Abstract
A consensus linkage map has been developed in the chicken that combines all of the genotyping data from the three available chicken mapping populations. Genotyping data were contributed by the laboratories that have been using the East Lansing and Compton reference populations and from the Animal Breeding and Genetics Group of the Wageningen University using the Wageningen/Euribrid population. The resulting linkage map of the chicken genome contains 1889 loci. A framework map is presented that contains 480 loci ordered on 50 linkage groups. Framework loci are defined as loci whose order relative to one another is supported by odds greater then 3. The possible positions of the remaining 1409 loci are indicated relative to these framework loci. The total map spans 3800 cM, which is considerably larger than previous estimates for the chicken genome. Furthermore, although the physical size of the chicken genome is threefold smaller then that of mammals, its genetic map is comparable in size to that of most mammals. The map contains 350 markers within expressed sequences, 235 of which represent identified genes or sequences that have significant sequence identity to known genes. This improves the contribution of the chicken linkage map to comparative gene mapping considerably and clearly shows the conservation of large syntenic regions between the human and chicken genomes. The compact physical size of the chicken genome, combined with the large size of its genetic map and the observed degree of conserved synteny, makes the chicken a valuable model organism in the genomics as well as the postgenomics era. The linkage maps, the two-point lod scores, and additional information about the loci are available at web sites in Wageningen (http://www.zod.wau.nl/vf/research/chicken/frame_chicken.html) and East Lansing (http://poultry.mph.msu.edu/).
The chicken is increasingly becoming of great interest as an intermediate evolutionary model organism, ideally placed between mammals and more distant vertebrates as the pufferfish and zebrafish. There are a number of different reasons for this increasing interest in the chicken genome. First, the genome size is only one-third that of mammals (Tiersch and Wachtel 1991) mainly because of its low amount of repetitive sequences and reduced intron sizes (Hughes and Hughes 1995). Furthermore, It has an interesting complex genomic structure with two chromosomal subtypes—macrochromosomes and microchromosomes (Bloom et al. 1993)—with the microchromosomes appearing to be somewhat more gene dense then the macrochromosomes, reaching densities comparable to that of the Fugu genome (McQueen et al. 1998; Clark et al. 1999). Second, the level of conserved synteny between chicken and humans appears to be very high (Burt et al. 1995; Hu et al. 1995; Klein et al. 1996; Jones et al. 1997; Groenen et al. 1999; Nanda et al. 1999). Third, the chicken is being studied intensively for genes affecting polygenic traits (quantitative trait loci or QTL), which drive international efforts toward detailed physical and linkage mapping in the chicken.
Although the first genetic linkage map in chicken was published >60 years ago (Hutt 1936), it was not until the development of large numbers of molecular markers in the last decade that the generation of linkage maps in chicken increased. In chicken, three different linkage maps were developed using three different mapping populations. The first genetic map, based completely on DNA markers, was published by Bumstead and Palyga (1992). This map, based on the Compton (C) reference population, consisted solely of restriction fragment length polymorphism (RFLP) markers. The second genetic map to be published (Levin et al. 1993, 1994) was based on the East Lansing (EL) reference population and consisted primarily of RFLPs, random amplified polymorphic DNA (RAPD) markers, and chicken repeat element 1 (CRI) markers. Since then, both populations have been used to map a considerable number of microsatellite markers (Cheng et al. 1995; Crooijmans et al. 1997; Gibbs et al. 1997) and AFLP markers (Knorr et al. 1999) as well. The third map (Groenen et al. 1998, Herbergs et al. 1999) was based on a large F2 population and consisted solely of microsatellite and amplified fragment length polymorphism (AFLP) markers. Increasing marker densities and increased initiatives in physical mapping in chicken have necessitated the need of a single consensus linkage map in chicken. Because all three maps have many markers in common, this goal has become feasible for the large and intermediate-sized chromosomes.
In this paper we describe the integration of all available data of the three mapping populations, resulting in a consensus linkage map of the chicken genome comprised of 50 linkage groups, with a total of 1889 loci.
