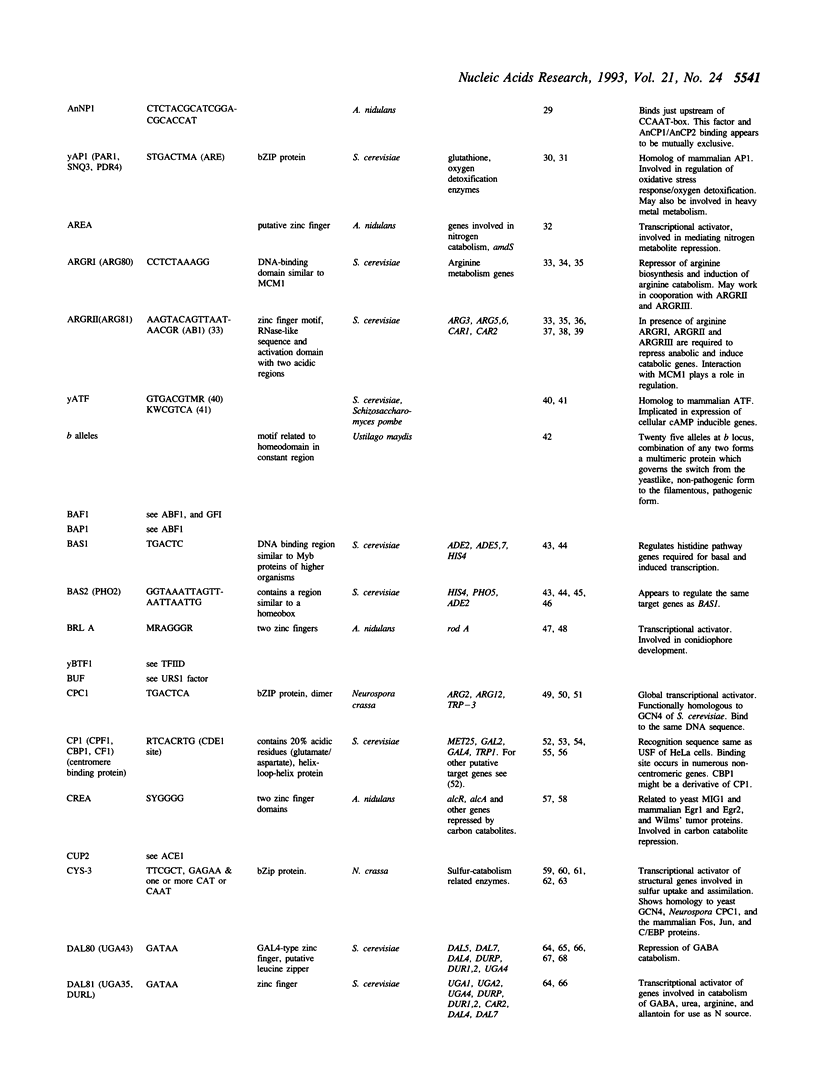
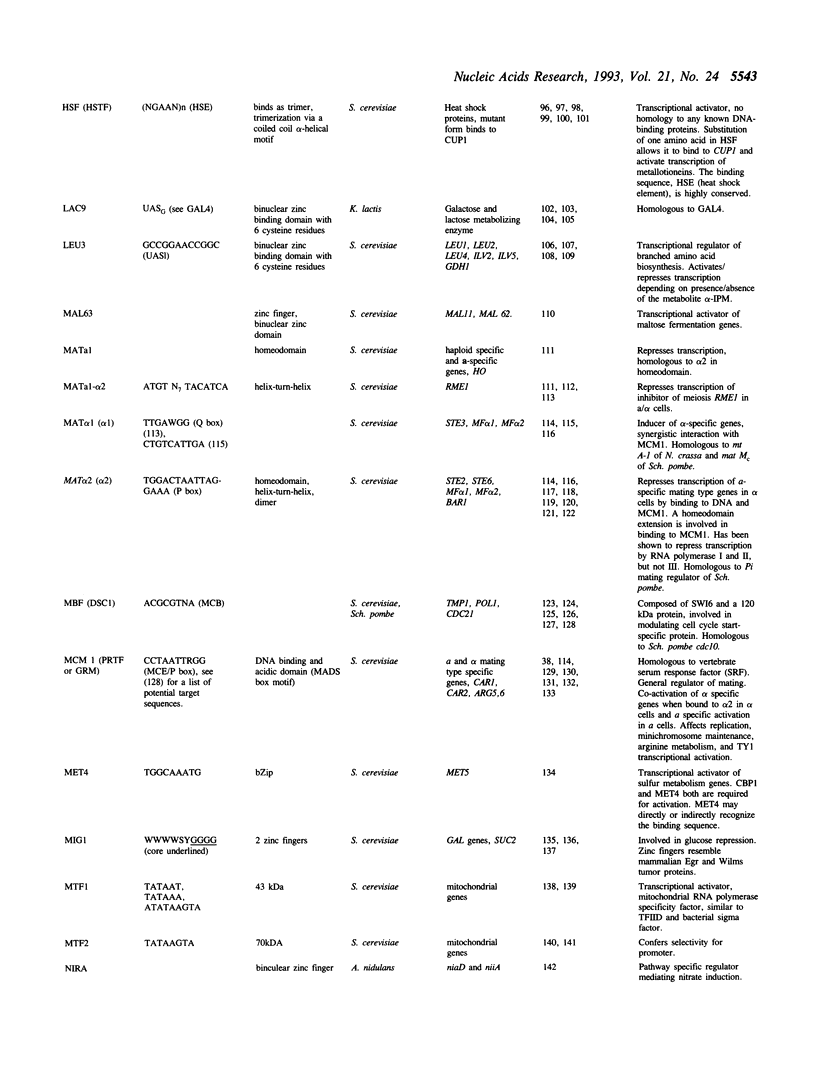
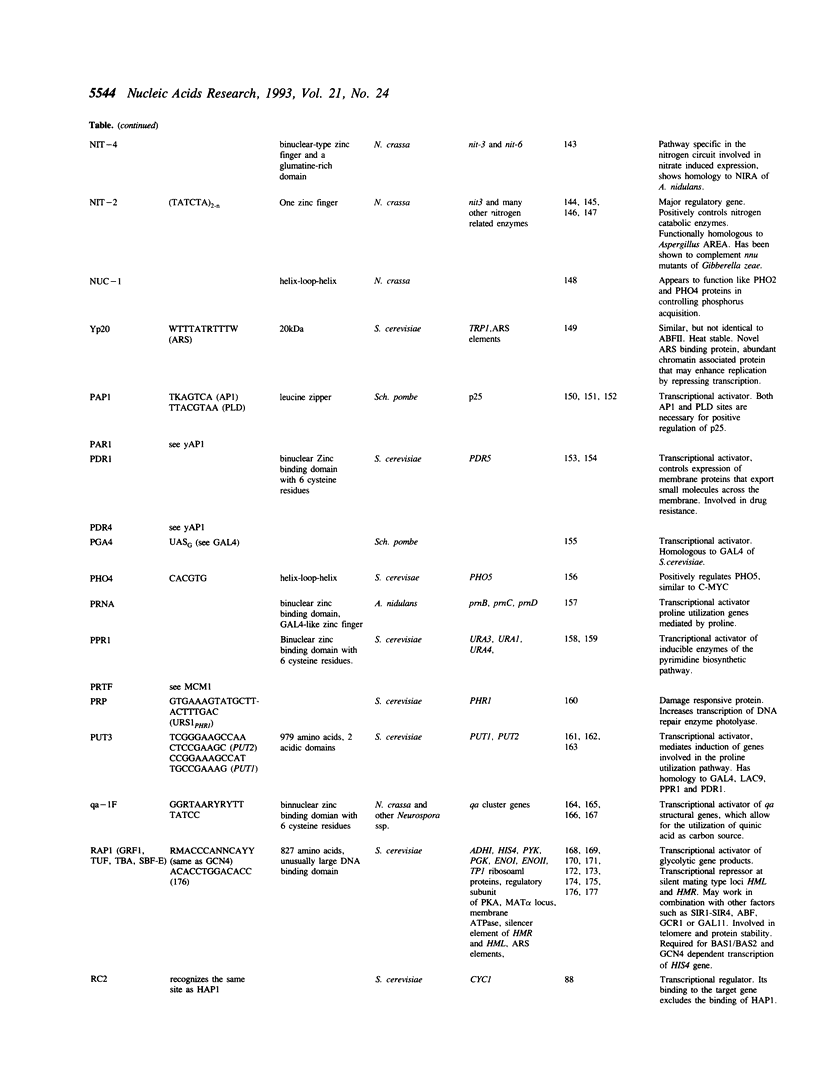
Full text
PDF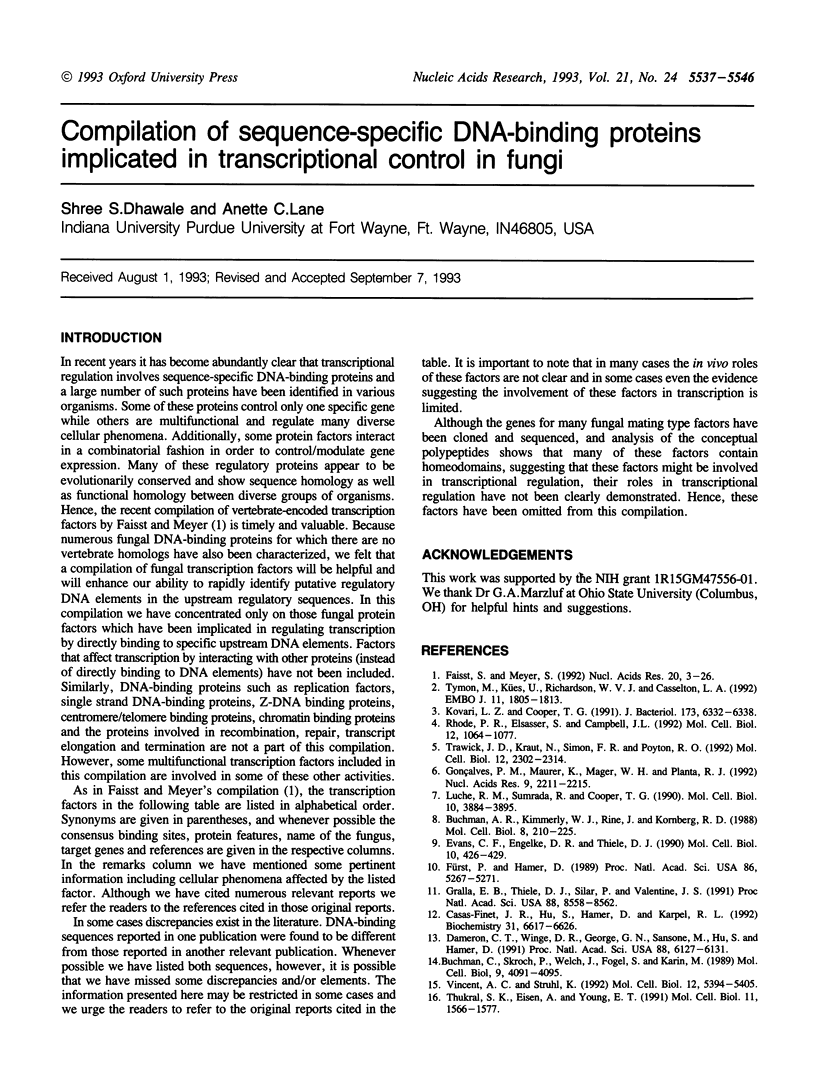





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams T. H., Deising H., Timberlake W. E. brlA requires both zinc fingers to induce development. Mol Cell Biol. 1990 Apr;10(4):1815–1817. doi: 10.1128/mcb.10.4.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerer G. Identification, purification, and cloning of a polypeptide (PRTF/GRM) that binds to mating-specific promoter elements in yeast. Genes Dev. 1990 Feb;4(2):299–312. doi: 10.1101/gad.4.2.299. [DOI] [PubMed] [Google Scholar]
- Andrianopoulos A., Hynes M. J. Cloning and analysis of the positively acting regulatory gene amdR from Aspergillus nidulans. Mol Cell Biol. 1988 Aug;8(8):3532–3541. doi: 10.1128/mcb.8.8.3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- André B., Hein C., Grenson M., Jauniaux J. C. Cloning and expression of the UGA4 gene coding for the inducible GABA-specific transport protein of Saccharomyces cerevisiae. Mol Gen Genet. 1993 Feb;237(1-2):17–25. doi: 10.1007/BF00282779. [DOI] [PubMed] [Google Scholar]
- André B. The UGA3 gene regulating the GABA catabolic pathway in Saccharomyces cerevisiae codes for a putative zinc-finger protein acting on RNA amount. Mol Gen Genet. 1990 Jan;220(2):269–276. doi: 10.1007/BF00260493. [DOI] [PubMed] [Google Scholar]
- Archambault J., Milne C. A., Schappert K. T., Baum B., Friesen J. D., Segall J. The deduced sequence of the transcription factor TFIIIA from Saccharomyces cerevisiae reveals extensive divergence from Xenopus TFIIIA. J Biol Chem. 1992 Feb 15;267(5):3282–3288. [PubMed] [Google Scholar]
- Arndt K., Fink G. R. GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5' TGACTC 3' sequences. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8516–8520. doi: 10.1073/pnas.83.22.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asch D. K., Orejas M., Geever R. F., Case M. E. Comparative studies of the quinic acid (qa) cluster in several Neurospora species with special emphasis on the qa-x-qa-2 intergenic region. Mol Gen Genet. 1991 Dec;230(3):337–344. doi: 10.1007/BF00280289. [DOI] [PubMed] [Google Scholar]
- Baker H. V. GCR1 of Saccharomyces cerevisiae encodes a DNA binding protein whose binding is abolished by mutations in the CTTCC sequence motif. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9443–9447. doi: 10.1073/pnas.88.21.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R. E., Masison D. C. Isolation of the gene encoding the Saccharomyces cerevisiae centromere-binding protein CP1. Mol Cell Biol. 1990 Jun;10(6):2458–2467. doi: 10.1128/mcb.10.6.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzi E., Chen W., Ulaszewski S., Capieaux E., Goffeau A. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J Biol Chem. 1987 Dec 15;262(35):16871–16879. [PubMed] [Google Scholar]
- Bartholomew B., Kassavetis G. A., Braun B. R., Geiduschek E. P. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J. 1990 Jul;9(7):2197–2205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B., Kassavetis G. A., Geiduschek E. P. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol Cell Biol. 1991 Oct;11(10):5181–5189. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartuv J., Valansi C., Shalitin C. Characterization of DNA binding properties of Yp20: an abundant nuclear protein isolated from Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1993 Mar 15;191(2):750–758. doi: 10.1006/bbrc.1993.1281. [DOI] [PubMed] [Google Scholar]
- Baum J. A., Geever R., Giles N. H. Expression of qa-1F activator protein: identification of upstream binding sites in the qa gene cluster and localization of the DNA-binding domain. Mol Cell Biol. 1987 Mar;7(3):1256–1266. doi: 10.1128/mcb.7.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg H., Eisen A., Sledziewski A., Bader D., Young E. T. Two zinc fingers of a yeast regulatory protein shown by genetic evidence to be essential for its function. 1987 Jul 30-Aug 5Nature. 328(6129):443–445. doi: 10.1038/328443a0. [DOI] [PubMed] [Google Scholar]
- Blumberg H., Silver P. A split zinc-finger protein is required for normal yeast growth. Gene. 1991 Oct 30;107(1):101–110. doi: 10.1016/0378-1119(91)90302-r. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Isolation of a Saccharomyces cerevisiae centromere DNA-binding protein, its human homolog, and its possible role as a transcription factor. Mol Cell Biol. 1987 Jan;7(1):403–409. doi: 10.1128/mcb.7.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden L., Nasmyth K. Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell. 1987 Feb 13;48(3):389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Kimmerly W. J., Rine J., Kornberg R. D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jan;8(1):210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Lue N. F., Kornberg R. D. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol. 1988 Dec;8(12):5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman C., Skroch P., Welch J., Fogel S., Karin M. The CUP2 gene product, regulator of yeast metallothionein expression, is a copper-activated DNA-binding protein. Mol Cell Biol. 1989 Sep;9(9):4091–4095. doi: 10.1128/mcb.9.9.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G., Strauss J., Scazzocchio C., Lang B. F. nirA, the pathway-specific regulatory gene of nitrate assimilation in Aspergillus nidulans, encodes a putative GAL4-type zinc finger protein and contains four introns in highly conserved regions. Mol Cell Biol. 1991 Nov;11(11):5746–5755. doi: 10.1128/mcb.11.11.5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin T. R. The yeast regulatory gene PHO2 encodes a homeo box. Cell. 1988 May 6;53(3):339–340. doi: 10.1016/0092-8674(88)90153-5. [DOI] [PubMed] [Google Scholar]
- Cai M. J., Davis R. W. Purification of a yeast centromere-binding protein that is able to distinguish single base-pair mutations in its recognition site. Mol Cell Biol. 1989 Jun;9(6):2544–2550. doi: 10.1128/mcb.9.6.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M., Davis R. W. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990 May 4;61(3):437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- Campbell W. H., Kinghorn K. R. Functional domains of assimilatory nitrate reductases and nitrite reductases. Trends Biochem Sci. 1990 Aug;15(8):315–319. doi: 10.1016/0968-0004(90)90021-3. [DOI] [PubMed] [Google Scholar]
- Casas-Finet J. R., Hu S., Hamer D., Karpel R. L. Characterization of the copper- and silver-thiolate clusters in N-terminal fragments of the yeast ACE1 transcription factor capable of binding to its specific DNA recognition sequence. Biochemistry. 1992 Jul 21;31(28):6617–6626. doi: 10.1021/bi00143a036. [DOI] [PubMed] [Google Scholar]
