Abstract
Background
RNA interference (RNAi) is a powerful technique for functional genomics research in insects. Transgenic plants producing double-stranded RNA (dsRNA) directed against insect genes have been reported for lepidopteran and coleopteran insects, showing potential for field-level control of insect pests, but this has not been reported for other insect orders.
Methodology/Principal Findings
The Hemipteran insect brown planthopper (Nilaparvata lugens Stål) is a typical phloem sap feeder specific to rice (Oryza sativa L.). To analyze the potential of exploiting RNAi-mediated effects in this insect, we identified genes (Nlsid-1 and Nlaub) encoding proteins that might be involved in the RNAi pathway in N. lugens. Both genes are expressed ubiquitously in nymphs and adult insects. Three genes (the hexose transporter gene NlHT1, the carboxypeptidase gene Nlcar and the trypsin-like serine protease gene Nltry) that are highly expressed in the N. lugens midgut were isolated and used to develop dsRNA constructs for transforming rice. RNA blot analysis showed that the dsRNAs were transcribed and some of them were processed to siRNAs in the transgenic lines. When nymphs were fed on rice plants expressing dsRNA, levels of transcripts of the targeted genes in the midgut were reduced; however, lethal phenotypic effects after dsRNA feeding were not observed.
Conclusions
Our study shows that genes for the RNAi pathway (Nlsid-1 and Nlaub) are present in N. lugens. When insects were fed on rice plant materials expressing dsRNAs, RNA interference was triggered and the target genes transcript levels were suppressed. The gene knockdown technique described here may prove to be a valuable tool for further investigations in N. lugens. The results demonstrate the potential of dsRNA-mediated RNAi for field-level control of planthoppers, but appropriate target genes must be selected when designing the dsRNA-transgenic plants.
Introduction
RNA interference (RNAi), first characterized in Caenorhabditis elegans [1], has been developed as an effective gene-silencing tool in a wide variety of organisms [2], and double-stranded RNA (dsRNA) mediated RNAi has emerged as one of the most powerful strategies for the rapid analysis of gene function, particularly in organisms for which stable transgenesis is not available, such as insects [3]. dsRNA-mediated gene-silencing is a conserved mechanism in many eukaryotes [4], in which Dicer RNase III type enzymes bind and digest cytoplasmic dsRNAs into small interfering RNAs (siRNAs), duplexes composed of approximately 21 to 23 dsRNA nucleotides. These small RNA cleavage products then function as sequence-specific interfering RNA in transcript turnover, cleavage, and translational control [5]. Gene knockdown via dsRNA has been successfully demonstrated in several insect orders, including Diptera [6], Coleoptera [7], Hymenoptera [8], Orthoptera [9], Blattodea [10], Lepidoptera [11], [12] and Isoptera [13], and has been regularly applied in entomology to investigate RNAi mechanisms [14], and the function [15], expression and regulation [16] of gene cascades. RNAi might therefore serve as a new technique for the control of insect pests in agriculture. Nevertheless, the majority of these experiments have been carried out through dsRNA injection directly into the organisms, which is not practicable for insect pest control in the field.
For RNAi of a target gene by dsRNA to be used as an effective means of insect control, the targeted insects will have to take up dsRNA from the environment. The body of an insect is covered by a chitin exoskeleton, while the midgut of most insects is lined by the peritrophic membrane (PM), or, in Hemipterans [17], the perimicrovillar membrane (PMM). Hence, the midgut is the only portion of an insect's body that has an active interface with the physical environment. The cells of the midgut, which are responsible for nutrient absorption from the gut lumen, can take up dsRNA, and are the route through which RNAi effects would be achieved in insects [18]. In animals, the best-studied uptake mechanism of dsRNA is that of C. elegans. Studies with systemic RNAi defective mutants (sid) have resulted in the description of two proteins involved in non-cell autonomous RNAi. SID-1 is a multispan transmembrane protein that is expressed on the cell surface, thereby mediating a systemic RNAi effect [19]. It probably functions as a multimer, in cooperation and/or coordination with SID-2, transporting dsRNA passively into the C. elegans cells. Homologs of sid-1-like genes have been identified in some insect species such as Tribolium castaneum, Bombyx mori and Apis mellifera [3], and recently in aphids [20]. However, in the best characterized model insect, Drosophila melanogaster, no sid-1-like gene has been found.
Target regulation by siRNAs is mediated by the RNA-induced silencing complex (RISC). RISC-associated Argonaute (Ago) proteins play an essential role in mediating distinct assembly and cleavage steps of the RNA interference catalytic cycle. The Ago protein, as the principal factor in the RISC exhibiting RNA ‘slicer’ activity, is a key player in the RNAi mechanism, effecting transcriptional repression and post-transcriptional control in animals [21]. The Argonaute protein family can be divided into two classes: the Ago subfamily and the Piwi subfamily. Members of both subfamilies are defined by the presence of four domains: the N terminal domain of variable length, the central PAZ (Piwi-Argonaute-Zwille) domain that is important for binding single-stranded small RNA 3′ ends without sequence specificity, the mid domain that participates in 5′ end binding to small RNAs and binding to the 7-methylguanine (m7G) cap of target mRNAs, and the RNase H-like C-terminal Piwi domain [22]. The Piwi domain of some Ago proteins performs a crucial role in RNA slicer activity that is responsible for mRNA degradation in siRNA- and miRNA-induced gene-silencing pathways [23]. Ago proteins have been identified in several insects, including D. melanogaster [24] and A. mellifera [25].
Systemic RNAi in plants is based on the RNA-dependent RNA polymerase (RdRp) [26] and the spread of siRNAs through the plasmodesmata [27]. RdRp orthologs are present in nematodes, but probably not in insects [28]. The absence of dsRNA amplification and RdRP in insects suggests that gene knockdown effects exhibited by feeding dsRNA to insects would be temporary. Thus, RNAi effects achieved in the gut would require a continuous input of high levels of dsRNA to persist. Production of adequate dsRNA in a transgenic plant and its delivery to the insect as food would resolve this technical problem [29].
Plant-mediated RNAi has been reported in lepidopteran and coleopteran plant pests [30], [31]. However, there is a more urgent need to develop this technique for application in insects for which no effective Bt (Bacillus thuringiensis) toxins are known. Insects in this category include phloem-sucking hemipteran pests, such as planthoppers, aphids and whitefly, which are highly destructive agricultural pests worldwide, causing millions of dollars' worth of yield loss and control costs [32].
The brown planthopper, Nilaparvata lugens Stål (Hemiptera: Delphacidae), is the most destructive insect pest of rice crops. The brown planthopper damages rice plants by directly sucking the phloem sap [33] and by acting as a vector for the transmission of the rice grassy stunt virus [34]. Although insecticide control of N. lugens has been a convenient option, indiscriminate usage has resulted in resistance, leading to a resurgence of the insect [35], besides creating serious environment pollution. Hence, genetic improvement of rice host resistance is a preferred alternative. Plant-mediated RNAi is a potential approach for controlling this insect pest of rice.