RESULTS AND DISCUSSION
Linkage Maps
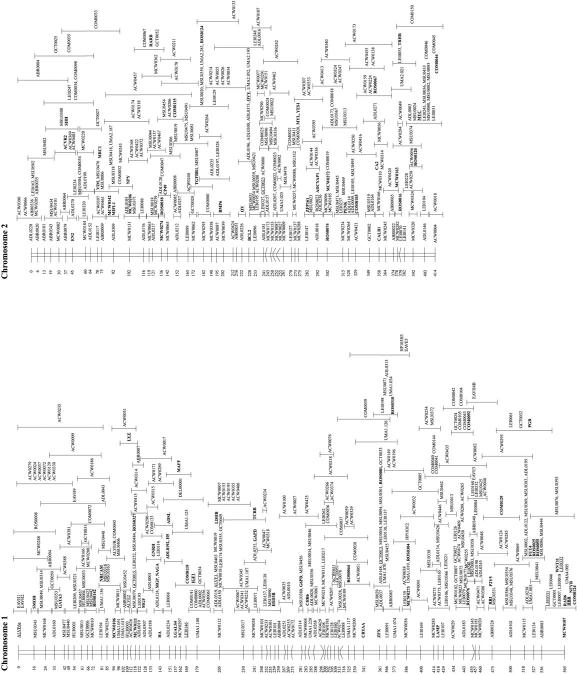
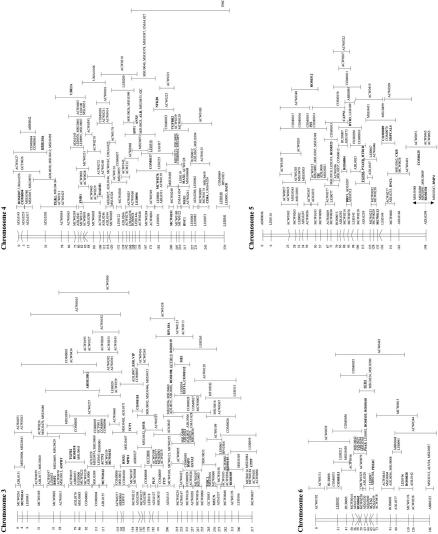
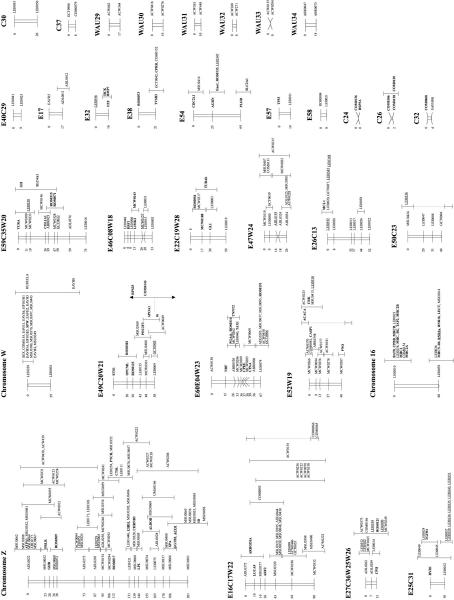
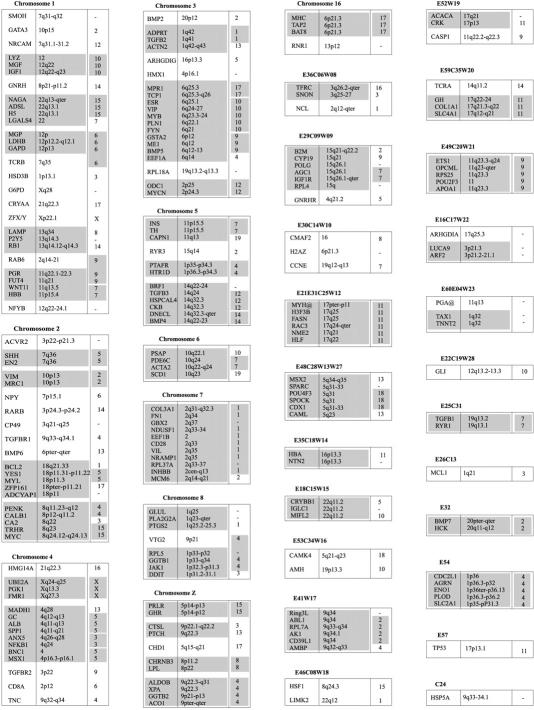
The genotyping data from the three chicken mapping populations were combined and analyzed simultaneously using the CRIMAP linkage program. Contributions of genotyping data were made from laboratories that have been using the EL and C reference populations and from the Animal Breeding and Genetics Group of the Wageningen University using the Wageningen/Euribrid (WAU) population. A complicating factor for the integration of all maps in chicken is the fact that not all types of markers are evenly distributed over the macro- and microchromosomes, particularly the low abundance of microsatellites on the microchromosomes (Primmer et al. 1997). Consequently, many of the small linkage groups do not have a marker in common, making the integration impossible at present. Furthermore, linkage groups C15 and C20 had only one marker in common with the corresponding linkage groups in the WAU and EL data sets, and as a consequence, the other loci from C15 and C20 could not be positioned very precisely (Fig. 1, linkage groups E18C15W15 and E49C20W21).
