Abstract
Traditional high throughput screening (HTS) is carried out in centralized facilities that require extensive robotic liquid and plate handling equipment. This model of HTS is restrictive as such facilities are not accessible to many researchers. We have designed a simple microarray platform for cell-based screening that can be carried out at the bench top. The device creates a microarray of 2100 individual cell-based assays in a standard microscope slide format. A microarray of chemical-laden hydrogels addresses a matching array of cell-laden microwells thus creating a microarray of sealed microscale cell cultures each with unique conditions. We demonstrate the utility of the device by screening the extent of apoptosis and necrosis in MCF-7 breast cancer cells in response to exposure to a small library of chemical compounds. From a set of screens we produced a rank order of chemicals that preferentially induce apoptosis over necrosis in MCF-7 cells. Treatment with doxorubicin induced high levels of apoptosis in comparison with staurosporine, ethanol, and hydrogen peroxide, while treatment with 100 μM ethanol induced minimal apoptosis with high levels of necrosis. We anticipate broad application of the device for various research and discovery applications as it is easy to use, scalable and can be fabricated and operated with minimal peripheral equipment.
Introduction
The drug discovery pipeline has produced many successful drug treatments and therapies; however, these successes have come at a high failure rate.1 This failure rate persists despite the maturation of proteomics and genomics and the consequent identification of an increasing number of screenable targets.2 One approach to enhancing the current drug discovery process is to increase access to high throughput screening (HTS) with technologies that enable cell-based drug screening in the common laboratory. Currently, such screening experiments are conducted in centralized facilities that require high capital costs for robotic liquid handling equipment and high throughput imaging systems.3 Technologies that bring HTS to the common laboratory will help drive down the costs of screening by reducing equipment needs as well as allowing for wider array of HTS experiments.
A number of combinatorial, microarray, and microfluidic screening devices have demonstrated the potential of microscale engineering to produce a significant change in the way that drug screening is carried out.3 For example, small molecule and siRNA screening has been carried out with live-cell microarrays4, 5, and cell-laden hydrogel microarrays have been used to screen the cytotoxicity of metabolic products6, 7. Microfluidic devices have been developed for investigating cell-microenvironment interactions in three-dimensional (3D) cell culture arrays8, drug toxicity testing9, drug metabolite toxicity assays10, creating multi-phenotype cell arrays11, and monitoring real-time gene expression across arrayed microscale cell cultures12. Microfluidic gradient generators have also been used to simultaneously screen a wide range of concentration effects.13, 14
These new screening technologies represent significant progress towards more accessible HTS but peripheral equipment, such as liquid pumps and liquid handling equipment, are still required to operate these systems. In this regard, the development of pressure-driven flow and flow-powered gating are important advances.14,15 Using such techniques, it has been possible to create standalone devices for parallel screening of chemicals in array format that are easy to use and operate.16, 17
Plate-based screening (384 and 1536-well plates) is the industry standard for high throughput (HT) cell-based screening. These technologies have proven successful, but are limited as they require extensive robotic liquid handling equipment and can suffer from inconsistencies due to uncontrolled evaporation of dispensed liquids.3, 18 Successful replication of HTS at the bench top requires devices that, i) require minimal expertise to operate, ii) are amenable to many different cell-based assays, iii) can easily generate quantifiable read-outs, and iv) are inexpensive to manufacture and operate.
Here, we demonstrate a microarray device for cell-based chemical screening that can be operated at the bench top and can be easily fabricated. The design improves on a previously developed device of a sandwiched microarray platform for cell-based high throughput screening at the bench top.19 We advance this technology to include the ability to screen chemical-induced apoptosis, the potential to control chemical release from arrays of chemical-laden hydrogels, and significant advancement in device alignment and operation. The microarray device is simple to operate, portable, inexpensive to fabricate, and has the potential to be commercialized for use in various fields of biology and pharmacology. The platform aims to address a number of limitations in HTS of cells including the need for large sample volumes, and largely eliminates the need for expensive robotics. We demonstrate the platform by screening a small library of chemicals for chemical-induced apoptosis in arrays of isolated microscale breast cancer cell cultures.