In this work, we cloned the sid-1 and Argonaute genes and verified their expression in N. lugens. These genes could play important roles in the RNAi pathway in this insect. We also isolated genes encoding a hexose transporter (NlHT1), a carboxypeptidase (Nlcar) and a trypsin-like serine protease (Nltry), which are highly expressed in the midgut of N. lugens. dsRNA constructs targeting these genes were developed and transformed into rice plants. The results demonstrate that expression of the target genes was significantly suppressed in insects that fed on the transgenic plants.
Results
Cloning and characterization of the sid-1 gene and the Argonaute gene in N. lugens
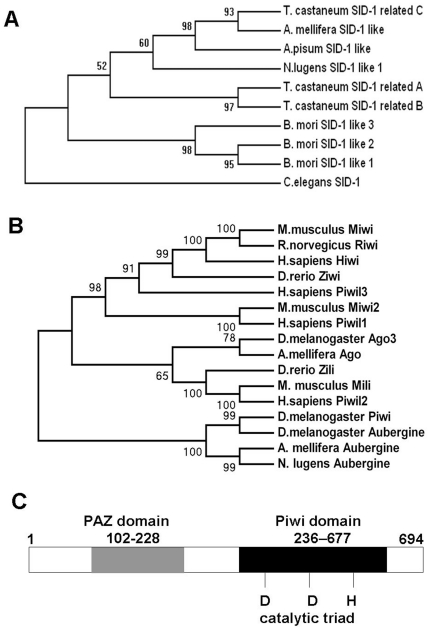
Because of the probable role of the SID protein in the uptake of dsRNA from the environment in C. elegans, we cloned the sid-1 gene from N. lugens. The full-length cDNA of the sid-1 gene is 2,119 bp long and contains an open reading frame (ORF) of 1,875 bp (GenBank accession no. JF915743), encoding a protein of 624 amino acids with a calculated molecular mass of 70.8 kDa and an isolectric point (pI) of 6.67 (Figure S1). Multiple alignment and phylogenetic analysis of the deduced amino acid sequences confirmed that this gene is a sid-1 like gene, hence we named it Nlsid-1 (Nilaparvata lugens sid-1). The phylogenetic tree of deduced amino acid sequences showed that the N. lugens SID-1 protein is most closely related to the A. mellifera SID-1 like protein (40% identity, Figure 1A).
Figure 1. Phylogenetic relationships of the SID and Argonaute proteins from N. lugens and other species, and the structure of the N. lugens Aubergine protein.
Phylogenetic analysis of (A) SID protein sequences, and (B) Argonaute protein sequences from N. lugens and other animals were conducted using MEGA 4.0; nodes with a bootstrap similarity of at least 50% are shown. (C) Conserved domains of Aubergine in N. lugens. It has the PAZ and Piwi domains that are typical motifs of Argonaute family proteins. The catalytic triad DDH is also shown. Sequence motifs were predicted by the NCBI Conserved Domains Server.
To determine the potential for RNAi effects in N. lugens, we first obtained a sequence that is part of an Argonaute gene by random screening of a midgut cDNA library of N. lugens, then cloned the full-length cDNA corresponding to this Argonaute gene (Figure S2). The full-length cDNA of this N. lugens Argonaute gene is 3,446 bp long, including a 2,085 bp ORF (GenBank accession no. JF915742). The deduced protein was composed of 694 amino acid residues, with a molecular mass of 79.27 kDa and an isolectric point (pI) of 9.27. We compared 16 genes of the Argonaute family, including those previously reported from Homo sapiens, Mus musculus, Rattus norvegicus, Danio rerio, D. melanogaster, Bombyx mori and A. mellifera. As expected, the Argonaute gene in the brown planthopper belonged to an insect cluster and was closely related to Aubergine from A. mellifera (Figure 1B). Based on the phylogenetic analysis, we named the N. lugens gene Nlaub (Nilaparvata lugens Aubergine). Sequence motif analysis using the NCBI Conserved Domains Server identified the presence of typical PAZ and Piwi domains (Figure 1C), which are signature motifs of the Argonaute family proteins [36]. The predicted PAZ domain of Nlaub was 127 aa long, located at positions 102 to 228, while the Piwi domain of this protein was identified at residues 236–677. Degenerate catalytic motifs of the type Asp-Asp-Asp/His/Glu/Lys (known as DDH motifs), essential for slicer-mediated cleavage, were also identified in this protein.
Analysis of the expression of Nlsid-1 and Nlaub in N. lugens
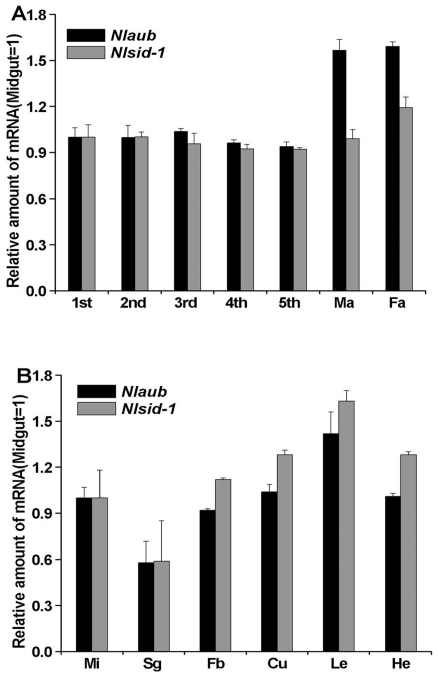
To probe the functions of the Nlsid-1 and Nlaub genes, their mRNA levels were analyzed, using qRT-PCR, at various developmental stages of N. lugens insects, including nymphs from 1st to 5th instars, female adults and male adults. The developmental expression pattern revealed that Nlsid-1 and Nlaub transcripts were present at all development stages. For both genes, mRNA levels were highest in female adults, but Nlaub mRNA had lower expression compared to that of Nlsid-1 (Figure 2A). In order to further assess the tissue distribution of the transcripts, qRT-PCR was used to amplify Nlsid-1 and Nlaub from cDNA of midgut, salivary gland, fat body, cuticle, leg, and head tissues. The results indicated that Nlsid-1 and Nlaub were expressed in all six tissues tested (Figure 2B).
Figure 2. Expression of Nlsid-1 and Nlaub in N. lugens.
(A) Developmental expression of Nlsid-1 and Nlaub in N. lugens from 1st nymph to male adult (Ma) and female adult (Fa). (B) Tissue distribution of Nlsid-1 and Nlaub in N. lugens 3rd instar nymph. qRT-PCR analyses were performed using total RNA from midgut (Mi), salivary gland (Sg), fat body (Fb), cuticle (Cu), leg (Le), and head (He). Data shown are means ± standard errors (N = 3).
Expression Profiles of NlHT1, Nltry, and Nlcar
Genes that are expressed in the N. lugens midgut were chosen as targets. Full length cDNA sequences of the transmembrane transporter gene NlHT1 [37], the carboxypeptidase gene Nlcar and the trypsin-like serine protease gene Nltry were isolated from the N. lugens cDNA library. The sequences of Nltry and Nlcar are shown in Figure S3 and S4. The cDNA sequence of Nltry (GenBank accession no. JF915745) and the cDNA sequence of Nlcar (GenBank accession no. JF915744) have been deposited in GenBank.