Figure 1.
Framework consensus linkage map of the chicken genome. The framework loci (loci whose order relative to one another is supported by odds >3, Keats et al. 1991) have been ordered and their position is indicated by the number at left. The possible location of the loci whose order is not supported by odds >3 is indicated by an error bar. Loci that have been mapped cytogenetically are underlined; those known to represent expressed sequences (identified genes and ESTs) are shown in boldface type.
The total number of different loci that have been typed on at least one of the three mapping populations was 2019. However, a relatively large proportion of these markers was either unlinked (95) or could not be positioned clearly on the linkage maps (35) and therefore were omitted from the final map shown in Figure 1. A large proportion of the omitted markers (89) are AFLP markers typed on the WAU population (Herbergs et al. 1999).
The resulting linkage map of the chicken genome contains 1889 loci (Table 1). The framework map contains 480 loci ordered on 50 linkage groups (Fig. 1). The possible positions of the remaining 1409 loci are indicated relative to these framework loci. When all linkage groups are taken into account, the total length of the linkage map is 4000 cM. However, it is expected that several of the smaller EL, C, and WAU linkage groups belong to the same chromosomes. If we correct for this fact, then the minimal length of the chicken consensus linkage map is ∼3800 cM, which still is considerably larger than the previous estimates 2600–3000 cM for the chicken genome (Rodionov et al. 1992; Burt et al. 1995).
Table 1.
Number and Type of Loci on Chicken Linkage Maps
| WAU | EL | C | Consensus Map | |
|---|---|---|---|---|
| A. Loci | ||||
| WAU | 1011 | 290 | 119 | 923 |
| EL | 290 | 1068 | 195 | 1050 |
| C | 119 | 195 | 447 | 428 |
| Type I loci | 93 | 252 | 107 | 350 |
| Linkage groups | 34 | 42 | 36 | 50 |
| B. Markers | ||||
| Microsatellite | 573 | 479 | 190 | 801 |
| Minisatellite | — | 34 | 30 | 40 |
| RFLP | — | 92 | 191 | 244 |
| AFLP | 350 | 202 | — | 552 |
| SSCP | — | 50 | 15 | 59 |
| ASO | — | 71 | 1 | 71 |
| RAPD | — | 65 | — | 65 |
| CR1 | — | 47 | — | 47 |
| Classical | — | 10 | 2 | 10 |
(A) The total number of different loci analyzed on the Wageningen (WAU), East Lansing (EL), and Compton (C) linkage maps, as well as the number of loci that are shared among the different maps. (B) The different types of markers on the different chicken linkage maps. Included are loci located on the consensus linkage map, as in Fig. 1.
Although there are large differences in length between many of the EL (male) and the C linkage groups (female), these variations are most likely the result of differences between the lines used and typing errors in some of the RFLP markers in the C map. The differences in length between the male and female maps based on the WAU population generally are small with an overall difference between male and female maps of only 1.15%. Because of the much larger number of informative meioses within the WAU population, the framework of the consensus map is mainly built up by microsatellites typed on this population. Therefore, for the consensus map, the differences seen between the size of the male and female maps are similar as those described for the WAU map (Groenen et al. 1998).
Discrepancies Between the Different Maps
Discepancies for seven loci were observed in map locations in the original maps from the three populations used. (1) G6PD has been mapped on E1 and CW, and (2) EIF4A2 has been mapped on E36 and C3. As these are both RFLPs, it is possible that different bands were scored in each population. With regard to EIF4A2, it is noteworthy that its location within the EL data is supported by the comparative mapping data (Fig. 2). In the EL data EIF4A2 maps to E36, close to the SNON and TFRC genes. In human, all three genes are located on the q arm of chromosome 3. However, given the discrepancies in the data, these two loci have not been included on the map. (3) LEI0144 has been mapped on chromosome 4 (WAU) and chromosome Z (EL); (4) MCW0066 has been mapped on chromosome 2 (C), E30 and W10; (5) MCW0166 has been mapped on chromosome 2 (WAU) and chromosome 4 (EL).(6) The BAT8 gene was mapped by a single laboratory (Spike and Lamont 1995) to the end of chromosome 4 (EL) and chromosome 16, the chromosome to which the major histocompatibility complex (MHC) class I and II genes have been mapped. Finally, (7) The α-tubulin gene (TUBA) was mapped to C15 and to E22. Because this gene has been mapped by two different laboratories using different methods, it is likely that two different loci of the α-tubulin gene family have been mapped. These two genes have been included in the map as TUBAa and TUBAb, respectively.