Experimental Methods
Microwell fabrication
Arrayed poly(ethlyene glycol) (PEG, MW 258) microwells (400 μm in diameter, 300 μm in depth) were fabricated by photolithography and micromolding with a poly(dimethylsiloxane) (PDMS) template.20 The PDMS template was created from a silicon wafer. Briefly, a layer of photoresist (SU-8, Microchem) was photopatterned on a 3″-silicon wafer to produce a master mold with a negative relief microwell array pattern. PDMS (10:1, PDMS:curing agent; Sylgard 184, Dow Corning) was cast onto the master, cured at 80 °C for 2 hours and peeled away. Microwells of low molecular weight (MW 258) PEG-diacrylate (PEGDA) polymer (Sigma-Aldrich Co., St. Louis, MO) with 1% (w/w) of photoinitiator 2-hydroxy-2-methyl propiophenone (Sigma-Aldrich Co., St. Louis, MO) were molded from a PDMS template upon exposure to 10 mW cm−2 of UV light (OmniCure Series 2000, EXFO, Mississauga, Canada) for 600 seconds. On each standard microscope glass slide (25 × 75 mm) an array of 2100 microwells was fabricated in the same format as the chemical-laden PEGDA hydrogel arrays described below. Arrayed microwells were molded onto 3-(trimethoxysilyl) propylmethacrylate (TMSPMA) modified glass slides thus allowing for cells and/or proteins to adhere and absorb to the bottom of each well.21
Fabrication of chemical-laden hydrogel arrays
A MicroGridTAS contact printer (Digilab) was use to create arrays of photocrosslinked PEGDA hydrogels (20% PEGDA, MW 2000, with 1% (w/w photoinitiator) in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen). Twenty-percent glycerol was added to slow evaporation of the printed spot. Up to 2100 spots (110 μM in diameter) were printed on PDMS (75×25×1 mm). Arrayed PEGDA hydrogels on PDMS comprise the top of the microarray sandwich device used to seal microwell cultures in array format. Each hydrogel is loaded with the desired screening compound at the time of printing. Each hydrogel has a volume of 0.35 nL resulting in a 100-fold dilution of loaded chemical concentration to final microwell concentration.
Microarray device alignment
Arrayed chemical-laden hydrogels were aligned with arrayed microwells with the aid of 2× magnification and alignment features integrated into the device. In this way each hydrogel addresses a single cell-laden microwell resulting in an array of isolated cell-based assays. The bottom of the device was laid flat on a microscope stage and two glass slides, one at either end of the device, were used to separate the top from the bottom during alignment. The top was gently moved relative to the bottom until the alignment features were matched. Once aligned the glass slides separating the top from the bottom were removed sequentially, and the PDMS top with arrayed hydrogels was gently pressed down on to the PEGDA microwells.
Cell culture and cell seeding
MCF-7 cells were cultured in DMEM containing 10% fetal bovine serum. All cells were cultured at 37°C, 5% CO2 in a humidified incubator. Microwells were seeded by pipetting 1 mL of media containing 1×106 cells on to the arrayed microwells resulting in 70±10 cells per well. Uniform seeding was accomplished by slowly sliding the edge of a cover slip across the microwell array as previously described.22 Cells were allowed to settle in the microwells for 15 minutes prior to culturing in excess media for 24 hours before being used.
Analysis of apoptosis
Evaluations of viability and apoptosis were carried out after 12 hours of exposure to chemicals unless otherwise mentioned. After chemical exposure, the PDMS substrate was carefully peeled from the microwells, and the microwells were gently washed with PBS three times. To determine the extent of apoptosis and necrosis, cells were incubated with SYTOX-Orange and allophycocyanin (APC)-conjugated annexin V (Invitrogen). Annexin V-APC binds to exposed phosphatidylserine on the membrane of apoptotic cells. SYTOX-Orange binds to nucleic acids in cells with compromised membranes such as late apoptotic and necrotic cells, and does not permeate the membranes of early apoptotic cells. Both annexin V-APC and SYTOX-Orange were used as directed by manufacture’s instructions. Fluorescent images were acquired and analyzed with a GenePix 4100a microarray scanner and GenePix pro software (Axon Instruments, Union City, CA). Herein annexin V-APC fluorescence is shown in red (Ex:Em 635:675/25), and SYTOX-Orange and Rhodamine B fluorescence are shown in green (Ex:Em 532:575/15).