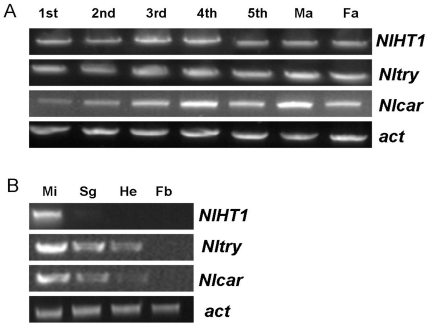
As shown in Figure 3, each gene is transcribed in all developmental stages from 1st to 5th instars, female adults and male adults. The three genes show primary expression in midgut tissues with limited transcription in salivary glands, fat body and head.
Figure 3. Transcription profiles of NlHT1, Nltry, and Nlcar at different developmental stages and in different tissues.
(A) Transcript levels of the three genes in N. lugens developmental stages from 1st nymph to male adult (Ma) and female adult (Fa). (B) Transcript levels in tissues of N. lugens 3rd instar nymphs: midgut (Mi), salivary gland (Sg), fat body (Fb), and head (He).
Development of dsRNA-transgenic rice plants
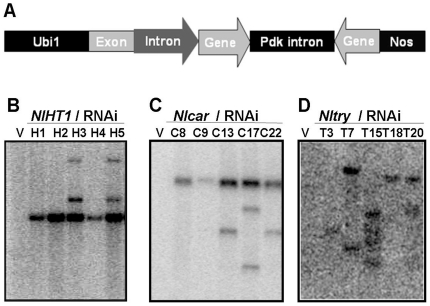
Double strand RNA interference (dsRNAi) constructs NlHT1-RNAi, Nlcar-RNAi and Nltry-RNAi were developed for the three N. lugens genes NlHT1, Nlcar and Nltry, under the control of the Ubi1 promoter and the nopaline synthase (nos) terminator cassette [38], [39]. The genes were cloned at the BamHI site of the hygromycin gene expression cassette in the pCU vector of Agrobacterium (Figure 4A). Transgenic plants were generated by introducing the constructs into the japonica rice variety Hejiang 19 by Agrobacterium tumefaciens–mediated transformation, and genomic DNA was isolated from the hygromycin-tolerant transgenic rice plants. Positive transformants were detected by PCR of the hygromycin resistance gene, while wild type (WT) plants failed to show such amplification (data not shown). No significant morphological differences were found in these transgenic lines compared with WT plants (Figure 5). Southern blot analysis showed that the PCR-positive plants had 1–3 copies of the target coding sequences (Figure 4B–4D). Conversely, genomic DNA from transgenic plants carrying an empty vector failed to show any hybridization with the probes. Two independently transformed lines (H2 and H4, C8 and C9, T3 and T18) with a single-copy-insertion were chosen for NlHT1-RNAi, Nlcar-RNAi and Nltry-RNAi,respectively, for further analyses. Resistance and non-resistance to hygromycin were observed in progenies of these plants in the ratio 3∶1 (Table S2), confirming integration of the respective transgenes at a single locus in each case.
Figure 4. Structure of constructs for rice transformation and Southern blot analyses.
(A) dsRNAi constructs (NlHT1-RNAi, Nlcar-RNAi and Nltry-RNAi) of target genes were developed under the control of the Ubi1 promoter and the nopaline synthase (Nos) terminator cassette. (B) Southern blot analysis of DraI-digested genomic DNA from leaves of NlHT1-RNAi-T0 transformants probed with part of the NlHT1 coding sequence (lane V, DNA from the empty transformation vector plant; lanes 1–5, DNA from the H1, H2, H3, H4 and H5 transgenic lines). (C) Southern blot analysis of DraI-digested genomic DNA from leaves of Nlcar- RNAi-T0 transformants probed with part of the Nlcar coding sequence (lane V, DNA from the empty transformation vector plant; lanes 1–5, DNA from the C8, C9, C13, C17 and C22 transgenic lines). (D) Southern blot analysis of DraI-digested genomic DNA from leaves of Nltry- RNAi-T0 transformants probed with part of the Nltry coding sequence (lane V, DNA from the empty transformation vector plant; lanes 1–5, DNA from the T3, T7, T15, T18 and T20 transgenic lines).
Figure 5. Growth phenotypes of wild type (WT) plants, empty transformation vector plants and transgenic lines.
(A) Two-week-old seedlings. (B) Plants at the four leaf stage. (C) Mature plants.
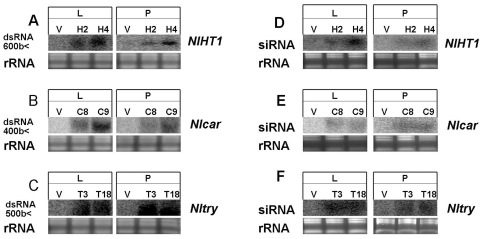
RNA blot analysis showed that the NlHT1, Nlcar and Nltry dsRNAs were transcribed in the transgenic lines and some of them were processed to siRNAs (Figure 6). The expression and processing of dsRNAs for the target N. lugens genes in the transgenic rice lines provided dsRNA and siRNA molecules for potential ingestion by the insects.
Figure 6. Northern blot analysis of the expression pattern of NlHT1, Nlcar and Nltry dsRNAs in different transgenic rice lines.
RNA blots for the expression of (A) NlHT1, (B) Nlcar and (C) Nltry dsRNA in T1 lines homozygous for the dsRNA transgenes (H2 and H4, C8 and C9, T3 and T18 respectively) and in the absence of dsRNA in the T1 line homozygous for the empty transformation vector homozygous T1 line (V): L, leaf; P, phloem sap. (D) NlHT1, (E) Nlcar and (F) Nltry siRNAs were detected in the three dsRNA transgenic lines, respectively. Loading of equal amounts of RNA was confirmed by ethidium bromide staining.