- Cavallini B., Faus I., Matthes H., Chipoulet J. M., Winsor B., Egly J. M., Chambon P. Cloning of the gene encoding the yeast protein BTF1Y, which can substitute for the human TATA box-binding factor. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9803–9807. doi: 10.1073/pnas.86.24.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challice J. M., Segall J. Transcription of the 5 S rRNA gene of Saccharomyces cerevisiae requires a promoter element at +1 and a 14-base pair internal control region. J Biol Chem. 1989 Nov 25;264(33):20060–20067. [PubMed] [Google Scholar]
- Chambers A., Tsang J. S., Stanway C., Kingsman A. J., Kingsman S. M. Transcriptional control of the Saccharomyces cerevisiae PGK gene by RAP1. Mol Cell Biol. 1989 Dec;9(12):5516–5524. doi: 10.1128/mcb.9.12.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C., Timberlake W. E. Identification of Aspergillus brlA response elements (BREs) by genetic selection in yeast. Genetics. 1993 Jan;133(1):29–38. doi: 10.1093/genetics/133.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ C., Tye B. K. Functional domains of the yeast transcription/replication factor MCM1. Genes Dev. 1991 May;5(5):751–763. doi: 10.1101/gad.5.5.751. [DOI] [PubMed] [Google Scholar]
- Coornaert D., Vissers S., André B., Grenson M. The UGA43 negative regulatory gene of Saccharomyces cerevisiae contains both a GATA-1 type zinc finger and a putative leucine zipper. Curr Genet. 1992 Apr;21(4-5):301–307. doi: 10.1007/BF00351687. [DOI] [PubMed] [Google Scholar]
- Coornaert D., Vissers S., André B. The pleiotropic UGA35(DURL) regulatory gene of Saccharomyces cerevisiae: cloning, sequence and identity with the DAL81 gene. Gene. 1991 Jan 15;97(2):163–171. doi: 10.1016/0378-1119(91)90048-g. [DOI] [PubMed] [Google Scholar]
- Covitz P. A., Herskowitz I., Mitchell A. P. The yeast RME1 gene encodes a putative zinc finger protein that is directly repressed by a1-alpha 2. Genes Dev. 1991 Nov;5(11):1982–1989. doi: 10.1101/gad.5.11.1982. [DOI] [PubMed] [Google Scholar]
- Covitz P. A., Herskowitz I., Mitchell A. P. The yeast RME1 gene encodes a putative zinc finger protein that is directly repressed by a1-alpha 2. Genes Dev. 1991 Nov;5(11):1982–1989. doi: 10.1101/gad.5.11.1982. [DOI] [PubMed] [Google Scholar]
- Cunningham T. S., Cooper T. G. Expression of the DAL80 gene, whose product is homologous to the GATA factors and is a negative regulator of multiple nitrogen catabolic genes in Saccharomyces cerevisiae, is sensitive to nitrogen catabolite repression. Mol Cell Biol. 1991 Dec;11(12):6205–6215. doi: 10.1128/mcb.11.12.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daignan-Fornier B., Fink G. R. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):6746–6750. doi: 10.1073/pnas.89.15.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dameron C. T., Winge D. R., George G. N., Sansone M., Hu S., Hamer D. A copper-thiolate polynuclear cluster in the ACE1 transcription factor. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6127–6131. doi: 10.1073/pnas.88.14.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty J. R., Rai R., el Berry H. M., Cooper T. G. Regulatory circuit for responses of nitrogen catabolic gene expression to the GLN3 and DAL80 proteins and nitrogen catabolite repression in Saccharomyces cerevisiae. J Bacteriol. 1993 Jan;175(1):64–73. doi: 10.1128/jb.175.1.64-73.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. A., Hynes M. J. Regulatory genes in Aspergillus nidulans. Trends Genet. 1989 Jan;5(1):14–19. doi: 10.1016/0168-9525(89)90006-1. [DOI] [PubMed] [Google Scholar]
- De Rijcke M., Seneca S., Punyammalee B., Glansdorff N., Crabeel M. Characterization of the DNA target site for the yeast ARGR regulatory complex, a sequence able to mediate repression or induction by arginine. Mol Cell Biol. 1992 Jan;12(1):68–81. doi: 10.1128/mcb.12.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin C., Tice-Baldwin K., Shore D., Arndt K. T. RAP1 is required for BAS1/BAS2- and GCN4-dependent transcription of the yeast HIS4 gene. Mol Cell Biol. 1991 Jul;11(7):3642–3651. doi: 10.1128/mcb.11.7.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickman M. B., Leslie J. F. The regulatory gene nit-2 of Neurospora crassa complements a nnu mutant of Gibberella zeae (Fusarium graminearum). Mol Gen Genet. 1992 Nov;235(2-3):458–462. doi: 10.1007/BF00279394. [DOI] [PubMed] [Google Scholar]
- Dirick L., Moll T., Auer H., Nasmyth K. A central role for SWI6 in modulating cell cycle Start-specific transcription in yeast. Nature. 1992 Jun 11;357(6378):508–513. doi: 10.1038/357508a0. [DOI] [PubMed] [Google Scholar]
- Dolan J. W., Kirkman C., Fields S. The yeast STE12 protein binds to the DNA sequence mediating pheromone induction. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5703–5707. doi: 10.1073/pnas.86.15.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald R. G., Schindler U., Batschauer A., Cashmore A. R. The plant G box promoter sequence activates transcription in Saccharomyces cerevisiae and is bound in vitro by a yeast activity similar to GBF, the plant G box binding factor. EMBO J. 1990 Jun;9(6):1727–1735. doi: 10.1002/j.1460-2075.1990.tb08296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsman J. C., van Heeswijk W. C., Grivell L. A. Yeast general transcription factor GFI: sequence requirements for binding to DNA and evolutionary conservation. Nucleic Acids Res. 1990 May 11;18(9):2769–2776. doi: 10.1093/nar/18.9.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowzer C. E., Kelly J. M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol Cell Biol. 1991 Nov;11(11):5701–5709. doi: 10.1128/mcb.11.11.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois E., Bercy J., Descamps F., Messenguy F. Characterization of two new genes essential for vegetative growth in Saccharomyces cerevisiae: nucleotide sequence determination and chromosome mapping. Gene. 1987;55(2-3):265–275. doi: 10.1016/0378-1119(87)90286-1. [DOI] [PubMed] [Google Scholar]
- Dubois E., Bercy J., Messenguy F. Characterization of two genes, ARGRI and ARGRIII required for specific regulation of arginine metabolism in yeast. Mol Gen Genet. 1987 Apr;207(1):142–148. doi: 10.1007/BF00331501. [DOI] [PubMed] [Google Scholar]
- Dubois E., Messenguy F. In vitro studies of the binding of the ARGR proteins to the ARG5,6 promoter. Mol Cell Biol. 1991 Apr;11(4):2162–2168. doi: 10.1128/mcb.11.4.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbole D. J., Paluh J. L., Plamann M., Sachs M. S., Yanofsky C. cpc-1, the general regulatory gene for genes of amino acid biosynthesis in Neurospora crassa, is differentially expressed during the asexual life cycle. Mol Cell Biol. 1991 Feb;11(2):928–934. doi: 10.1128/mcb.11.2.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble R., Tye B. K. Both activation and repression of a-mating-type-specific genes in yeast require transcription factor Mcm1. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10966–10970. doi: 10.1073/pnas.88.23.10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errede B., Ammerer G. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 1989 Sep;3(9):1349–1361. doi: 10.1101/gad.3.9.1349. [DOI] [PubMed] [Google Scholar]