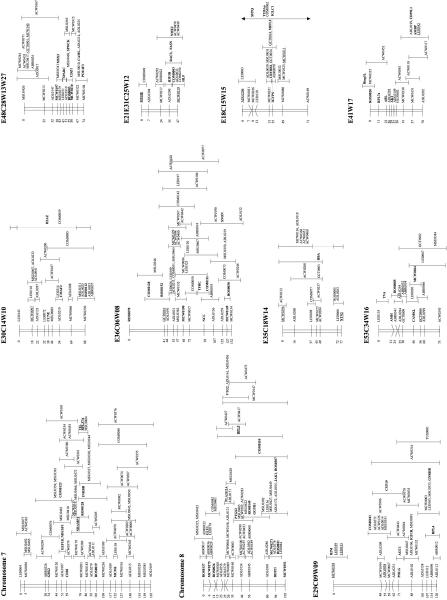
Figure 2.
Comparative mapping results among chicken, man, and mouse. The order of the loci is according to the linkage map shown in Fig. 1. The second column in each linkage group shows the location of the loci on the human cytogenetic map according to Genome Data Base (http://www.gdb.org/); the third column shows the map location in the mouse. Blocks of conserved synteny between chicken and man and between chicken and mouse are shaded.
To resolve the discrepancies between the microsatellites, these markers were retyped. Typing of LEI0144 on the EL population by M. Groenen and coworkers, (unpubl.) showed that this gene also mapped to chromosome 4, which is in agreement with the WAU data. Similarly, retyping of this marker by T. Burke and coworkers, (unpubl.) confirmed their previous results of LEI0144 mapping to the Z chromosome. A possible explanation for this discrepancy is that the typings are done by the two groups using different primers [Crooijmans et al. (1997) and Gibbs et al. (1997), respectively]. This marker has therefore been included on both locations on the map as LEI0144a and LEI0144b. The new typing results showed clearly that the previous assignment of MCW0066 to C chromosome 2 and of MCW0166 to EL chromosome 4 are not correct and that these two markers are on linkage group E30C14W10 and chromosome 2, respectively.
In addition, although there were no conflicting data between them, the mapping results of two loci for the three different populations were rather unexpected or somewhat unlikely. Marker LEI0192 appeared to be unlinked in the EL population, whereas it is mapped to chromosome 6 using the WAU population and to linkage group C21 using the C population. Similarly, marker HUJ0005 has been mapped to chromosome 6 on the WAU map and it is unlinked in the other two maps. Moreover, both markers are linked tightly to each other and located at the end of chromosome 6 of the WAU map, whereas they are unlinked in both of the other maps. Flipping of the typing phase of HUJ0005 in the original MAPMANAGER file of the EL population results in linkage to several markers on chromosome 6 but not to LEI0192. Flipping of the phase had no effect in these markers in the C data set. Physical mapping of a BAC clone containing LEI0192 confirmed its location at the end of chromosome 6 (V. Fillon and A. Vignal, pers. comm.).
Two additional discrepancies are observed between the consensus linkage map and the cytogenetic map. The CYP19 gene was mapped on linkage group E029C09W09, whereas it was mapped to chromosome 1 on the cytogenetic map (Tereba et al. 1991). Comparative mapping data support the location of this gene on linkage group E29C09W09. On the linkage map the MAX gene maps to chromosome 4, whereas cytogenetically it was mapped to chromosome 5p (Nanda et al. 1997). In this case, the comparative mapping data support the cytogenetic location of this gene on chromosome 5, where several other human genes have been mapped that are located on chromosome 14q.
Finally, LEI0229 was mapped to both the Z chromosome (EL) and the W chromosome (C). The most likely explanation is that LEI0229 maps to one of the pseudoautosomal regions of the chicken Z chromosome (Fridolfsson et al. 1998).
Anchoring Linkage Groups to Chromosomes
In addition to the integration of the three linkage maps, eventually these maps will have to be integrated with the physical map in chicken. However, the integration of the physical and genetic maps presents considerable difficulties and has proceeded at a slower pace, as a result (Morisson et al. 1998). The chicken karyotype is composed of 2n = 78 chromosomes which, according to their size, are classified as macro- and microchromosomes (Bloom et al. 1993). Due to the presence of microchromosomes in chicken, a standard karyotype could only be established for the eight large macrochromosomes and the two sex chromosomes (International Committee for the Standardization of the Avian Karyotype).