Suppression of target gene expression in N. lugens by ingestion of dsRNA- transgenic rice plants
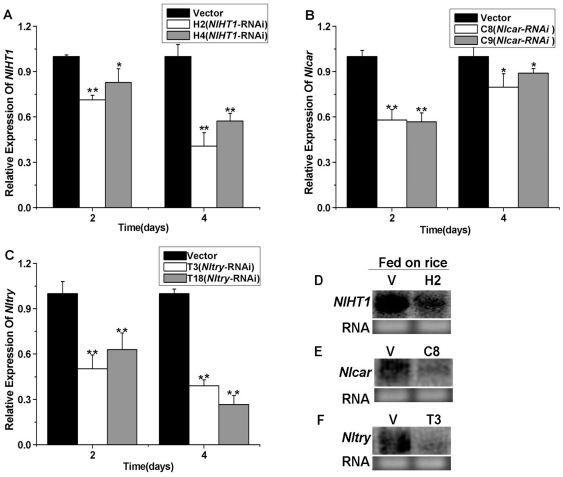
Third instar nymphs, previously reared on susceptible rice (cultivar Taichung Native 1, TN1) plants, were transferred onto the dsRNA-transgenic plants and allowed to feed for 2 to 4 days. The transcript levels of the target genes in N. lugens were then detected using qRT-PCR. In nymphs fed on the H2 and H4 lines, NlHT1 transcript levels began to decrease on day 2 and were reduced by 59.3% in H2 and 42.7% in H4 on day 4 (Figure 7A). A similar experiment was performed on the Nlcar and Nltry dsRNA-transgenic plants. The qRT-PCR results revealed that the mRNA transcripts of Nlcar and Nltry after ingestion of the respective dsRNAs were reduced on days 2 and 4. The maximum reductions in the Nlcar transcript level, of 42.1% for the C8 line and 43.3% for the C9 line, occurred on day 2 (Figure 7B). The maximum reductions in the Nltry transcript levels, of 61% for the T3 line and 73.3% for the T18 line, occurred on day 4 (Figure 7C). The northern blotting results confirm that the brown planthopper nymphs fed on H2 plants had less NlHT1 transcript compared to nymphs on the plants transformed with an empty transformation vector (Figure 7D). Similarly, the nymphs from fed on C8 and T3 plants had lower Nlcar and Nltry expression levels than those on the plants transformed with an empty transformation vector (Figure 7E–7F). Figure 7A–7C also shows that the RNAi efficiency for Nltry was significantly higher (p<0.01, t-test) than for NlHT1 and Nlcar on day 4. The results show that ingestion of dsRNA-transgenic rice plants expressing dsRNA and siRNA is an effective way to trigger RNA interference in the Hemipteran insect Nilaparvata lugens.
Figure 7. NlHT1, Nlcar and Nltry suppression in nymphs fed on transgenic plants.
(A) NlHT1 mRNA levels in the empty transformation vector and NlHT1-RNAi transgenic lines after feeding; nymphs were monitored by qRT-PCR after feeding on H2 and H4 phloem sap for 2 d and 4 d. (B) Nlcar mRNA levels in the empty transformation vector and Nlcar-RNAi transgenic lines after feeding; nymphs were monitored by qRT-PCR after feeding on C8 and C9 phloem sap for 2 d and 4 d. (C) Nltry mRNA levels in the empty transformation vector and Nltry-RNAi transgenic lines after feeding; nymphs were monitored by qRT-PCR after feeding on T3 and T18 phloem sap for 2 d and 4 d. Statistical analysis of mRNA levels was performed with student t-test.(*, P<0.05; **, P <0.01.) (D) Northern blot of NlHT1 transcripts of 3rd instar nymphs fed on the empty transformation vector and H2 transgenic plants for 4 d. (E) Northern blot of Nlcar transcripts of 3rd instar nymphs fed on the empty transformation vector and C8 transgenic line for 4 d. (F) Northern blot of Nltry transcripts of 3rd instar nymphs fed on the empty transformation vector and T3 transgenic line for 4 d.
We also assessed the performance of BPH on the transgenic plants(NlHT1-RNAi, Nlcar-RNAi and Nltry-RNAi and an empty transformation vector). The significant lethal phenotype was not observed (Figure S5).
Discussion
RNAi occurs widely in eukaryotic organisms and previous studies have demonstrated that feeding-based RNAi can specifically induce an RNAi response in several insect species, such as S. exigua, H. armigera and D. virgifera virgifera. In Lepidoptera, trends that are found are that RNAi is particularly successful in the family Saturniidae [12]. In this study, we showed that the target genes in the midgut of the hemipteran N. lugens can be suppressed by dsRNA transgenic plant-mediated RNA interference.
Identification of SID-1 and Argonaute genes in N. lugens
SID-1 is required for the uptake and spread of gene-silencing signals between tissues, thereby mediating a systemic RNAi effect in C. elegans [40], [41]. In this study, we have identified a sid-1 gene (Nlsid-1) in N. lugens. Sid-1-like genes have been found in many insects [3], and expression of these genes has been confirmed in Tribolium castaneum, Apis mellifera and Schistocerca americana [7], [42], [43]. In Hemiptera, a sid-1-like gene of the cotton aphid (Aphis gossypii) has been cloned and the topological structure of the SID-1 protein suggested a possible role in dsRNA uptake [20]. To determine whether SID-1 exists in N. lugens, we cloned a cDNA sequence of the sid-1 gene from the brown planthopper and qRT-PCR results showed that Nlsid-1 was expressed in various tissues of nymphs and adult insects. The deduced protein showed the closest relationship with the honey bee (Apis mellifera) SID-1 protein (40% identity). The honey bee SID-1, a putative transmembrane protein encoded by AmSid-1, is necessary for the uptake of systemically administered dsRNA and subsequent gene silencing [42]. Consistent with conservation and expression of the dsRNA channel protein gene sid-1 in grasshopper, systemic application of dsRNA leads to specific, long-term and phenotypically visible reduction of endogenous grasshopper v gene activity [43].Nlsid-1 may therefore perform an essential role in the uptake of dsRNA in the midgut of N. lugens and in any RNAi effect which is subsequently induced. Further research should establish whether there is a direct correlation between increased Nlsid-1 expression and the silencing effect in N. lugens.
Members of the Argonaute protein family have demonstrated activity in both transcriptional and post-transcriptional gene silencing. Ago proteins play important roles in RNAi because they can bind siRNAs as well as miRNAs and are involved in the suppression of specific target RNAs by either mRNA breakdown or inhibition of translation [44], [45]. In insects, the Ago and Piwi subfamilies both belong to the Argonaute protein family. Phylogenetic analysis indicated that the Piwi subfamily could be divided into three clusters, Piwi, Aubergine (Aub), and Ago3. Aubergine has been demonstrated to be required in RISC assembly in vitro [46] and in RNAi in oocytes in vivo [47]. In Drosophila melanogaster, the overexpression of AUB protein interferes with some crucial component or function of the RNAi pathway in somatic tissues [48]. Interestingly, AUB was thought to have a major role in silencing repetitive elements at the germ-tissue level [49]. In this study, we isolated an N. lugens Piwi subfamily gene Nlaub, which shares high identity (50%) with the A. mellifera Aubergine gene (GenBank accession no. ACV84377). The deduced amino acid sequence of Nlaub demonstrated the presence of the PAZ and Piwi domains. The Piwi domain is essential for slicing events in RNAi [36]. The presence of Nlaub gene transcripts in different developmental stages and tissues of N. lugens suggests a possible role in dsRNA-processing for the RNAi pathway in this insect.