- Errede B. MCM1 binds to a transcriptional control element in Ty1. Mol Cell Biol. 1993 Jan;13(1):57–62. doi: 10.1128/mcb.13.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch F. The yeast putative transcriptional repressor RGM1 is a proline-rich zinc finger protein. Nucleic Acids Res. 1991 Sep 25;19(18):4873–4877. doi: 10.1093/nar/19.18.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans C. F., Engelke D. R. Binding of yeast TFIIIC to tRNA gene bipartite internal promoters: analysis of physical effects on the intervening DNA. Biochem Biophys Res Commun. 1991 Apr 30;176(2):826–832. doi: 10.1016/s0006-291x(05)80260-8. [DOI] [PubMed] [Google Scholar]
- Evans C. F., Engelke D. R., Thiele D. J. ACE1 transcription factor produced in Escherichia coli binds multiple regions within yeast metallothionein upstream activation sequences. Mol Cell Biol. 1990 Jan;10(1):426–429. doi: 10.1128/mcb.10.1.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faisst S., Meyer S. Compilation of vertebrate-encoded transcription factors. Nucleic Acids Res. 1992 Jan 11;20(1):3–26. doi: 10.1093/nar/20.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaver W. J., Gileadi O., Kornberg R. D. Purification and characterization of yeast RNA polymerase II transcription factor b. J Biol Chem. 1991 Oct 5;266(28):19000–19005. [PubMed] [Google Scholar]
- Fikes J. D., Becker D. M., Winston F., Guarente L. Striking conservation of TFIID in Schizosaccharomyces pombe and Saccharomyces cerevisiae. Nature. 1990 Jul 19;346(6281):291–294. doi: 10.1038/346291a0. [DOI] [PubMed] [Google Scholar]
- Fisher F., Jayaraman P. S., Goding C. R. C-myc and the yeast transcription factor PHO4 share a common CACGTG-binding motif. Oncogene. 1991 Jul;6(7):1099–1104. [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989 Aug;3(8):1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- Fu Y. H., Marzluf G. A. Characterization of nit-2, the major nitrogen regulatory gene of Neurospora crassa. Mol Cell Biol. 1987 May;7(5):1691–1696. doi: 10.1128/mcb.7.5.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. H., Marzluf G. A. cys-3, the positive-acting sulfur regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. J Biol Chem. 1990 Jul 15;265(20):11942–11947. [PubMed] [Google Scholar]
- Fu Y. H., Marzluf G. A. nit-2, the major nitrogen regulatory gene of Neurospora crassa, encodes a protein with a putative zinc finger DNA-binding domain. Mol Cell Biol. 1990 Mar;10(3):1056–1065. doi: 10.1128/mcb.10.3.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. H., Marzluf G. A. nit-2, the major positive-acting nitrogen regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5331–5335. doi: 10.1073/pnas.87.14.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. H., Paietta J. V., Mannix D. G., Marzluf G. A. cys-3, the positive-acting sulfur regulatory gene of Neurospora crassa, encodes a protein with a putative leucine zipper DNA-binding element. Mol Cell Biol. 1989 Mar;9(3):1120–1127. doi: 10.1128/mcb.9.3.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita A., Kikuchi Y., Kuhara S., Misumi Y., Matsumoto S., Kobayashi H. Domains of the SFL1 protein of yeasts are homologous to Myc oncoproteins or yeast heat-shock transcription factor. Gene. 1989 Dec 28;85(2):321–328. doi: 10.1016/0378-1119(89)90424-1. [DOI] [PubMed] [Google Scholar]
- Fürst P., Hamer D. Cooperative activation of a eukaryotic transcription factor: interaction between Cu(I) and yeast ACE1 protein. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5267–5271. doi: 10.1073/pnas.86.14.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner K. H., Pan T., Narula S., Rivera E., Coleman J. E. Structure of the binuclear metal-binding site in the GAL4 transcription factor. Biochemistry. 1991 Nov 26;30(47):11292–11302. doi: 10.1021/bi00111a015. [DOI] [PubMed] [Google Scholar]
- Geever R. F., Huiet L., Baum J. A., Tyler B. M., Patel V. B., Rutledge B. J., Case M. E., Giles N. H. DNA sequence, organization and regulation of the qa gene cluster of Neurospora crassa. J Mol Biol. 1989 May 5;207(1):15–34. doi: 10.1016/0022-2836(89)90438-5. [DOI] [PubMed] [Google Scholar]
- Glass N. L., Grotelueschen J., Metzenberg R. L. Neurospora crassa A mating-type region. Proc Natl Acad Sci U S A. 1990 Jul;87(13):4912–4916. doi: 10.1073/pnas.87.13.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves P. M., Maurer K., Mager W. H., Planta R. J. Kluyveromyces contains a functional ABF1-homologue. Nucleic Acids Res. 1992 May 11;20(9):2211–2215. doi: 10.1093/nar/20.9.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutte C., Johnson A. D. a1 protein alters the DNA binding specificity of alpha 2 repressor. Cell. 1988 Mar 25;52(6):875–882. doi: 10.1016/0092-8674(88)90429-1. [DOI] [PubMed] [Google Scholar]
- Goyer C., Lee H. S., Malo D., Sonenberg N. Isolation of a yeast gene encoding a protein homologous to the human Tat-binding protein TBP-1. DNA Cell Biol. 1992 Oct;11(8):579–585. doi: 10.1089/dna.1992.11.579. [DOI] [PubMed] [Google Scholar]
- Gralla E. B., Thiele D. J., Silar P., Valentine J. S. ACE1, a copper-dependent transcription factor, activates expression of the yeast copper, zinc superoxide dismutase gene. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8558–8562. doi: 10.1073/pnas.88.19.8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., Buratowski S., Sharp P. A., Guarente L. Identification of a yeast protein homologous in function to the mammalian general transcription factor, TFIIA. EMBO J. 1989 Nov;8(11):3379–3382. doi: 10.1002/j.1460-2075.1989.tb08501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanas J. S., Gaskins C. J., Smith J. F., Ogilvie M. K. Structure, function, evolution of transcription factor IIIA. Prog Nucleic Acid Res Mol Biol. 1992;43:205–239. doi: 10.1016/s0079-6603(08)61048-x. [DOI] [PubMed] [Google Scholar]
- Hardy C. F., Balderes D., Shore D. Dissection of a carboxy-terminal region of the yeast regulatory protein RAP1 with effects on both transcriptional activation and silencing. Mol Cell Biol. 1992 Mar;12(3):1209–1217. doi: 10.1128/mcb.12.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy C. F., Sussel L., Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992 May;6(5):801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- Harshman K. D., Moye-Rowley W. S., Parker C. S. Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4. Cell. 1988 Apr 22;53(2):321–330. doi: 10.1016/0092-8674(88)90393-5. [DOI] [PubMed] [Google Scholar]
- Henry Y. A., Chambers A., Tsang J. S., Kingsman A. J., Kingsman S. M. Characterisation of the DNA binding domain of the yeast RAP1 protein. Nucleic Acids Res. 1990 May 11;18(9):2617–2623. doi: 10.1093/nar/18.9.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschbach B. M., Johnson A. D. The yeast alpha 2 protein can repress transcription by RNA polymerases I and II but not III. Mol Cell Biol. 1993 Jul;13(7):4029–4038. doi: 10.1128/mcb.13.7.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S., Huse K., Valentin E., Mbonyi K., Thevelein J. M., Zimmermann F. K. Glucose-induced regulatory defects in the Saccharomyces cerevisiae byp1 growth initiation mutant and identification of MIG1 as a partial suppressor. J Bacteriol. 1992 Jun;174(12):4183–4188. doi: 10.1128/jb.174.12.4183-4188.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi M., Wang C. K., Fujii H., Cromlish J. A., Weil P. A., Roeder R. G. Cloning and structure of a yeast gene encoding a general transcription initiation factor TFIID that binds to the TATA box. Nature. 1989 Sep 28;341(6240):299–303. doi: 10.1038/341299a0. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Yamamoto T., Ohkuma Y., Weil P. A., Roeder R. G. Analysis of structure-function relationships of yeast TATA box binding factor TFIID. Cell. 1990 Jun 29;61(7):1171–1178. doi: 10.1016/0092-8674(90)90681-4. [DOI] [PubMed] [Google Scholar]