Many markers on the consensus map have been mapped cytogenetically as well, allowing the integration of the linkage map with the physical map. These loci are underlined in Figure 1. Because only a standard karyotype has been established for the macrochromosomes, only the linkage groups of these larger chromosomes could be assigned to their corresponding chromosomes. For historical reasons, the microchromosome containing the MHC is named chromosome 16.
To enable identification of the microchromosomes, a set of large insert clones is being developed that can be used as tags in two-color fluorescence in situ hybridization (Fillon et al. 1998). Polymorphic markers have been developed for many of these large insert clones (Morisson et al. 1998; P.A. Thomson and T. Burke, unpubl.), which will allow the assignment of the linkage groups to the corresponding microchromosomes as well. Furthermore, additional large insert BAC clones have been isolated using microsatellites from the small linkage groups, which provides additional probes for the identification of the microchromosomes (R. Crooijmans, V. Fillon, M. Groenen, and A. Vignal, unpubl.).
Comparative Gene Mapping
Genetic markers within or adjacent to known genes have been classified as type I markers (O'Brien 1991). The inclusion of type I markers on the linkage map makes it possible to access the mapping information that is available in densely mapped species such as humans and mice. Currently, the consensus map contains 350 markers within expressed sequences, 235 of which represent identified genes or sequences that have significant sequence identity to known genes. These loci are shown in boldface type in Figure 1. The orthologs of 204 of these 235 genes have also been mapped in human (Fig. 2). The comparative mapping data based on the consensus linkage map show a considerable amount of chromosomal conservation retained between man and chicken during evolution. This is in sharp contrast with the comparative mapping data between chicken and mouse, in which the amount of chromosomal conservation is considerably lower. Similar results are obtained for the comparisons between the genomes of different mammals, indicating that there have been extensive rearrangements during the evolution of the mouse genome and at a much higher rate than in birds or the other mammals (Andersson et al. 1996). Based only on the linkage data presented in this paper, at least 87 different chromosomal regions can be identified between man and chicken (Fig. 2). For many of these chromosomal regions, physically mapped genes provide additional evidence for the observed conservation of linkage (Burt et al. 1995; Andersson et al. 1996; Nanda et al. 1999; Burt et al. 1999). Furthermore, the physically mapped genes have identified additional conserved regions between the genomes of chicken and man. Based on this number of conserved chromosomal regions, it has been calculated that the number of autosomal conserved segments shared between the chicken and human genomes is probably <100 (Burt et al. 1999). This level of conservation of synteny between chicken and human, in combination with the threefold more compact genome of the chicken, makes it an excellent evolutionary model organism in addition to Fugu, mouse, and rat. This is particularly true for the microchromosomes, which appear to be somewhat more gene dense than the macrochromosomes, thereby reaching gene densities close to that of Fugu (Angrist 1998; McQueen et al. 1998; Clarke et al. 1999). Furthermore, the higher level of conservation of genome organization (Gilley et al. 1997; Reboul et al. 1999) and the easy accessibility of the chicken as an experimental animal in studies regarding complex polygenic traits are additional features favoring the chicken over other models such as Fugu.
Although the number of loci that are available for comparative mapping are still too limited to draw detailed conclusions, it is noteworthy that several of the small linkage groups in chicken, which most likely represent different microchromosomes, seem to represent, in almost their entirety, large fragments of specific human chromosomes (e.g., E29C09W09, E21E31C25W12, E48C28W13W27, E41W17, E54, E49C20W21, and chromosome 7).
Future Directions
The current map contains 801 microsatellite markers, which are the markers of choice for whole genome scans. However, the marker density is only sufficiently high for the macrochromosomes and a subset of the microchromosomes. Therefore, many more microsatellites are still needed to obtain (near) complete genome coverage in these kinds of studies. The integration of all the linkage maps and the cytological map in chicken is the first necessary step toward achieving this goal by identifying those regions that are particularly devoid of microsatellite markers. The major drawback, however, is the relatively low abundance of microsatellites on many of the microchromosomes (Primmer et al. 1997). Currently, increasing efforts are being put into the development of physical maps for several regions of the chicken genome, for example, Chromosome 16 (N. Bumstead, unpubl.) and linkage groups E29C09W09 and E53C34W16 (R. Crooijmans and M. Groenen, unpubl.). This has become feasible through an increased number of loci on the linkage map and because of the development of publicly available chicken YAC (Toye et al. 1997) and BAC (R.P.M.A. Crooijmans, J. Vrebalor, R.J.M. Dijkhof, J.J. van der Poel, and M.A.M. Groenen, in prep.; J. Dodgson, unpubl.) libraries. It is to be expected that physical maps eventually will become available for all chicken chromosomes. This in turn will make the targeted development of microsatellite markers possible for those regions that currently lack any such markers, thereby allowing the characterization of these regions in QTL studies as well.