Results and Discussion
Device design, fabrication, and operation
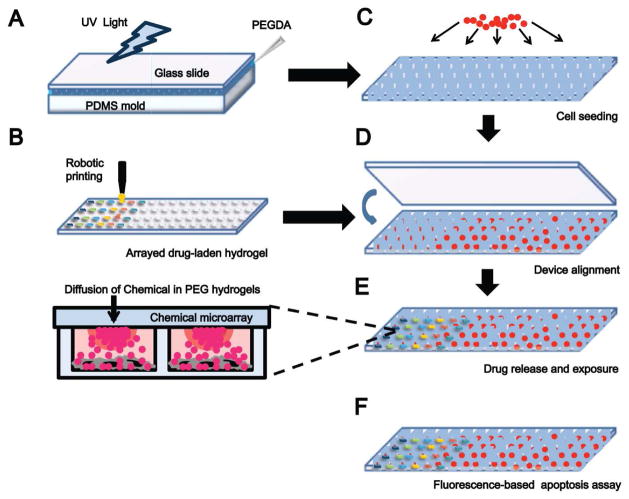
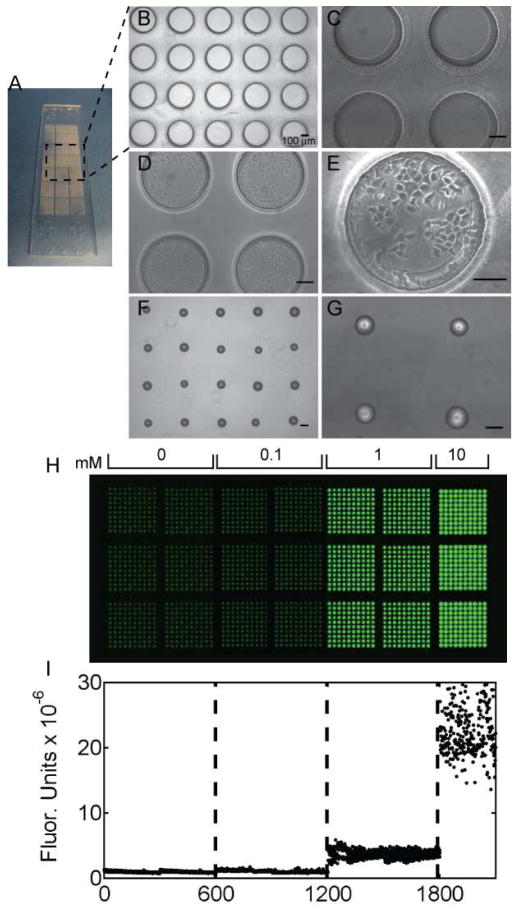
The fabrication and operation of a microarray chemical screening device including micromolding and printing of the top and bottom of the device, respectively, cell seeding, device alignment, chemical screening, and analysis are schematically shown in Figure 1. The bottom of the device, arrayed PEGDA microwells, was micromolded using a PDMS template. Each set of 2100 arrayed microwells was fabricated on a standard microscope slide (75 × 25 mm; Figure 2A). Each microwell is 400 μm in diameter and 300 μm deep (Figure 2B,C), and was seeded with 70±10 cells per well (Figure 2D,E). In all experiments herein microwells were seeded with MCF-7 breast cancer cells. The top of the device, arrayed PEGDA hydrogels, was fabricated by contact printing onto a thin slab of PDMS (1 mm thick), followed by UV exposure. Figures 2H and I show the loading of a range of Rhodamine B concentrations (0.1, 1, and 10 mM) in arrayed hydrogels. Differences in chemical-loading concentrations were observed by fluorescent imaging (Figure 2H) and by quantification of the mean fluorescent intensity of each hydrogel (Figure 2I). Fluorescent scanning images were acquired using a standard microarray scanner, and quantification was done using GenePix software.