- Huie M. A., Scott E. W., Drazinic C. M., Lopez M. C., Hornstra I. K., Yang T. P., Baker H. V. Characterization of the DNA-binding activity of GCR1: in vivo evidence for two GCR1-binding sites in the upstream activating sequence of TPI of Saccharomyces cerevisiae. Mol Cell Biol. 1992 Jun;12(6):2690–2700. doi: 10.1128/mcb.12.6.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S. H., Jaehning J. A. The yeast mitochondrial RNA polymerase specificity factor, MTF1, is similar to bacterial sigma factors. J Biol Chem. 1991 Nov 25;266(33):22671–22677. [PubMed] [Google Scholar]
- Jarvis E. E., Hagen D. C., Sprague G. F., Jr Identification of a DNA segment that is necessary and sufficient for alpha-specific gene control in Saccharomyces cerevisiae: implications for regulation of alpha-specific and a-specific genes. Mol Cell Biol. 1988 Jan;8(1):309–320. doi: 10.1128/mcb.8.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Herskowitz I. A repressor (MAT alpha 2 Product) and its operator control expression of a set of cell type specific genes in yeast. Cell. 1985 Aug;42(1):237–247. doi: 10.1016/s0092-8674(85)80119-7. [DOI] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. H., Jones N. C. Mammalian cAMP-responsive element can activate transcription in yeast and binds a yeast factor(s) that resembles the mammalian transcription factor ANF. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2176–2180. doi: 10.1073/pnas.86.7.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. H., Moreno S., Nurse P., Jones N. C. Expression of the SV40 promoter in fission yeast: identification and characterization of an AP-1-like factor. Cell. 1988 May 20;53(4):659–667. doi: 10.1016/0092-8674(88)90581-8. [DOI] [PubMed] [Google Scholar]
- Ju Q. D., Morrow B. E., Warner J. R. REB1, a yeast DNA-binding protein with many targets, is essential for growth and bears some resemblance to the oncogene myb. Mol Cell Biol. 1990 Oct;10(10):5226–5234. doi: 10.1128/mcb.10.10.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer B., Guyonvarch A., Hubert J. C. Yeast regulatory gene PPR1. I. Nucleotide sequence, restriction map and codon usage. J Mol Biol. 1984 Dec 5;180(2):239–250. doi: 10.1016/s0022-2836(84)80002-9. [DOI] [PubMed] [Google Scholar]
- Kanaan M. N., Marzluf G. A. Mutational analysis of the DNA-binding domain of the CYS3 regulatory protein of Neurospora crassa. Mol Cell Biol. 1991 Sep;11(9):4356–4362. doi: 10.1128/mcb.11.9.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Metzenberg R. L. Molecular analysis of nuc-1+, a gene controlling phosphorus acquisition in Neurospora crassa. Mol Cell Biol. 1990 Nov;10(11):5839–5848. doi: 10.1128/mcb.10.11.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Bartholomew B., Blanco J. A., Johnson T. E., Geiduschek E. P. Two essential components of the Saccharomyces cerevisiae transcription factor TFIIIB: transcription and DNA-binding properties. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7308–7312. doi: 10.1073/pnas.88.16.7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Braun B. R., Nguyen L. H., Geiduschek E. P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990 Jan 26;60(2):235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Riggs D. L., Negri R., Nguyen L. H., Geiduschek E. P. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989 Jun;9(6):2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher C. A., Goutte C., Johnson A. D. The yeast cell-type-specific repressor alpha 2 acts cooperatively with a non-cell-type-specific protein. Cell. 1988 Jun 17;53(6):927–936. doi: 10.1016/s0092-8674(88)90449-7. [DOI] [PubMed] [Google Scholar]
- Kelly M., Burke J., Smith M., Klar A., Beach D. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J. 1988 May;7(5):1537–1547. doi: 10.1002/j.1460-2075.1988.tb02973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keng T. HAP1 and ROX1 form a regulatory pathway in the repression of HEM13 transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Jun;12(6):2616–2623. doi: 10.1128/mcb.12.6.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Michels C. A. The MAL63 gene of Saccharomyces encodes a cysteine-zinc finger protein. Curr Genet. 1988 Oct;14(4):319–323. doi: 10.1007/BF00419988. [DOI] [PubMed] [Google Scholar]
- Kirkman-Correia C., Stroke I. L., Fields S. Functional domains of the yeast STE12 protein, a pheromone-responsive transcriptional activator. Mol Cell Biol. 1993 Jun;13(6):3765–3772. doi: 10.1128/mcb.13.6.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klekamp M. S., Weil P. A. Partial purification and characterization of the Saccharomyces cerevisiae transcription factor TFIIIB. J Biol Chem. 1986 Feb 25;261(6):2819–2827. [PubMed] [Google Scholar]
- Kobayashi N., McEntee K. Identification of cis and trans components of a novel heat shock stress regulatory pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Jan;13(1):248–256. doi: 10.1128/mcb.13.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovari L. Z., Cooper T. G. Participation of ABF-1 protein in expression of the Saccharomyces cerevisiae CAR1 gene. J Bacteriol. 1991 Oct;173(20):6332–6338. doi: 10.1128/jb.173.20.6332-6338.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuddus R., Schmidt M. C. Effect of the non-conserved N-terminus on the DNA binding activity of the yeast TATA binding protein. Nucleic Acids Res. 1993 Apr 25;21(8):1789–1796. doi: 10.1093/nar/21.8.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla B., Caddick M. X., Langdon T., Martinez-Rossi N. M., Bennett C. F., Sibley S., Davies R. W., Arst H. N., Jr The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 1990 May;9(5):1355–1364. doi: 10.1002/j.1460-2075.1990.tb08250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkens T., Riggs D. L., Heck J. D., Planta R. J., Nomura M. The yeast RNA polymerase I promoter: ribosomal DNA sequences involved in transcription initiation and complex formation in vitro. Nucleic Acids Res. 1991 Oct 11;19(19):5363–5370. doi: 10.1093/nar/19.19.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulmburg P., Judewicz N., Mathieu M., Lenouvel F., Sequeval D., Felenbok B. Specific binding sites for the activator protein, ALCR, in the alcA promoter of the ethanol regulon of Aspergillus nidulans. J Biol Chem. 1992 Oct 15;267(29):21146–21153. [PubMed] [Google Scholar]
- Kulmburg P., Mathieu M., Dowzer C., Kelly J., Felenbok B. Specific binding sites in the alcR and alcA promoters of the ethanol regulon for the CREA repressor mediating carbon catabolite repression in Aspergillus nidulans. Mol Microbiol. 1993 Mar;7(6):847–857. doi: 10.1111/j.1365-2958.1993.tb01175.x. [DOI] [PubMed] [Google Scholar]