METHODS
Mapping Populations
The three mapping populations have been described in detail previously. Briefly, the EL population (Crittenden et al. 1993) consists of 52 BC1 animals derived from a backcross between a partially inbred jungle fowl line and a highly inbred white leghorn line. The C population (Bumstead and Palyga 1992) consists of 56 BC1 animals derived from a backcross between two inbred white leghorn lines that differed in their disease resistance. The WAU population (Groenen et al. 1998) consists of 456 F2 animals from a cross between two broiler dam lines originating from the white Plymouth Rock breed.
Markers
A detailed description of all individual loci, including their references and the number of informative meioses, is available at the web site of the Animal Breeding and Genetics Group in Wageningen (http://www.zod.wau.nl/vf/research/chicken/frame_chicken.html) and East Lansing (http://poultry.mph.msu.edu/)
Linkage Analysis
For each of the linkage groups, the genotyping data of the three populations were combined into a single file. To analyze the genotyping data of the backcross populations, together with data from the WAU population, the genotypes for these two populations were recoded as either being 1:1 (homozygous) or 1:2 (heterozygous). The combined data therefore consisted of 12 individual families, 1 EL, 1C, and 10 W. Linkage analysis was performed using CRIMAP version 2.4 (Green et al. 1990). Initially, a two-point linkage analysis was performed in which all markers were analyzed against each other. Tables containing all two-point lod scores for all markers are available at the web site of the Animal Breeding and Genetics Group in Wageningen. When possible, markers that had been typed on all three maps were used to start building the map using the CRIMAP-BUILD option. Finally, the order of the framework loci was checked using the CRIMAP-flips5 function.
Acknowledgments
We thank our many colleagues that have contributed to the mapping of the loci that are described in this paper, including R. Acar, S. Ambady, R. Bergé, D. Burke, R. Crooijmans, D. Dawson, E. Dufour, R. Dijkhof, M. Gibbs, O. Hanotte, L. Heltemes, J. Herbergs, B. Van Hest, J. Hillel, H. Khatib, C. Knorr, I. Levin, S. McConnell, C. Moran, M. Morisson, R. Okimoto, J. Palyga, F. Pitel, B.J. Sheldon, E.J. Smith, M. Siwek, C. Spike, S. Suchyta, P. Thomson, A. Toye, R.J. Vallejo, T. Veenendaal, B.J. van Hest, and A. Wardle.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL martien.groenen@alg.vf.wau.nl; FAX (0031) 317 483929.
REFERENCES
- Andersson L, Archibald A, Ashburner M, Audun S, Barendse W, Bitgood J, Bottema C, Broad T, Brown S, Burt D, et al. Comparative genome organization of vertebrates. The First International Workshop on Comparative Genome Organization. Mamm Genome. 1996;7:717–734. doi: 10.1007/s003359900222. [DOI] [PubMed] [Google Scholar]
- Angrist M. Less is more: Compact genomes pay dividends. Genome Res. 1998;8:683–685. doi: 10.1101/gr.8.7.683. [DOI] [PubMed] [Google Scholar]
- Bloom S E, Delaney M E, Muscarella D E. Constant and variable features of avian chromosomes. In: Etches RJ, Gibbins AMV, editors. Manipulation of the avian genome. Boca Raton, FL: CRC Press; 1993. pp. 39–60. [Google Scholar]
- Bumstead N, Palyga J. A preliminary linkage map of the chicken genome. Genomics. 1992;13:690–697. doi: 10.1016/0888-7543(92)90143-g. [DOI] [PubMed] [Google Scholar]
- Burt DW, Bruley C, Dunn IC, Jones CT, Ramage A, Law AS, Morrice DR, Paton IR, Smith J, Windsor D, Sazanov A, Fries R, Waddington D. The dynamics of chromosome evolution in birds and mammals. Nature. 1999;402:411–413. doi: 10.1038/46555. [DOI] [PubMed] [Google Scholar]
