRII are mainly clustered tandemly in telomeric and centromeric regions (Zamir and Tanksley 1988), which might be difficult to clone, though there are four STCs (e.g., toxb0001cE11f) that are similar to Lycopersicon pennellii paracentromeric sequence (GenBank accession no. AF07252).
By extrapolating the sequence and library characterization analysis above, if the insert ends of the entire BAC library of 121,744 clones (243,488 ends) were sequenced, there would be, on average, one STC every 3.73 kb DNA across the tomato genome. With a moderate DNA sequencing success rate (∼80%) and an average of 360 high-quality bases per sequence, the STC database would represent 7.4% of the tomato genome. Such an STC database will no doubt provide an indispensable tool for the generation of a complete tomato genome sequence in the future. In addition, even during the early stages of developing such a database, end sequences and contig information from fingerprinting together with integrated genetic markers will facilitate local physical mapping, gene isolation, and gene discovery. Finally, an STC database will provide a vision of the organization of the tomato genome before the entire genome sequence becomes known. For example, assuming that 7% of the STCs represent putative genes, based on BLASTX and TBLASTX similarity searching, sequencing the entire tomato BAC library could produce the partial sequence of 17,044 genes.
METHODS
BAC Vector Isolation
pBeloBAC 11 DNA was isolated using an alkaline lysis method (Sambrook et al. 1989). Vector DNA was purified by two rounds of CsCl density gradient centrifugation, completely linearized with HindIII (New England Biolabs), and dephosphorylated with heat-killable thermolabile phosphatase (Epicenter Technologies). The extent of digestion, dephosphorylation, and integrity of vector DNA were assayed by comparing several ligation reactions with HindIII-cut λ DNA (New England Biolabs) on agarose gels and tested by transformation into E. coli DH10B (Research Genetics).
Preparation of Partially Digested DNA of Tomato
High-molecular-weight DNA was prepared from young leaf nuclei of L. esculentum cv. Heinz 1706 grown under greenhouse conditions. Leaf tissue (30 gm) was harvested and kept at −80°C or used directly. Nuclei were extracted by grinding the tissue to a smooth powder in liquid nitrogen and processed (De Scenzo and Wise 1996). The nuclei were embedded in 1% low-melting agarose plugs (Ganal and Tanksley 1989). The use of nuclei preparation, instead of the protoplast preparation, should reduce the contribution of organellar DNA such as chloroplast and mitochondria genomes. Chopped plugs were serially digested with HindIII (0, 0.5, 1.0, 2.5, 5.0, and 50 units) in a total volume of 70 μl at 37°C for 20 min. After inactivating the restriction enzyme, partially digested fragments were separated by PFGE (CHEF DR II, Bio-Rad) and three DNA fractions of 100–150, 150–200, and 200–250 kb were excised. Gel pieces were washed three times with 1 ml of cold TE buffer on ice for 10 min each and stored at 4°C.
BAC Library Construction
DNA fragments in 100 mg of gel slices were electroeluted (Strong et al. 1997) and the concentrations of the eluant were assayed on an agarose gel. Ligation was performed in a final volume of 100 μl with 50–150 ng of eluted DNA and 20 ng of dephosphorylated linearized pBeloBAC11 vector. Before adding the T4 ligase (Promega), the ligation mixture was incubated at 55°C for 10 min and then cooled down to room temperature. Ligation was performed at 16°C overnight. One microliter of desalted ligation mixture was transformed with 20 μl of E. coli DH10B (Research Genetics) using a BRL electroporator with the following settings: 320 V, 330 μF capacitance, low ohms impedance, fast charge rate, and 4000 Ω voltage booster resistance. Transformed cells were spread onto LB media containing 12.5 μg of chloramphenicol, X-gal, and IPTG and grown at 37°C for 18 hr. The plates were then transferred to the dark and stored at room temperature for an additional 1–2 days to allow the nonrecombinant colonies to turn dark blue, thus making automated colony picking with the Genetix Q-Bot (Genetix LTD) more efficient (∼1500–2000 colonies per 500-cm2 plate). White clones were picked and stored in 384-well microtiter master plates containing LB freezing media [36 mm K2HPO4, 13.2 mm KH2PO4, 1.7 mm sodium citrate, 0.4 mm MgSO4, 6.8 mm (NH4)2SO4, 4.4% glycerol (vol/vol), 12.5 μg/ml chloramphenicol].