- Kulmburg P., Sequeval D., Lenouvel F., Mathieu M., Felenbok B. Identification of the promoter region involved in the autoregulation of the transcriptional activator ALCR in Aspergillus nidulans. Mol Cell Biol. 1992 May;12(5):1932–1939. doi: 10.1128/mcb.12.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrion G., Boakye K. A., Lustig A. J. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Nov;12(11):5159–5173. doi: 10.1128/mcb.12.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre O., Carles C., Conesa C., Swanson R. N., Bouet F., Riva M., Sentenac A. TFC3: gene encoding the B-block binding subunit of the yeast transcription factor IIIC. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10512–10516. doi: 10.1073/pnas.89.21.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. S., Green M. R. Identification and purification of a Saccharomyces cerevisiae protein with the DNA binding specificity of mammalian activating transcription factor. Proc Natl Acad Sci U S A. 1989 Jan;86(1):109–113. doi: 10.1073/pnas.86.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisowsky T. Molecular analysis of the mitochondrial transcription factor mtf2 of Saccharomyces cerevisiae. Mol Gen Genet. 1990 Jan;220(2):186–190. doi: 10.1007/BF00260480. [DOI] [PubMed] [Google Scholar]
- Littlejohn T. G., Hynes M. J. Analysis of the site of action of the amdR product for regulation of the amdS gene of Aspergillus nidulans. Mol Gen Genet. 1992 Oct;235(1):81–88. doi: 10.1007/BF00286184. [DOI] [PubMed] [Google Scholar]
- Lowndes N. F., Johnson A. L., Breeden L., Johnston L. H. SWI6 protein is required for transcription of the periodically expressed DNA synthesis genes in budding yeast. Nature. 1992 Jun 11;357(6378):505–508. doi: 10.1038/357505a0. [DOI] [PubMed] [Google Scholar]
- Lowndes N. F., Johnson A. L., Johnston L. H. Coordination of expression of DNA synthesis genes in budding yeast by a cell-cycle regulated trans factor. Nature. 1991 Mar 21;350(6315):247–250. doi: 10.1038/350247a0. [DOI] [PubMed] [Google Scholar]
- Lowndes N. F., McInerny C. J., Johnson A. L., Fantes P. A., Johnston L. H. Control of DNA synthesis genes in fission yeast by the cell-cycle gene cdc10+. Nature. 1992 Jan 30;355(6359):449–453. doi: 10.1038/355449a0. [DOI] [PubMed] [Google Scholar]
- Lowry C. V., Cerdán M. E., Zitomer R. S. A hypoxic consensus operator and a constitutive activation region regulate the ANB1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1990 Nov;10(11):5921–5926. doi: 10.1128/mcb.10.11.5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche R. M., Smart W. C., Cooper T. G. Purification of the heteromeric protein binding to the URS1 transcriptional repression site in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7412–7416. doi: 10.1073/pnas.89.16.7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche R. M., Sumrada R., Cooper T. G. A cis-acting element present in multiple genes serves as a repressor protein binding site for the yeast CAR1 gene. Mol Cell Biol. 1990 Aug;10(8):3884–3895. doi: 10.1128/mcb.10.8.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai B., Lipp M. Identification of a protein from Saccharomyces cerevisiae with E2F-like DNA-binding and transactivating properties. FEBS Lett. 1993 Apr 26;321(2-3):153–158. doi: 10.1016/0014-5793(93)80098-f. [DOI] [PubMed] [Google Scholar]
- Marczak J. E., Brandriss M. C. Analysis of constitutive and noninducible mutations of the PUT3 transcriptional activator. Mol Cell Biol. 1991 May;11(5):2609–2619. doi: 10.1128/mcb.11.5.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Arias A., Yost H. J., Casadaban M. J. Role of an upstream regulatory element in leucine repression of the Saccharomyces cerevisiae leu2 gene. Nature. 1984 Feb 23;307(5953):740–742. doi: 10.1038/307740b0. [DOI] [PubMed] [Google Scholar]
- McNeil J. B., Dykshoorn P., Huy J. N., Small S. The DNA-binding protein RAP1 is required for efficient transcriptional activation of the yeast PYK glycolytic gene. Curr Genet. 1990 Dec;18(5):405–412. doi: 10.1007/BF00309909. [DOI] [PubMed] [Google Scholar]
- Mei B., Budde A. D., Leong S. A. sid1, a gene initiating siderophore biosynthesis in Ustilago maydis: molecular characterization, regulation by iron, and role in phytopathogenicity. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):903–907. doi: 10.1073/pnas.90.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J., Rathjen J., Jiang W., Barnes C. A., Dowell S. J. DNA binding of CPF1 is required for optimal centromere function but not for maintaining methionine prototrophy in yeast. Nucleic Acids Res. 1991 Jun 11;19(11):2961–2969. doi: 10.1093/nar/19.11.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F., Dubois E., Boonchird C. Determination of the DNA-binding sequences of ARGR proteins to arginine anabolic and catabolic promoters. Mol Cell Biol. 1991 May;11(5):2852–2863. doi: 10.1128/mcb.11.5.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F., Dubois E. Genetic evidence for a role for MCM1 in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Apr;13(4):2586–2592. doi: 10.1128/mcb.13.4.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers S., Schauer W., Balzi E., Wagner M., Goffeau A., Golin J. Interaction of the yeast pleiotropic drug resistance genes PDR1 and PDR5. Curr Genet. 1992 May;21(6):431–436. doi: 10.1007/BF00351651. [DOI] [PubMed] [Google Scholar]
- Miller A. M., MacKay V. L., Nasmyth K. A. Identification and comparison of two sequence elements that confer cell-type specific transcription in yeast. Nature. 1985 Apr 18;314(6012):598–603. doi: 10.1038/314598a0. [DOI] [PubMed] [Google Scholar]
- Miller S. M., Magasanik B. Role of the complex upstream region of the GDH2 gene in nitrogen regulation of the NAD-linked glutamate dehydrogenase in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Dec;11(12):6229–6247. doi: 10.1128/mcb.11.12.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minehart P. L., Magasanik B. Sequence and expression of GLN3, a positive nitrogen regulatory gene of Saccharomyces cerevisiae encoding a protein with a putative zinc finger DNA-binding domain. Mol Cell Biol. 1991 Dec;11(12):6216–6228. doi: 10.1128/mcb.11.12.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow B. E., Ju Q., Warner J. R. Purification and characterization of the yeast rDNA binding protein REB1. J Biol Chem. 1990 Dec 5;265(34):20778–20783. [PubMed] [Google Scholar]
- Mösch H. U., Scheier B., Lahti R., Mäntsäla P., Braus G. H. Transcriptional activation of yeast nucleotide biosynthetic gene ADE4 by GCN4. J Biol Chem. 1991 Oct 25;266(30):20453–20456. [PubMed] [Google Scholar]
- Nagai K., Nakaseko Y., Nasmyth K., Rhodes D. Zinc-finger motifs expressed in E. coli and folded in vitro direct specific binding to DNA. Nature. 1988 Mar 17;332(6161):284–286. doi: 10.1038/332284a0. [DOI] [PubMed] [Google Scholar]