Ingestion of dsRNA-transgenic rice plants induces target gene repression in N. lugens nymphs
Most experiments on RNAi in insects reported to date have involved injecting dsRNA directly into the body, although development of a robust dsRNA feeding technique is a prerequisite for application of RNAi to crop protection against insect pests. However, there have been some exceptions, including the following examples: Turner et al. [11] demonstrated RNAi effects after oral delivery of dsRNA to Epiphyas postvittana to silence a gut carboxylesterase gene (EposCXE1); only a local RNAi effect was required for repression. Zhou et al. found that knockdown of a gene encoding a caste-regulatory hexamerin storage protein could be achieved through voluntary feeding of high-dose dsRNA to Reticulitermes flavipes [13]. In contrast to these positive results, in an earlier study attempts to feed dsRNA to the lepidopteran Spodoptera litura to knockdown the midgut aminopeptidase-N gene in larvae did not generate an RNAi response [50]. Application of RNAi technology through transgenic plants to deliver dsRNA to herbivorous insects has only recently been realized in two chewing insects, cotton bollworm (Helicoverpa armigera; Lepidoptera) and western corn rootworm (WCR; Diabrotica virgifera virgifera; Coleoptera) [30], [31].
N. lugens feeds on plants by sucking phloem sap through its stylet mouthparts and causes substantial physiological damage to rice [51]. Sugars and amino acids are present in rice phloem sap at high concentrations. Some of the nutrients are absorbed into the planthopper body through the midgut cells, and the rest are excreted as honeydew [52]. Price et al. [37] reported that NlHT1, which is expressed at highest levels in the midgut, plays an important role in glucose transport from the gut, and in carbon nutrition in vivo. Digestion by serine proteases also plays an important role in N. lugens nutrition [53], since proteins are major components of plant phloem sap too; for example, cucurbit exudate contains high concentrations of proteins, up to 100 mg ml−1 [54]. The carboxypeptidase gene Nlcar, the trypsin-like serine protease gene Nltry and the NlHT1 gene are primarily expressed in the midgut of N. lugens (Figure 3). In this study, these three genes were selected for the development of dsRNA constructs which were transferred into rice plants. In the transgenic plants, some long dsRNAs were processed into siRNAs and both dsRNA and siRNA were present in the phloem sap (Figure 6). Ingestion of transgenic rice plants producing NlHT1, Nlcar and Nltry dsRNA was shown to suppress the expression of the three genes in N. lugens. Reduction of mRNA expression by 40% to 70% was observed (Figure 7), demonstrating the effectiveness of delivering dsRNA through the feeding of transgenic plants for the brown planthopper. Two research groups have demonstrated that artificial diet-feeding [55] and direct injection [56] of dsRNA can specifically induce an RNAi response in N. lugens. By injecting dsRNAs into N. lugens, Liu et al. [56] obtained a decrease of 25% to 40% of the expression of calreticulin, cathepsin-B and Nlß2 genes. The results reported in the present paper show that, in addition to injection and artificial diet-feeding, ingestion of dsRNA in host plants can induce inhibition of target gene expression in N. lugens.
Interestingly, although the transcription levels of the target genes were suppressed (Figure 7), we did not observe a significant lethal phenotype (Figure S5). In root knot nematode (RKN; Meloidogyne spp.), a splicing factor and integrase have been successfully down-regulated by transgenic plants expressing dsRNAs, resulting in almost complete resistance [57]. In another study, transgenic Arabidopsis expressing dsRNAs specific to genes encoding a nematode secretory peptide (16D10) that stimulates root growth successfully knocked down expression of genes in four major root knot nematodes, resulting in effective levels of resistance against the RKN species [58]. A further study has shown the feasibility of suppressing a root knot nematode transcription factor, MjTis11, in vivo through plant-expressed dsRNA. Those transgenic plants that induced the silencing of MjTis11, however, did not result in a lethal phenotype [59]. For N. lugens in our study, one possible reason for such a result is that the amount of dsRNA-uptake by the insects was not sufficient. Alternatively, the experimental system was not sufficient for maximum penetrance of the RNAi effects to cause a lethal phenotype. Another possible explanation is that the genes of interest may belong to multi-gene families in which other members may compensate for the suppression of one member [52], [60]. It is also possible that the target genes are not physiologically significant enough to induce mortality when silenced. The susceptibility of different target genes to RNAi in N. lugens shows considerable variation and further investigation is required. Although possible systems for dsRNA uptake and RNAi are present and target genes can be knocked down, many factors must be optimized in order to develop an RNAi approach for countering insect pest species such as N. lugens. This study has demonstrated the presence of a system for dsRNA uptake and the potential for knockdown of target gene expression by RNAi, but other factors to consider include the dsRNA fragment length, the relative susceptibilities of different insect developmental stages, and the nucleotide sequences selected as targets. It has been proved in C. elegans that RNAi effects are species-specific because knockdown experiments and identification of lethal phenotypes has not led to a universal set of ‘nematode target genes’ that are helpful for protection against plant parasitic nematodes [29]. Therefore, in order for the transgenic plant approach to be successful, appropriate target genes must be selected individually for each insect pest species. In addition, the specificity of ideal genes for inhibition in transgenic plants is an important consideration for the use of this technology in practical applications, since effects on non-target insects should be minimized.
Materials and Methods
Plant and Insect Material
The rice brown planthopper (BPH; Nilaparvata lugens) was obtained from rice fields in Zhejiang Province, China, and reared on two to three month old rice plants of the susceptible variety TN1, under controlled environmental conditions (70–80% r.h., 25±2°C, 16 h light/8 h dark). Four week old plants were used for the feeding experiments. For each feeding experiment, synchronous nymphs were selected and divided into groups, each group containing 30–40 individuals. After feeding on transgenic plants for the number of days indicated in the figures, midguts of nymphs were taken for further analysis.
Cloning of N. lugens genes
A cDNA library of N. lugens was constructed with RNA isolated from dissected midguts using the Creator SMART cDNA Library Construction Kit (CLONTECH) according to the manufacturer's recommendations. The cDNA was size-fractionated to remove fragments of <500 bp, directionally ligated into Lambda TriplEx2 vectors (BD-Clontech) and packaged into lambda phages (Stratagene). Individual clones were randomly selected and sequenced over 5′ and 3′ ends. Sequences were annotated by BLAST similarity searches against the GenBank database. Genes with similarity to the carboxypeptidase gene Nlcar, the trypsin-like serine protease gene Nltry and the Argonaute gene Nlaub were identified and the entire coding sequence was reamplified by PCR from N. lugens midgut-specific first-strand cDNA. For the sid-1 like gene, a TBLASTN search of the BPH expressed sequence tags (ESTs) database, using the Acyrthosiphon pisum similar to SID1-like protein [GenBank: XP_001951907] as query, revealed one SID1-like EST [GenBank: S_NLNA6932].
To obtain the full-length sequence of each truncated sequence from N. lugens, 5′ and 3′ RACE amplifications were performed using a 5′-Full RACE Kit and 3′-Full RACE Core Set Ver. 2.0 (TaKaRa) following the manufacturer's instructions. For 5′-RACE, gene-specific primers were designed based on sequencing, and an external reverse and a nested primer (Table S1) were used. For 3′-RACE, the cDNA was then amplified by nested PCR with the external forward primer and nested forward primer (Table S1) with ExTaq DNA polymerase (TaKaRa). The first round PCR cycling conditions were: 94°C for 3 min, followed by 20 cycles of 94°C for 30 sec, annealing at 55°C for 30 sec, and 72°C for 2 min, then an additional final elongation step of 72°C for 10 min. Nested PCR amplification was performed under the same conditions for 25 cycles. Purified PCR products were ligated to pGEM®-T Easy Vector Systems (Promega) and 3 to 5 positive colonies were sequenced.