- Burt DW, Bumstead N, Bitgood JJ, Ponce de Leon FA, Crittenden LB. Chicken genome mapping: A new era in avian genetics. Trends Genet. 1995;11:190–194. doi: 10.1016/s0168-9525(00)89042-3. [DOI] [PubMed] [Google Scholar]
- Cheng HH, Levin I, Vallejo RL, Khatib H, Dodgson JB, Crittenden LB, Hillel J. Development of a genetic map of the chicken with markers of high utility. Poultry Sci. 1995;74:1855–1874. doi: 10.3382/ps.0741855. [DOI] [PubMed] [Google Scholar]
- Clark MS, Edwards YJK, McQueen HA, Meek SE, Smith S, Umrania Y, Warner S, Williams G, Elgar G. Sequence scanning chicken cosmids: A methodology for genome screening. Gene. 1999;227:223–230. doi: 10.1016/s0378-1119(98)00610-6. [DOI] [PubMed] [Google Scholar]
- Crittenden LB, Provencher L, Levin I, Abplanalp H, Briles RW, Briles WE, Dodgson JB. Characterization of a red jungle fowl by white leghorn backcross reference population for molecular mapping of the chicken genome. Poultry Sci. 1993;72:334–348. [Google Scholar]
- Crooijmans RPMA, Dijkhof RJM, van der Poel JJ, Groenen MAM. New microsatellite markers in chicken optimised for automated fluorescent genotyping. Anim Genet. 1997;28:427–437. doi: 10.1111/j.1365-2052.1997.00205.x. [DOI] [PubMed] [Google Scholar]
- Fillon V, Morisson M, Zoorob R, Auffray C, Douaire M, Gellin J, Vignal A. Identification of 16 chicken microchromosomes by molecular markers using two-colour fluorescence in situ hybridization (FISH) Chrom Res. 1998;6:307–313. doi: 10.1023/a:1009274925051. [DOI] [PubMed] [Google Scholar]
- Fridolfsson A-K, Cheng H, Copeland NG, Jenkins NA, Liu H-C, Raudsepp T, Woodage T, Chowdhary B, Halverson J, Ellegren H. Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc Natl Acad Sci. 1998;95:8147–8152. doi: 10.1073/pnas.95.14.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs M, Dawson DA, McCamley C, Wardle AF, Armour JAL, Burke T. Chicken microsatellite markers isolated from libraries enriched for simple tandem repeats. Anim Genet. 1997;28:401–417. [PubMed] [Google Scholar]
- Gilley J, Armes N, Fried M. Fugu genome is not a good mammalian model. Nature. 1997;385:305–306. doi: 10.1038/385305a0. [DOI] [PubMed] [Google Scholar]
- Green P, Falls K, Crooks S. Documentation for CRI-MAP, version 2.4. St. Louis, MO: Washington School of Medicine; 1990. [Google Scholar]
- Groenen MAM, Crooijmans RPMA, Veenendaal A, Cheng HH, Siwek M, Van der Poel JJ. A comprehensive microsatellite linkage map of the chicken genome. Genomics. 1998;49:265–274. doi: 10.1006/geno.1998.5225. [DOI] [PubMed] [Google Scholar]
- Groenen MAM, Crooijmans RPMA, Dijkhof RJM, Acar R, van der Poel JJ. Extending the chicken-human comparative map by placing 15 genes on the chicken linkage map. Anim Genet. 1999;30:418–422. doi: 10.1046/j.1365-2052.1999.00527.x. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. Sex ratio and unisexual sterility in hybrid animals. J Genet. 1922;12:101–109. [Google Scholar]
- Herbergs J, Siwek M, Crooijmans RPMA, van der Poel JJ, Groenen MAM. Multicolour fluorescent detection and mapping of AFLP markers in chicken (Gallus domesticus) Anim Genet. 1999;30:274–285. doi: 10.1046/j.1365-2052.1999.00494.x. [DOI] [PubMed] [Google Scholar]
- Hu J, Bumstead N, Burke D, Ponce de Leon FA, Skamene E, Gros P, Malo D. Genetic and physical mapping of the natural resistance-associated macrophage protein 1 (NRAMP1) in chicken. Mamm Genome. 1995;6:809–815. doi: 10.1007/BF00539010. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Hughes MK. Small genomes for better flyers. Nature. 1995;377:391. doi: 10.1038/377391a0. [DOI] [PubMed] [Google Scholar]
- Hutt FB. Genetics of the fowl VI. A tentative chromosome map. Neue Forschungen in Tierzucht und Abstammungslehre. 1936. (Duerst Festschrift) pp. 105–112. [Google Scholar]