- Nagata O., Takashima T., Tanaka M., Tsukagoshi N. Aspergillus nidulans nuclear proteins bind to a CCAAT element and the adjacent upstream sequence in the promoter region of the starch-inducible Taka-amylase A gene. Mol Gen Genet. 1993 Feb;237(1-2):251–260. doi: 10.1007/BF00282807. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Adolf G., Lydall D., Seddon A. The identification of a second cell cycle control on the HO promoter in yeast: cell cycle regulation of SW15 nuclear entry. Cell. 1990 Aug 24;62(4):631–647. doi: 10.1016/0092-8674(90)90110-z. [DOI] [PubMed] [Google Scholar]
- Nehlin J. O., Carlberg M., Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991 Nov;10(11):3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlin J. O., Carlberg M., Ronne H. Yeast SKO1 gene encodes a bZIP protein that binds to the CRE motif and acts as a repressor of transcription. Nucleic Acids Res. 1992 Oct 25;20(20):5271–5278. doi: 10.1093/nar/20.20.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlin J. O., Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 1990 Sep;9(9):2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki N., Okazaki K., Tanaka K., Okayama H. The ste4+ gene, essential for sexual differentiation of Schizosaccharomyces pombe, encodes a protein with a leucine zipper motif. Nucleic Acids Res. 1991 Dec;19(25):7043–7047. doi: 10.1093/nar/19.25.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen J. T., Fikes J. D., Guarente L. The Schizosaccharomyces pombe homolog of Saccharomyces cerevisiae HAP2 reveals selective and stringent conservation of the small essential core protein domain. Mol Cell Biol. 1991 Feb;11(2):611–619. doi: 10.1128/mcb.11.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen J., Hahn S., Guarente L. Yeast HAP2 and HAP3 activators both bind to the CYC1 upstream activation site, UAS2, in an interdependent manner. Cell. 1987 Dec 24;51(6):953–961. doi: 10.1016/0092-8674(87)90582-4. [DOI] [PubMed] [Google Scholar]
- Olive M. G., Daugherty J. R., Cooper T. G. DAL82, a second gene required for induction of allantoin system gene transcription in Saccharomyces cerevisiae. J Bacteriol. 1991 Jan;173(1):255–261. doi: 10.1128/jb.173.1.255-261.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paietta J. V., Akins R. A., Lambowitz A. M., Marzluf G. A. Molecular cloning and characterization of the cys-3 regulatory gene of Neurospora crassa. Mol Cell Biol. 1987 Jul;7(7):2506–2511. doi: 10.1128/mcb.7.7.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paietta J. V. Production of the CYS3 regulator, a bZIP DNA-binding protein, is sufficient to induce sulfur gene expression in Neurospora crassa. Mol Cell Biol. 1992 Apr;12(4):1568–1577. doi: 10.1128/mcb.12.4.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluh J. L., Orbach M. J., Legerton T. L., Yanofsky C. The cross-pathway control gene of Neurospora crassa, cpc-1, encodes a protein similar to GCN4 of yeast and the DNA-binding domain of the oncogene v-jun-encoded protein. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3728–3732. doi: 10.1073/pnas.85.11.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluh J. L., Yanofsky C. Characterization of Neurospora CPC1, a bZIP DNA-binding protein that does not require aligned heptad leucines for dimerization. Mol Cell Biol. 1991 Feb;11(2):935–944. doi: 10.1128/mcb.11.2.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T., Halvorsen Y. D., Dickson R. C., Coleman J. E. The transcription factor LAC9 from Kluyveromyces lactis-like GAL4 from Saccharomyces cerevisiae forms a Zn(II)2Cys6 binuclear cluster. J Biol Chem. 1990 Dec 15;265(35):21427–21429. [PubMed] [Google Scholar]
- Park H. D., Luche R. M., Cooper T. G. The yeast UME6 gene product is required for transcriptional repression mediated by the CAR1 URS1 repressor binding site. Nucleic Acids Res. 1992 Apr 25;20(8):1909–1915. doi: 10.1093/nar/20.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons M. C., Weil P. A. Purification and characterization of Saccharomyces cerevisiae transcription factor TFIIIC. Polypeptide composition defined with polyclonal antibodies. J Biol Chem. 1990 Mar 25;265(9):5095–5103. [PubMed] [Google Scholar]
- Passmore S., Elble R., Tye B. K. A protein involved in minichromosome maintenance in yeast binds a transcriptional enhancer conserved in eukaryotes. Genes Dev. 1989 Jul;3(7):921–935. doi: 10.1101/gad.3.7.921. [DOI] [PubMed] [Google Scholar]
- Peteranderl R., Nelson H. C. Trimerization of the heat shock transcription factor by a triple-stranded alpha-helical coiled-coil. Biochemistry. 1992 Dec 8;31(48):12272–12276. doi: 10.1021/bi00163a042. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Arcangioli B., Guarente L. Yeast HAP1 activator competes with the factor RC2 for binding to the upstream activation site UAS1 of the CYC1 gene. Cell. 1987 Apr 10;49(1):9–18. doi: 10.1016/0092-8674(87)90750-1. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Kim K. S., Kogan S., Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989 Jan 27;56(2):291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Prezant T., Guarente L. Yeast HAP1 activator binds to two upstream activation sites of different sequence. Cell. 1987 Apr 10;49(1):19–27. doi: 10.1016/0092-8674(87)90751-3. [DOI] [PubMed] [Google Scholar]
- Qui H. F., Dubois E., Messenguy F. Dissection of the bifunctional ARGRII protein involved in the regulation of arginine anabolic and catabolic pathways. Mol Cell Biol. 1991 Apr;11(4):2169–2179. doi: 10.1128/mcb.11.4.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhode P. R., Elsasser S., Campbell J. L. Role of multifunctional autonomously replicating sequence binding factor 1 in the initiation of DNA replication and transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Mar;12(3):1064–1077. doi: 10.1128/mcb.12.3.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemen G., Michaelis G. A point mutation in the core subunit gene of yeast mitochondrial RNA polymerase is suppressed by a high level of specificity factor MTF1. Mol Gen Genet. 1993 Feb;237(1-2):49–57. doi: 10.1007/BF00282783. [DOI] [PubMed] [Google Scholar]
- Roy A., Exinger F., Losson R. cis- and trans-acting regulatory elements of the yeast URA3 promoter. Mol Cell Biol. 1990 Oct;10(10):5257–5270. doi: 10.1128/mcb.10.10.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruden D. M. Identification of Schizosaccharomyces pombe transcription factor PGA4, which binds cooperatively to Saccharomyces cerevisiae GAL4-binding sites. Mol Cell Biol. 1990 Apr;10(4):1432–1438. doi: 10.1128/mcb.10.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron J. M., Jr, Johnston S. A. Analysis of the Kluyveromyces lactis positive regulatory gene LAC9 reveals functional homology to, but sequence divergence from, the Saccharomyces cerevisiae GAL4 gene. Nucleic Acids Res. 1986 Oct 10;14(19):7767–7781. doi: 10.1093/nar/14.19.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell N., Krems B., Entian K. D. The PAR1 (YAP1/SNQ3) gene of Saccharomyces cerevisiae, a c-jun homologue, is involved in oxygen metabolism. Curr Genet. 1992 Apr;21(4-5):269–273. doi: 10.1007/BF00351681. [DOI] [PubMed] [Google Scholar]