BAC Insert Size Analysis
BAC DNA was prepared by a standard alkaline lysis method (Sambrook et al. 1989) from a 3-ml overnight culture using the Autogen 740 automated DNA isolation system (Integrated Separation System). DNA was digested with NotI (New England Biolabs) to completion and separated by PFGE (CHEF DR III, Bio-Rad) on a 1% agarose gel in 0.5× TBE with a linear pulse from 5 to 15 sec for 14 hr at 14°C along with a mid-range PFGE marker I (New England Biolabs).
High Density Filter Production and Hybridization
The entire library containing 129,024 clones was gridded onto seven 22.5 × 22.5-cm nylon filters (Amersham N+) using the Genetix Q-Bot (Genetix Ltd.). Each filter contained 18,432 individual clones that were doubly spotted. The high-density hybridization filters were hybridized as described (Church and Gilbert 1984) except that BSA was omitted. Gel-purified DNA fragments were labeled by random priming (Feinberg and Vogelstein 1984) with [32P] dCTP (NEN). After hybridization, the filters were washed at 65°C twice in 1.0× SSC, 0.1% SDS, and twice in 0.5× SSC, 0.1% SDS, for 20 min each time and exposed to X-ray film (Kodak X-Omat). Three barley chloroplast-specific probes were obtained from Dr. J. Mullet (Texas A&M University, College Station): ndhA of plasmid pBHP20, rbcL of pBPH134, and psbA of pBHE319. Tomato chromosome 12 RFLP markers were obtained from S. Tanksley (Cornell University, Ithaca, NY).
BAC End Sequencing and Bioinformatics
Four microliters of BAC culture in LB freezing media was inoculated into 4 ml of LB media containing chloramphenicol and incubated for 20 hr at 37°C. BAC DNA was isolated using the Autogen 740. DNA pellets were resuspended in 25 μl of 1 mm Tris-HCl (pH 7.5), and 20 μl was used as the template for sequencing reactions in a total volume of 30 μl [5 μl of ABI Big Dye (Perkin Elmer), 50 pmoles of primer, 1.75 μl of sequencing buffer containing 800 mm Tris-HCl (pH 9.0), 20 mm MgCl2, 2.25 μl dH2O]. PCR reactions were performed as follows: one cycle for 4 min at 95°C; 2) 70 cycles of 15 sec at 95°C, followed by 4 min at 60°C and 10 sec at 51°C. PCR products were precipitated with ethanol containing one-third volume of 7.5 m NH4OAc and run on ABI377 automatic sequencers. Base-calling was performed automatically by PHRED (Ewing and Green 1998), and vector sequences were removed by CROSS-MATCH (http://www. genome.washington.edu). High-quality BAC end sequences (defined as those having >75 nonvector bases with PHRED-quality value >20) were used as queries in BLASTX and BLASTN (Altschul et al. 1990) searches of 77,977 protein sequences of SwissProt version 37 (Bairoch and Apweiler 1999) and a subset of 46,221 plant DNA sequences from GenBank version 111 (Benson et al. 1999). All software was run locally on a Sun Ultra30 workstation running Solaris 2.6.
Acknowledgments
We thank Dr. David Frisch, Edward Bishop, and Michael Atkins of the CUGI BAC Resource Center for their help in using the Q-bot for robotic picking and filter gridding. We also thank Yeisoo Yu for his BAC end sequencing protocol and Maciek Sasinowski and Robert Kingsbury III for their bioinformatics assistance. This work was supported by the U.S. Department of Agriculture (NRICGP grant no. 9701388), the National Science Foundation (NSF) MRI grant no. 9724557 and NSF Plant Genome grant no. DBI-987276), and the Coker Endowed Chair to R.A.W.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rwing@clemson.edu: FAX (864) 656-4293.