- Jones CT, Morrice DR, Paton IR, Burt DW. Gene homologs on human chromosome 15q21–q26 and a chicken microchromosome identify a new conserved segment. Mamm Genome. 1997;8:436–440. doi: 10.1007/s003359900463. [DOI] [PubMed] [Google Scholar]
- Keats BJB, Sherman SL, Morton NE, Robson EB, Buetow KH, Cartwright PE, Chakravarti A, Francke U, Green PP, Ott J. Guidelines for human linkage maps; an international system for human linkage maps. Genomics. 1991;9:557–560. doi: 10.1016/0888-7543(91)90426-f. [DOI] [PubMed] [Google Scholar]
- Klein S, Morrice DR, Sang H, Crittenden LB, Burt DW. Genetic and physical mapping of the chicken IGF1 gene to chromosome 1 and conservation of synteny with other vertebrate genomes. J Hered. 1996;87:10–14. doi: 10.1093/oxfordjournals.jhered.a022946. [DOI] [PubMed] [Google Scholar]
- Knorr C, Cheng HH, Dodgson JB. Application of AFLP markers to genome mapping in poultry. Anim Genet. 1999;30:28–35. doi: 10.1046/j.1365-2052.1999.00411.x. [DOI] [PubMed] [Google Scholar]
- Levin I, Crittenden LB, Dodgsen JB. Genetic map of the chicken Z chromosome using random amplified polymorphic DNA (RAPD) markers. Genomics. 1993;16:224–230. doi: 10.1006/geno.1993.1163. [DOI] [PubMed] [Google Scholar]
- Levin I, Santagelo L, Cheng H, Crittenden LB, Dodgsen JB. An autosomal genetic linkage map of the chicken. J Hered. 1994;85:79–85. doi: 10.1093/oxfordjournals.jhered.a111427. [DOI] [PubMed] [Google Scholar]
- McQueen HA, Siriaco G, Bird AP. Chicken microchromosomes are hyperacetylated, early replicating, and gene rich. Genome Res. 1998;8:621–630. doi: 10.1101/gr.8.6.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisson M, Pitel F, Fillon V, Pouzadoux A, Bergé R, Vit JP, Zoorob R, Auffray C, Gellin J, Vignal A. Integration of chicken cytogenetic and genetic maps:18 new polymorphic markers isolated from BAC and PAC clones. Anim Genet. 1998;29:348–355. doi: 10.1046/j.1365-2052.1998.295348.x. [DOI] [PubMed] [Google Scholar]
- Nanda I, Shan Z, Schartl M, Burt DW, Koehler M, Nothwang H-G, Grutzner F, Paton IR, Windsor D, Dunn I, et al. 300 million years of conserved synteny between chicken Z and human chromosome 9. Nat Genet. 1999;21:258–259. doi: 10.1038/6769. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ. Mammalian genome mapping: Lessons and prospects. Curr Opin Genet Dev. 1991;1:105–111. doi: 10.1016/0959-437x(91)80050-v. [DOI] [PubMed] [Google Scholar]
- Primmer CR, Raudsepp T, Chowdhary BP, Moller AP, Ellegren H. Low frequency of microsatellites in the avian genome. Genome Res. 1997;7:471–482. doi: 10.1101/gr.7.5.471. [DOI] [PubMed] [Google Scholar]
- Reboul J, Gardiner K, Monneron D, Uzé G, Lutfalla G. Comparative genomic analysis of the interferon/interleukin-10 receptor gene cluster. Genome Res. 1999;9:242–250. [PMC free article] [PubMed] [Google Scholar]
- Rodionov AV, Myakoshina YA, Chelysheva LA, Solovei IV, Gaginskaya ER. Chiasmata in the lambrush chromosomes of Gallus Gallus domesticus: The cytogenetic study of recombination frequency and linkage map lengths. Genetika. 1992;28:53–63. [Google Scholar]
- Spike CS, Lamont SJ. Genetic analysis of three loci homologous to human G9a: Evidence for linkage of a class III gene with the chicken MHC. Anim Genet. 1995;26:185–187. doi: 10.1111/j.1365-2052.1995.tb03160.x. [DOI] [PubMed] [Google Scholar]
- Tereba A, McPhaul MJ, Wilson JD. The gene for aromatase (p450 arom) in the chicken is located on the long arm of chromosome 1. J Hered. 1991;82:80–81. doi: 10.1093/jhered/82.1.80. [DOI] [PubMed] [Google Scholar]
- Tiersch TR, Wachtel SS. On the evolution of genome size in birds. J Hered. 1991;82:363–368. doi: 10.1093/oxfordjournals.jhered.a111105. [DOI] [PubMed] [Google Scholar]
- Toye AA, Bumstead N, Moran C. Chicken solute carrier family 4 anion exchanger member 1 (SLC4A1): Microsatellite polymorphism and mapping. Anim Genet. 1997;28:316–317. [PubMed] [Google Scholar]