- Schulz B., Banuett F., Dahl M., Schlesinger R., Schäfer W., Martin T., Herskowitz I., Kahmann R. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell. 1990 Jan 26;60(2):295–306. doi: 10.1016/0092-8674(90)90744-y. [DOI] [PubMed] [Google Scholar]
- Scott E. W., Baker H. V. Concerted action of the transcriptional activators REB1, RAP1, and GCR1 in the high-level expression of the glycolytic gene TPI. Mol Cell Biol. 1993 Jan;13(1):543–550. doi: 10.1128/mcb.13.1.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian J., Sancar G. B. A damage-responsive DNA binding protein regulates transcription of the yeast DNA repair gene PHR1. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11251–11255. doi: 10.1073/pnas.88.24.11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengstag C., Hinnen A. The sequence of the Saccharomyces cerevisiae gene PHO2 codes for a regulatory protein with unusual aminoacid composition. Nucleic Acids Res. 1987 Jan 12;15(1):233–246. doi: 10.1093/nar/15.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987 Dec 4;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Siddiqui A. H., Brandriss M. C. A regulatory region responsible for proline-specific induction of the yeast PUT2 gene is adjacent to its TATA box. Mol Cell Biol. 1988 Nov;8(11):4634–4641. doi: 10.1128/mcb.8.11.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui A. H., Brandriss M. C. The Saccharomyces cerevisiae PUT3 activator protein associates with proline-specific upstream activation sequences. Mol Cell Biol. 1989 Nov;9(11):4706–4712. doi: 10.1128/mcb.9.11.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova J., Breeden L. Analysis of the SWI4/SWI6 protein complex, which directs G1/S-specific transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Feb;13(2):1069–1077. doi: 10.1128/mcb.13.2.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silar P., Butler G., Thiele D. J. Heat shock transcription factor activates transcription of the yeast metallothionein gene. Mol Cell Biol. 1991 Mar;11(3):1232–1238. doi: 10.1128/mcb.11.3.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sophianopoulou V., Suárez T., Diallinas G., Scazzocchio C. Operator derepressed mutations in the proline utilisation gene cluster of Aspergillus nidulans. Mol Gen Genet. 1993 Jan;236(2-3):209–213. doi: 10.1007/BF00277114. [DOI] [PubMed] [Google Scholar]
- Sorger P. K., Nelson H. C. Trimerization of a yeast transcriptional activator via a coiled-coil motif. Cell. 1989 Dec 1;59(5):807–813. doi: 10.1016/0092-8674(89)90604-1. [DOI] [PubMed] [Google Scholar]
- Sorger P. K., Pelham H. R. Purification and characterization of a heat-shock element binding protein from yeast. EMBO J. 1987 Oct;6(10):3035–3041. doi: 10.1002/j.1460-2075.1987.tb02609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman D. J., Bankier A. T., Seddon A., Groenhout E. G., Nasmyth K. A. Characterization of a transcription factor involved in mother cell specific transcription of the yeast HO gene. EMBO J. 1988 Feb;7(2):485–494. doi: 10.1002/j.1460-2075.1988.tb02836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Suckow M., von Wilcken-Bergmann B., Müller-Hill B. Identification of three residues in the basic regions of the bZIP proteins GCN4, C/EBP and TAF-1 that are involved in specific DNA binding. EMBO J. 1993 Mar;12(3):1193–1200. doi: 10.1002/j.1460-2075.1993.tb05760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto A., Iino Y., Maeda T., Watanabe Y., Yamamoto M. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 1991 Nov;5(11):1990–1999. doi: 10.1101/gad.5.11.1990. [DOI] [PubMed] [Google Scholar]
- Suárez T., Oestreicher N., Peñalva M. A., Scazzocchio C. Molecular cloning of the uaY regulatory gene of Aspergillus nidulans reveals a favoured region for DNA insertions. Mol Gen Genet. 1991 Dec;230(3):369–375. doi: 10.1007/BF00280293. [DOI] [PubMed] [Google Scholar]
- Swanson R. N., Conesa C., Lefebvre O., Carles C., Ruet A., Quemeneur E., Gagnon J., Sentenac A. Isolation of TFC1, a gene encoding one of two DNA-binding subunits of yeast transcription factor tau (TFIIIC). Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4887–4891. doi: 10.1073/pnas.88.11.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze J. Y., Kohlhaw G. B. Purification and structural characterization of transcriptional regulator Leu3 of yeast. J Biol Chem. 1993 Feb 5;268(4):2505–2512. [PubMed] [Google Scholar]
- Sze J. Y., Woontner M., Jaehning J. A., Kohlhaw G. B. In vitro transcriptional activation by a metabolic intermediate: activation by Leu3 depends on alpha-isopropylmalate. Science. 1992 Nov 13;258(5085):1143–1145. doi: 10.1126/science.1439822. [DOI] [PubMed] [Google Scholar]
- Taylor W. E., Young E. T. cAMP-dependent phosphorylation and inactivation of yeast transcription factor ADR1 does not affect DNA binding. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4098–4102. doi: 10.1073/pnas.87.11.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., Jacquemin I., Surdin-Kerjan Y. MET4, a leucine zipper protein, and centromere-binding factor 1 are both required for transcriptional activation of sulfur metabolism in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Apr;12(4):1719–1727. doi: 10.1128/mcb.12.4.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thukral S. K., Eisen A., Young E. T. Two monomers of yeast transcription factor ADR1 bind a palindromic sequence symmetrically to activate ADH2 expression. Mol Cell Biol. 1991 Mar;11(3):1566–1577. doi: 10.1128/mcb.11.3.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thukral S. K., Morrison M. L., Young E. T. Mutations in the zinc fingers of ADR1 that change the specificity of DNA binding and transactivation. Mol Cell Biol. 1992 Jun;12(6):2784–2792. doi: 10.1128/mcb.12.6.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thukral S. K., Tavianini M. A., Blumberg H., Young E. T. Localization of a minimal binding domain and activation regions in yeast regulatory protein ADR1. Mol Cell Biol. 1989 Jun;9(6):2360–2369. doi: 10.1128/mcb.9.6.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice-Baldwin K., Fink G. R., Arndt K. T. BAS1 has a Myb motif and activates HIS4 transcription only in combination with BAS2. Science. 1989 Nov 17;246(4932):931–935. doi: 10.1126/science.2683089. [DOI] [PubMed] [Google Scholar]
- Ticho B. S., Getz G. S. The characterization of yeast mitochondrial RNA polymerase. A monomer of 150,000 daltons with a transcription factor of 70,000 daltons. J Biol Chem. 1988 Jul 25;263(21):10096–10103. [PubMed] [Google Scholar]
- Toda T., Shimanuki M., Saka Y., Yamano H., Adachi Y., Shirakawa M., Kyogoku Y., Yanagida M. Fission yeast pap1-dependent transcription is negatively regulated by an essential nuclear protein, crm1. Mol Cell Biol. 1992 Dec;12(12):5474–5484. doi: 10.1128/mcb.12.12.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Shimanuki M., Yanagida M. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 1991 Jan;5(1):60–73. doi: 10.1101/gad.5.1.60. [DOI] [PubMed] [Google Scholar]