REFERENCES
- Altschul SF, Gish W, Miller W, Myiers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Alpert KB, Tanksley SD. High-resolution mapping and isolation of a yeast artificial chromosome contig containing fw2.2: A major fruit weight quantitative trait locus in tomato. Proc Natl Acad Sci. 1996;93:15503–15507. doi: 10.1073/pnas.93.26.15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 1999. GenBank. Nucleic Acids Res. 1999;27:49–54. doi: 10.1093/nar/27.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Boguski MS, Lipman DJ, Ostell J, Ouellette BF, Rapp BA, Wheeler DL. Nucleic Acids Res. 1999;27:12–17. doi: 10.1093/nar/27.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonierbale MW, Plaisted RL, Tanksley SD. RFLP maps based on a Common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics. 1988;120:1095–1103. doi: 10.1093/genetics/120.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnema G, Hontelelz J, Verkerk R, Zhang YQ, van Daelen R, van Kammen A, Zabel P. An improved method of partially digesting plant megabase DNA suitable for YAC cloning: Application to the construction of a 5.5 genome equivalent YAC library of tomato. Plant J. 1996;9:125–133. doi: 10.1046/j.1365-313x.1996.09010125.x. [DOI] [PubMed] [Google Scholar]
- Boysen C, Simon ML, Hood L. Analysis of the 1.1-Mb human a/d T-cell receptor locus with bacterial artificial chromosome clones. Genome Res. 1997;7:330–338. doi: 10.1101/gr.7.4.330. [DOI] [PubMed] [Google Scholar]
- Briza J, Carroll BJ, Klimyuk VI, Thomas CM, Jones DA, Jones JDG. Distribution of unlinked transposition of a Ds element from a T-DNA locus on tomato chromosome 4. Genetics. 1995;141:383–390. doi: 10.1093/genetics/141.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P, Tanksley SD. Characterization and genetic mapping of simple repeat sequences in the tomato genome. Mol & Gen Genet. 1996;250:39–49. doi: 10.1007/BF02191823. [DOI] [PubMed] [Google Scholar]
- Choi SD, Creelman R, Mullet J, Wing RA. Construction and characterization of a bacterial artificial chromosome library from Arabidopsis thaliana. Weeds World. 1995;2:17–20. [Google Scholar]
- Church G, Gilbert W. Genomic sequencing. Proc Natl Acad Sci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesh D, Peneula S, Mudge J, Denny RL, Nordstrom H, Martinez JP, Young ND. A bacterial artificial chromosome library for soybean and identification of clones near a major cyst nematode resistance gene. Theor Appl Genet. 1998;96:196–202. [Google Scholar]
- De Scenzo RA, Wise RP. Variation in the ratio of physical to genetic distance in intervals adjacent to the Mla locus on barley chromosome 1H. Mol & Gen Genet. 1996;251:472–482. doi: 10.1007/BF02172376. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein SA. A technique for radio-labeling DNA fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Ganal MW, Lapitan NLV, Tanksley SD. A molecular and cytogenetics survey of major repeated DNA sequences in tomato. Mol & Gen Genet. 1988;213:262–268. [Google Scholar]
- Ganal MW, Tanksley SD. Analysis of tomato DNA by pulsed filed gel electrophoresis. Plant Mol Biol. 1989;7:17–28. [Google Scholar]
- Gould WA. Tomato production, processing and technology. 3rd ed. Baltimore, MD: CTI Publications; 1992. [Google Scholar]
- Green ED, Riethman HC, Dutchik JE, Olson MV. Detection and characterization of chimeric yeast artificial-chromosome clones. Genomics. 1991;11:658–669. doi: 10.1016/0888-7543(91)90073-n. [DOI] [PubMed] [Google Scholar]
- Hamilton CM, Frary A, Xu Y, Tanksley SD, Zhang H-B. Construction of tomato genomic DNA libraries in a binary BAC (BIBAC) vector. Plant J. 1999;18:223–229. [Google Scholar]
- Marek FL, Shoemaker RC. BAC contig development by fingerprint analysis in soybean. Genomics. 1997;40:420–427. doi: 10.1139/g97-056. [DOI] [PubMed] [Google Scholar]
- Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein D, Wilson RK, McDonald KM, Hillier LW, McPherson DL, Waterston RH. High throughput fingerprint analysis of large-insert clones. Genome Res. 1997;7:1072–1082. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M, Kucaba T, Sekhon M, Hillier L, Martienssen R, Chinwalla A, Crockett I, Fedele J, Grover H, Gund C, McCombie WR, McDonald K, McPherson J, Mudd N, Parnell L, Schein J, Seim R, Shelby P, Waterston R, Wilson R. ZA map for sequence analysis of the Arabidopsis thaliana genome. Nat Genet. 1999;22:265–270. doi: 10.1038/10327. [DOI] [PubMed] [Google Scholar]
- Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- Martin GB, Ganal MW, Tanksley SD. Construction of a yeast artificial chromosome library of tomato and identification of cloned segments linked to two disease resistance loci. Mol & Gen Genet. 1992;233:25–32. doi: 10.1007/BF00587557. [DOI] [PubMed] [Google Scholar]
- McCormick S, Niedermeyer J, Fry J, Barnason A, Horsch R, Fraley R. Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Reports. 1986;5:81–84. doi: 10.1007/BF00269239. [DOI] [PubMed] [Google Scholar]
- Mozo T, Dewar K, Dunn P, Ecker JR, Fischer S, Kloska S, Lehrach H, Marra M, Martienssen R, Meier-Ewert S, Altmann T. A complete BAC-based physical map of the Arabidopsis thaliana genome. Nat Genet. 1999;22:271–275. doi: 10.1038/10334. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Asakawa S, Ohmido N, Fukui K, Shimizu N, Kawasaki S. Construction of an 800-kb contig in the near-centromeric region of the rice blast resistance gene Pi-ta2 using a highly representative rice BAC library. Mol & Gen Genet. 1997;254:611–620. doi: 10.1007/s004380050459. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Lander ES, Hewitt JD, Peterson S, Lincoln SE, Tanksley SD. Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature. 1988;335:721–726. doi: 10.1038/335721a0. [DOI] [PubMed] [Google Scholar]
- Philouze J. Description of isogenic lines, except for one, or two, monogenically controlled morphological traits in tomato, Lycopersicon esculentum Mill. Euphytica. 1991;56:121–131. [Google Scholar]
- Rick CM, Yoder JT. Classical and molecular genetics of tomato: Highlights and perspectives. Annu Rev Genet. 1988;22:281–300. doi: 10.1146/annurev.ge.22.120188.001433. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Strong SJ, Ohta Y, Litman GW, Amemiya CT. Marked improvement of PAC and BAC cloning is achieved using electroelution of pulsed-field gel-separated partial digests of genomic DNA. Nucleic Acids Res. 1997;25:3959–3961. doi: 10.1093/nar/25.19.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, Bernatzky R, Lapitan NJ, Prince JP. Conservation of gene repertoire but not gene order in pepper and tomato. Proc Natl Acad Sci. 1988;85:6419–6423. doi: 10.1073/pnas.85.17.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, Ganal MW, Prince JP, de Vicente MC, Bonierbale MW, Brown P, Fulton TM, Giovanonni JJ, Grandillo S, Martin GB, et al. High density molecular linkage maps of the tomato and potato genomes. Genetics. 1992;132:1141–1160. doi: 10.1093/genetics/132.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkins JP, Mahalingam R, Smith H, Goicoechea JL, Knap HT, Wing RA. A bacterial artificial chromosome library for soybean PI 437654 and identification of clones associated with cyst nematode resistance. Plant Mol Biol. 1999;41:2532. doi: 10.1023/a:1006277417789. [DOI] [PubMed] [Google Scholar]
- Venter JC, Smith HO, Hood L. A new strategy for genome sequencing. Nature. 1996;381:364–366. doi: 10.1038/381364a0. [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ. An ethylene inducible component of signal transduction encoded by never-ripe. Science. 1995;270:1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- Woo S-S, Jiang J, Gill BS, Paterson AH, Wing RA. Construction and characterization of a bacterial artificial chromosome library of Sorghum bicolor. Nucleic Acids Res. 1994;22:4922–4931. doi: 10.1093/nar/22.23.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir D, Tanksley SD. Tomato genome is comprised largely of fast-evolving, low copy-number sequences. Mol & Gen Genet. 1988;213:254–261. [Google Scholar]
- Zhang HB, Choi SD, Woo S-S, Li Z, Wing RA. Construction and characterization of two rice bacterial artificial chromosome libraries from the parents of a permanent recombinant inbred mapping population. Mol Breeding. 1996;2:11–24. [Google Scholar]
- Zhang, H.B., M.A. Budiman, and R.A. Wing. 1999. Genetic mapping of jointless-2 to tomato chromosome 12 using RFLP and RAPD markers. Theor. Appl. Genet. (in press). [DOI] [PubMed]