Vector construction and plant transformation
For the RNA interference (RNAi) vectors of NlHT1, Nlcar and Nltry, DNA fragments with different restriction enzyme sites at both ends were amplified by PCR using RNAi1 and RNAi2 primer pairs (Table S1). The two PCR fragments were inserted at inverted repeats into the pKANNIBAL vector to generate a hairpin RNAi construct, which was then cloned into the binary vector pCU BamHI site [61]. The final RNAi vector was introduced into Agrobacterium tumefaciens (strain EHA105) by electroporation. Transgenic rice plants were generated by Agrobacterium-mediated transformation of rice calli, as previously described [62]. Transformed calli were selected by hygromycin resistance, and the transgenic plants were regenerated from the transformed calli. Regenerated transgenic rice plants were grown in a greenhouse.
Southern blot analysis
Genomic DNA was isolated from hygromycin-tolerant and empty transformation vector plants using the method of Chakraborti et al. [63]. The DNA from the empty transformation vector plants was used as a negative control. Fifteen microgram portions of the genomic DNA were digested with DraI, and the digested DNA was subjected to electrophoresis on a 0.8% agarose gel, then transferred to a Hybond N+ membrane (Amersham). The probe was labeled with [α-32P]dCTP using the Prime-a-Gene labeling system (Promega), and the membranes were hybridized for at least 10 h at 65°C with the labeled probe. Following incubation, the blot was washed with 2× SSC/0.5% SDS for 15 min at 65°C followed by 1× SSC/0.1% SDS for 15 min at 65°C.
Northern blot analysis
Total RNAs were isolated from N. lugens or plant tissues by Trizol reagent (Invitrogen).About 20 µg of total RNA was separated on a denaturing 1.5% formaldehyde agarose gel. Loading of equal amounts of RNA was confirmed by ethidium bromide staining. The formaldehyde gels were blotted onto a Hybond N+ membrane (Amersham) and hybridized with [α-32P] dCTP labeled N. lugens genes probe. The membranes were washed with 2× SSC/0.1% SDS for 15 min at 65°C followed by 1× SSC/0.1% SDS for 15 min at 65°C.
Small RNA analysis
siRNA detection was performed as previously described [64]. Electrophoresis was performed in 3% agarose gels in the presence of formaldehyde. Blots of siRNAs were prehybridized at 58°C for 2 h, and hybridizations were performed at 42°C for 16 h. The blots were washed with 2× SSC/0.5% SDS for 3×10min at 37°C followed by 2× SSC/0.2% SDS for 10 min at 42°C. The primers (18 and 21 nucleotides) were used as size standard and hybridization controls.
Quantitative real-time PCR analysis
Total RNA was isolated from dissected tissues by Trizol reagent (Invitrogen). First-strand cDNA was synthesized at 42°C from total RNA using M-MLV reverse transcriptase (Fermentas). Expression of selected N. lugens genes and ß-Actin (EU179846) [65] was quantified by quantitative RT-PCR, using an RG-6000 rotary analyzer (Corbett Research) and appropriate primers (Table S1) together with 1 µg of RNA per reaction. The following pair of primers was used for ß-Actin (which served as an internal control to normalize differences in recovery between samples: 5′-TGGACTTCGAGCAGGAAATGG-3′ and 5′- ACGTCGCACTTCATGATCGAG-3′).
Phylogenetic Analysis
The deduced amino acid sequences of SID-1 and Argonaute were aligned using ClustalW. Accession numbers of SID-1s used for sequence alignment and phylogenetic analyses were as follows: N. lugens SID-1 like, JF915743; B. mori SID-1 like 1, BAF95805; B. mori SID-1 like 2, BAF95807; B. mori SID-1 like 3, BAF95806; T. castaneum SID-1 related A, NP_001099012; T. castaneum SID-1 related B, NP_001103253; T. castaneum SID-1 related C, NP_001099128; A. mellifera SID-1 like, XP_395167; A. pisum SID-1 like, XP_001951907; and C. elegans SID-1, AAL78657. Accession numbers of Argonaute used for sequence alignment and phylogenetic analyses are as follows: N. lugens Aubergine, JF915742; M. musculus Mili, BAA93706; M. musculus Miwi, BAA93705; M. musculus Miwi2, AAN75583; D. rerio Ziwi, NP_899181; D. rerio Zili, ACH96370; R. norvegicus Riwi, NP_001102323; H. sapiens Hiwi, AAC97371; H. sapiens Piwil1, BAC81341; Homo sapiens Piwil3, BAC81343; H. sapiens Piwil2, BAC81342; D. melanogaster Piwi, AAD08705; D. melanogaster Ago3, ABO26294; D. melanogaster Aubergine, NP_476734; A. mellifera Aubergine, ACV84377; A. mellifera Argonaute, ACV84372. Phylogenetic analyses were conducted by the neighbor-joining method using MEGA 4.0. Bootstrap values were assessed with 1000 replicates.
BPH survival rate determination
For the plant feeding experiment, synchronous nymphs were selected and divided into groups. Pots, each containing one plant, were individually covered with plastic cages (diameter 12 cm, height 35 cm) into which 20–30 individuals were released. After feeding on plant diets for the indicated number of days, midguts of nymphs were taken for further analysis. Tests of statistical significance were performed with Student's t-test in Excel.
The survival rates of BPH nymphs on the transgenic plants(transformed with NlHT1-RNAi, Nlcar-RNAi and Nltry-RNAi constructs and with the empty transformation vector)were determined. Pots, each containing one plant, were individually covered with plastic cages (diameter 9 cm, height 12 cm) into each of which 10 newly hatched BPH nymphs were released. The number of surviving BPH nymphs on each plant was recorded every day until 12 days after the introduction of the herbivore. The experiment was repeated 10 times.
Supporting Information
Nlsid-1 nucleotide and deduced amino acid sequences. Transmembrane regions, as predicted by TMHMM, are underlined (TM 1–7).
(TIF)
Nlaub nucleotide and deduced amino acid sequences. The deduced amino acid sequence is shown below the cDNA sequence. The PAZ and Piwi domains are underlined.
(TIF)
The nucleotide and deduced amino acid sequences of the N. lugens trypsin-like serine protease gene (Nltry).
(TIF)
The nucleotide and deduced amino acid sequences of the N. lugens carboxypeptidase gene (Nlcar).
(TIF)
Mean survival rates of BPH nymphs fed on transgenic plants transformed with empty transformation vector (Vector), NlHT1 -RNAi (H2), Nlcar -RNAi (C8), and Nltry -RNAi (T3).
(TIF)
Primer sequences used in the present study.
(DOC)
Phenotypic segregation analysis in progeny (T1 generation) of six RNAi transgenic lines.