- Trawick J. D., Kraut N., Simon F. R., Poyton R. O. Regulation of yeast COX6 by the general transcription factor ABF1 and separate HAP2- and heme-responsive elements. Mol Cell Biol. 1992 May;12(5):2302–2314. doi: 10.1128/mcb.12.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueblood C. E., Poyton R. O. Identification of REO1, a gene involved in negative regulation of COX5b and ANB1 in aerobically grown Saccharomyces cerevisiae. Genetics. 1988 Nov;120(3):671–680. doi: 10.1093/genetics/120.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler B. M., Geever R. F., Case M. E., Giles N. H. Cis-acting and trans-acting regulatory mutations define two types of promoters controlled by the qa-1F gene of Neurospora. Cell. 1984 Feb;36(2):493–502. doi: 10.1016/0092-8674(84)90242-3. [DOI] [PubMed] [Google Scholar]
- Tymon A. M., Kües U., Richardson W. V., Casselton L. A. A fungal mating type protein that regulates sexual and asexual development contains a POU-related domain. EMBO J. 1992 May;11(5):1805–1813. doi: 10.1002/j.1460-2075.1992.tb05232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Türkel S., Farabaugh P. J. Interspersion of an unusual GCN4 activation site with a complex transcriptional repression site in Ty2 elements of Saccharomyces cerevisiae. Mol Cell Biol. 1993 Apr;13(4):2091–2103. doi: 10.1128/mcb.13.4.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura H., Jigami Y. GCR3 encodes an acidic protein that is required for expression of glycolytic genes in Saccharomyces cerevisiae. J Bacteriol. 1992 Sep;174(17):5526–5532. doi: 10.1128/jb.174.17.5526-5532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura H., Jigami Y. Role of GCR2 in transcriptional activation of yeast glycolytic genes. Mol Cell Biol. 1992 Sep;12(9):3834–3842. doi: 10.1128/mcb.12.9.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershon A. K., Johnson A. D. A short, disordered protein region mediates interactions between the homeodomain of the yeast alpha 2 protein and the MCM1 protein. Cell. 1993 Jan 15;72(1):105–112. doi: 10.1016/0092-8674(93)90054-t. [DOI] [PubMed] [Google Scholar]
- Vincent A. C., Struhl K. ACR1, a yeast ATF/CREB repressor. Mol Cell Biol. 1992 Dec;12(12):5394–5405. doi: 10.1128/mcb.12.12.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. K., Weil P. A. Purification and characterization of Saccharomyces cerevisiae transcription factor IIIA. J Biol Chem. 1989 Jan 15;264(2):1092–1099. [PubMed] [Google Scholar]
- Wang H., Nicholson P. R., Stillman D. J. Identification of a Saccharomyces cerevisiae DNA-binding protein involved in transcriptional regulation. Mol Cell Biol. 1990 Apr;10(4):1743–1753. doi: 10.1128/mcb.10.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. W., Hynes M. J., Davis M. A. Structural and functional analysis of the amdR regulatory gene of Aspergillus oryzae. Gene. 1992 Dec 1;122(1):147–154. doi: 10.1016/0378-1119(92)90042-n. [DOI] [PubMed] [Google Scholar]
- Wiederrecht G., Shuey D. J., Kibbe W. A., Parker C. S. The Saccharomyces and Drosophila heat shock transcription factors are identical in size and DNA binding properties. Cell. 1987 Feb 13;48(3):507–515. doi: 10.1016/0092-8674(87)90201-7. [DOI] [PubMed] [Google Scholar]
- Willett C. E., Gelfman C. M., Holland M. J. A complex regulatory element from the yeast gene ENO2 modulates GCR1-dependent transcriptional activation. Mol Cell Biol. 1993 Apr;13(4):2623–2633. doi: 10.1128/mcb.13.4.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte M. M., Dickson R. C. The C6 zinc finger and adjacent amino acids determine DNA-binding specificity and affinity in the yeast activator proteins LAC9 and PPR1. Mol Cell Biol. 1990 Oct;10(10):5128–5137. doi: 10.1128/mcb.10.10.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolberger C., Vershon A. K., Liu B., Johnson A. D., Pabo C. O. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991 Nov 1;67(3):517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- Woychik N. A., Young R. A. Genes encoding transcription factor IIIA and the RNA polymerase common subunit RPB6 are divergently transcribed in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992 May 1;89(9):3999–4003. doi: 10.1073/pnas.89.9.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray L. V., Jr, Witte M. M., Dickson R. C., Riley M. I. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Mar;7(3):1111–1121. doi: 10.1128/mcb.7.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne J., Treisman R. SRF and MCM1 have related but distinct DNA binding specificities. Nucleic Acids Res. 1992 Jul 11;20(13):3297–3303. doi: 10.1093/nar/20.13.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. M., Gahl W., Hamer D. Role of heat shock transcription factor in yeast metallothionein gene expression. Mol Cell Biol. 1991 Jul;11(7):3676–3681. doi: 10.1128/mcb.11.7.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G. F., Fu Y. H., Marzluf G. A. nit-4, a pathway-specific regulatory gene of Neurospora crassa, encodes a protein with a putative binuclear zinc DNA-binding domain. Mol Cell Biol. 1991 Nov;11(11):5735–5745. doi: 10.1128/mcb.11.11.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y. L., Fields S. Properties of the DNA-binding domain of the Saccharomyces cerevisiae STE12 protein. Mol Cell Biol. 1991 Dec;11(12):5910–5918. doi: 10.1128/mcb.11.12.5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L., Reinberg D. Initiation of transcription by RNA polymerase II: a multi-step process. Prog Nucleic Acid Res Mol Biol. 1993;44:67–108. doi: 10.1016/s0079-6603(08)60217-2. [DOI] [PubMed] [Google Scholar]
- Zhang L., Bermingham-McDonogh O., Turcotte B., Guarente L. Antibody-promoted dimerization bypasses the regulation of DNA binding by the heme domain of the yeast transcriptional activator HAP1. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2851–2855. doi: 10.1073/pnas.90.7.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K. M., Kohlhaw G. B. Transcriptional activator LEU3 of yeast. Mapping of the transcriptional activation function and significance of activation domain tryptophans. J Biol Chem. 1990 Oct 15;265(29):17409–17412. [PubMed] [Google Scholar]
- Zhou P., Szczypka M. S., Sosinowski T., Thiele D. J. Expression of a yeast metallothionein gene family is activated by a single metalloregulatory transcription factor. Mol Cell Biol. 1992 Sep;12(9):3766–3775. doi: 10.1128/mcb.12.9.3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Muller E. G., Amacher S. L., Northrop J. L., Davis T. N. A dosage-dependent suppressor of a temperature-sensitive calmodulin mutant encodes a protein related to the fork head family of DNA-binding proteins. Mol Cell Biol. 1993 Mar;13(3):1779–1787. doi: 10.1128/mcb.13.3.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitomer R. S., Lowry C. V. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol Rev. 1992 Mar;56(1):1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heeswijck R., Hynes M. J. The amdR product and a CCAAT-binding factor bind to adjacent, possibly overlapping DNA sequences in the promoter region of the Aspergillus nidulans amdS gene. Nucleic Acids Res. 1991 May 25;19(10):2655–2660. doi: 10.1093/nar/19.10.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]