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Fundamental Research Funds for the Central University of China (grant no. 1104002) and the Special Fund for Commonwealth Industry (grant no. 200803003) from the Ministry of Agriculture of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 3.Huvenne H, Smagghe G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol. 2010;56:227–235. doi: 10.1016/j.jinsphys.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Fire AZ. Gene silencing by double-stranded RNA (Nobel Lecture). Angew Chem Int Ed Engl. 2007;46:6966–6984. doi: 10.1002/anie.200701979. [DOI] [PubMed] [Google Scholar]
- 5.Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, et al. A systemic small RNA signaling system in plants. Plant Cell. 2004;16:1979–2000. doi: 10.1105/tpc.104.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dzitoyeva S, Dimitrijevic N, Manev H. Intra-abdominal injection of double-stranded RNA into anesthetized adult Drosophila triggers RNA interference in the central nervous system. Mol Psychiatry. 2001;6:665–670. doi: 10.1038/sj.mp.4000955. [DOI] [PubMed] [Google Scholar]
- 7.Tomoyasu Y, Miller SC, Tomita S, Schoppmeier M, Grossmann D, et al. Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol. 2008;9:R10. doi: 10.1186/gb-2008-9-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch JA, Desplan C. A method for parental RNA interference in the wasp Nasonia vitripennis. Nat Protoc. 2006;1:486–494. doi: 10.1038/nprot.2006.70. [DOI] [PubMed] [Google Scholar]
- 9.Meyering-Vos M, Muller A. RNA interference suggests sulfakinins as satiety effectors in the cricket Gryllus bimaculatus. J Insect Physiol. 2007;53:840–848. doi: 10.1016/j.jinsphys.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Martin D, Maestro O, Cruz J, Mane-Padros D, Belles X. RNAi studies reveal a conserved role for RXR in molting in the cockroach Blattella germanica. J Insect Physiol. 2006;52:410–416. doi: 10.1016/j.jinsphys.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Turner CT, Davy MW, MacDiarmid RM, Plummer KM, Birch NP, et al. RNA interference in the light brown apple moth, Epiphyas postvittana (Walker) induced by double-stranded RNA feeding. Insect Mol Biol. 2006;15:383–391. doi: 10.1111/j.1365-2583.2006.00656.x. [DOI] [PubMed] [Google Scholar]
- 12.Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, et al. RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol. 2011;57:231–245. doi: 10.1016/j.jinsphys.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Zhou X, Wheeler MM, Oi FM, Scharf ME. RNA interference in the termite Reticulitermes flavipes through ingestion of double-stranded RNA. Insect Biochem Mol Biol. 2008;38:805–815. doi: 10.1016/j.ibmb.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Roignant JY, Carre C, Mugat B, Szymczak D, Lepesant JA, et al. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA. 2003;9:299–308. doi: 10.1261/rna.2154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arakane Y, Li B, Muthukrishnan S, Beeman RW, Kramer KJ, et al. Functional analysis of four neuropeptides, EH, ETH, CCAP and bursicon, and their receptors in adult ecdysis behavior of the red flour beetle, Tribolium castaneum. Mech Dev. 2008;125:984–995. doi: 10.1016/j.mod.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Tomoyasu Y, Denell RE. Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev Genes Evol. 2004;214:575–578. doi: 10.1007/s00427-004-0434-0. [DOI] [PubMed] [Google Scholar]
- 17.Silva CP, Silva JR, Vasconcelos FF, Petretski MD, Damatta RA, et al. Occurrence of midgut perimicrovillar membranes in paraneopteran insect orders with comments on their function and evolutionary significance. Arthropod Struct Dev. 2004;33:139–148. doi: 10.1016/j.asd.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Wang P, Granados RR. Molecular structure of the peritrophic membrane (PM): identification of potential PM target sites for insect control. Arch Insect Biochem Physiol. 2001;47:110–118. doi: 10.1002/arch.1041. [DOI] [PubMed] [Google Scholar]
- 19.Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- 20.Xu W, Han Z. Cloning and phylogenetic analysis of sid-1-like genes from aphids. J Insect Sci. 2008;8:1–6. doi: 10.1673/031.008.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin H. piRNAs in the germ line. Science. 2007;316:397. doi: 10.1126/science.1137543. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Juranek S, Li H, Sheng G, Tuschl T, et al. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 24.Williams RW, Rubin GM. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc Natl Acad Sci U S A. 2002;99:6889–6894. doi: 10.1073/pnas.072190799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao Z, Jia Q, Li F, Han Z. Identification of two piwi genes and their expression profile in honeybee, Apis mellifera. Arch Insect Biochem Physiol. 2010;74:91–102. doi: 10.1002/arch.20362. [DOI] [PubMed] [Google Scholar]
- 26.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 27.Mlotshwa S, Voinnet O, Mette MF, Matzke M, Vaucheret H, et al. RNA silencing and the mobile silencing signal. Plant Cell. 2002;14(Suppl):S289–301. doi: 10.1105/tpc.001677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon KH, Waterhouse PM. RNAi for insect-proof plants. Nat Biotechnol. 2007;25:1231–1232. doi: 10.1038/nbt1107-1231. [DOI] [PubMed] [Google Scholar]
- 29.Price DR, Gatehouse JA. RNAi-mediated crop protection against insects. Trends Biotechnol. 2008;26:393–400. doi: 10.1016/j.tibtech.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, et al. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol. 2007;25:1307–1313. doi: 10.1038/nbt1352. [DOI] [PubMed] [Google Scholar]
- 31.Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol. 2007;25:1322–1326. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
- 32.Backus EA, Serrano MS, Ranger CM. Mechanisms of hopperburn: an overview of insect taxonomy, behavior, and physiology. Annu Rev Entomol. 2005;50:125–151. doi: 10.1146/annurev.ento.49.061802.123310. [DOI] [PubMed] [Google Scholar]
- 33.Cha YS, Ji H, Yun DW, Ahn BO, Lee MC, et al. Fine mapping of the rice Bph1 gene, which confers resistance to the brown planthopper (Nilaparvata lugens stal), and development of STS markers for marker-assisted selection. Mol Cells. 2008;26:146–151. [PubMed] [Google Scholar]
- 34.Falk BW, Tsai JH. Biology and molecular biology of viruses in the genus Tenuivirus. Annu Rev Phytopathol. 1998;36:139–163. doi: 10.1146/annurev.phyto.36.1.139. [DOI] [PubMed] [Google Scholar]
- 35.Senthil-Nathan S, Choi MY, Paik CH, Seo HY, Kalaivani K. Toxicity and physiological effects of neem pesticides applied to rice on the Nilaparvata lugens Stal, the brown planthopper. Ecotoxicol Environ Saf. 2009;72:1707–1713. doi: 10.1016/j.ecoenv.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 36.Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nat Chem Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 37.Price DR, Wilkinson HS, Gatehouse JA. Functional expression and characterisation of a gut facilitative glucose transporter, NlHT1, from the phloem-feeding insect Nilaparvata lugens (rice brown planthopper). Insect Biochem Mol Biol. 2007;37:1138–1148. doi: 10.1016/j.ibmb.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Christensen AH, Sharrock RA, Quail PH. Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol. 1992;18:675–689. doi: 10.1007/BF00020010. [DOI] [PubMed] [Google Scholar]
- 39.Christensen AH, Quail PH. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 1996;5:213–218. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- 40.Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 41.May RC, Plasterk RH. RNA interference spreading in C. elegans. Methods Enzymol. 2005;392:308–315. doi: 10.1016/S0076-6879(04)92018-6. [DOI] [PubMed] [Google Scholar]
- 42.Aronstein K, Pankiw T, Saldivar E. SID-1 is implicated in systemic gene silencing in the honey bee. Journal of Apicultural Research. 2006;45:20–24. [Google Scholar]
- 43.Dong Y, Friedrich M. Nymphal RNAi: systemic RNAi mediated gene knockdown in juvenile grasshopper. BMC Biotechnol. 2005;5:25. doi: 10.1186/1472-6750-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 45.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 46.Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, et al. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 47.Kennerdell JR, Yamaguchi S, Carthew RW. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes Dev. 2002;16:1884–1889. doi: 10.1101/gad.990802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Specchia V, Benna C, Mazzotta GM, Piccin A, Zordan MA, et al. Aubergine gene overexpression in somatic tissues of aubergine(sting) mutants interferes with the RNAi pathway of a yellow hairpin dsRNA in Drosophila melanogaster. Genetics. 2008;178:1271–1282. doi: 10.1534/genetics.107.078626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 50.Rajagopal R, Sivakumar S, Agrawal N, Malhotra P, Bhatnagar RK. Silencing of midgut aminopeptidase N of Spodoptera litura by double-stranded RNA establishes its role as Bacillus thuringiensis toxin receptor. J Biol Chem. 2002;277:46849–46851. doi: 10.1074/jbc.C200523200. [DOI] [PubMed] [Google Scholar]
- 51.Saha P, Majumder P, Dutta I, Ray T, Roy SC, et al. Transgenic rice expressing Allium sativum leaf lectin with enhanced resistance against sap-sucking insect pests. Planta. 2006;223:1329–1343. doi: 10.1007/s00425-005-0182-z. [DOI] [PubMed] [Google Scholar]
- 52.Kikuta S, Kikawada T, Hagiwara-Komoda Y, Nakashima N, Noda H. Sugar transporter genes of the brown planthopper, Nilaparvata lugens: A facilitated glucose/fructose transporter. Insect Biochem Mol Biol. 2010;40:805–813. doi: 10.1016/j.ibmb.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 53.Lee SI, Lee SH, Koo JC, Chun HJ, Lim CO, et al. Soybean Kunitz trypsin inhibitor (SKTI) confers resistance to the brown planthopper (Nilaparvata lugens Stål) in transgenic rice. Molecular Breeding. 1999;5:1–9. [Google Scholar]
- 54.Richardson PT, Baker DA, Ho LC. The chemical composition of cucurbit vascular exudates. Journal of Experimental Botany. 1982;33:202–209. [Google Scholar]
- 55.Chen J, Zhang D, Yao Q, Zhang J, Dong X, et al. Feeding-based RNA interference of a trehalose phosphate synthase gene in the brown planthopper, Nilaparvata lugens. Insect Mol Biol. 2010. [DOI] [PubMed]
- 56.Liu S, Ding Z, Zhang C, Yang B, Liu Z. Insect Biochem Mol Biol; 2010. Gene knockdown by intro-thoracic injection of double-stranded RNA in the brown planthopper, Nilaparvata lugens. [DOI] [PubMed] [Google Scholar]
- 57.Yadav BC, Veluthambi K, Subramaniam K. Host-generated double stranded RNA induces RNAi in plant-parasitic nematodes and protects the host from infection. Mol Biochem Parasitol. 2006;148:219–222. doi: 10.1016/j.molbiopara.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 58.Huang G, Allen R, Davis EL, Baum TJ, Hussey RS. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc Natl Acad Sci U S A. 2006;103:14302–14306. doi: 10.1073/pnas.0604698103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fairbairn DJ, Cavallaro AS, Bernard M, Mahalinga-Iyer J, Graham MW, et al. Host-delivered RNAi: an effective strategy to silence genes in plant parasitic nematodes. Planta. 2007;226:1525–1533. doi: 10.1007/s00425-007-0588-x. [DOI] [PubMed] [Google Scholar]
- 60.Yang Z, Xia X, Wang X, He G. cDNA cloning, heterogeneous expression and biochemical characterization of a novel trypsin-like protease from Nilaparvata lugens. Z Naturforsch C. 2010;65:109–118. doi: 10.1515/znc-2010-1-218. [DOI] [PubMed] [Google Scholar]
- 61.Chen R, Zhao X, Shao Z, Wei Z, Wang Y, et al. Rice UDP-Glucose Pyrophosphorylase1 Is Essential for Pollen Callose Deposition and Its Cosuppression Results in a New Type of Thermosensitive Genic Male Sterility. Plant Cell. 2007;19:847–861. doi: 10.1105/tpc.106.044123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 63.Chakraborti D, Sarkar S, Gupta S, Das S. Small and large scale genomic DNA isolation protocol for chickpea (Cicer arietinum L.), suitable for molecular marker and transgenic analyses. Afr J Biotechnol. 2006;5:585–589. [Google Scholar]
- 64.Lechtenberg B, Schubert D, Forsbach A, Gils M, Schmidt R. Neither inverted repeat T-DNA configurations nor arrangements of tandemly repeated transgenes are sufficient to trigger transgene silencing. Plant J. 2003;34:507–517. doi: 10.1046/j.1365-313x.2003.01746.x. [DOI] [PubMed] [Google Scholar]
- 65.Liu S, Yang B, Gu J, Yao X, Zhang Y, et al. Molecular cloning and characterization of a juvenile hormone esterase gene from brown planthopper, Nilaparvata lugens. J Insect Physiol. 2008;54:1495–1502. doi: 10.1016/j.jinsphys.2008.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nlsid-1 nucleotide and deduced amino acid sequences. Transmembrane regions, as predicted by TMHMM, are underlined (TM 1–7).
(TIF)
Nlaub nucleotide and deduced amino acid sequences. The deduced amino acid sequence is shown below the cDNA sequence. The PAZ and Piwi domains are underlined.
(TIF)
The nucleotide and deduced amino acid sequences of the N. lugens trypsin-like serine protease gene (Nltry).
(TIF)
The nucleotide and deduced amino acid sequences of the N. lugens carboxypeptidase gene (Nlcar).
(TIF)
Mean survival rates of BPH nymphs fed on transgenic plants transformed with empty transformation vector (Vector), NlHT1 -RNAi (H2), Nlcar -RNAi (C8), and Nltry -RNAi (T3).
(TIF)
Primer sequences used in the present study.
(DOC)
Phenotypic segregation analysis in progeny (T1 generation) of six RNAi transgenic lines.
(DOC)