Figure 1.
Design and fabrication of a controlled release microarray system for chemical screening. A) Micromolding of PEGDA by UV photopolymerization into arrayed microwells. B) Fabrication of a chemical-laden hydrogel microarray by robotic printing. C) Cell seeding in arrayed microwells. D) Alignment and sandwiching of arrayed chemical-laden hydrogels and cell-seeded microwells. E) Drug release and cell culturing for 6 – 24 hours. Close-up schematic shows the diffusion of chemicals from arrayed hydrogels into cell-seeded microwells. F) Analysis of apoptosis by fluorescence-based assay.
Figure 2.
Cell-seeded microwells and arrayed chemical-laden hydrogels. A) A photograph of arrayed microwells adhered to a standard glass microscope slide. B,C) Phase contrast images of microwells (400 μm in diameter and 300 μm deep) with (D,E) seeded MCF-7 breast cancer cells. F, G) Phase contrast images of arrayed PEGDA hydrogels on a PDMS substrate. All scale bars are 100 μm. H) Fluorescent scanner image of arrayed hydrogels with varying concentrations of Rhodamine B (Ex:Em, 532:575/25; green false color). The hydrogel microarray contains 600 spots of each of 0, 0.1, and 1 mM and 300 spots of 10 mM of Rhodamine B. I) Quantification of fluorescence in each arrayed hydrogel.
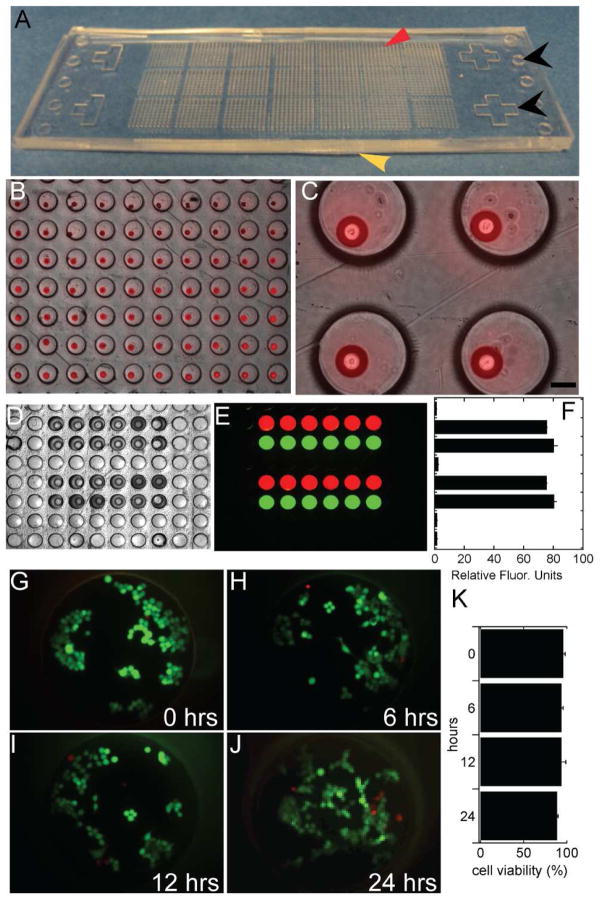
Cell-based chemical screening was carried out as the chemical-laden hydrogel array was aligned with, and sandwiched to the cell-laden microwells (Figure 3A). The sandwich microarray device operated as an array of individual chemical screens as each hydrogel addressed a single microwell and the PDMS top created a sealed chamber (Figure 3B). Compounds contained within each hydrogel were released into the cell culture media within a microwell. This concept is shown in Figure 3C–E where hydrogels containing FITC-labeled dextran and Rhodamine B were aligned and sandwiched to microwells containing culture media. Fluorescent imaging of the sandwiched device revealed that the fluorescent compounds diffused within the entire well and that each well was isolated from neighboring wells (Figure 3E,F). The release rate of chemicals contained in the arrayed hydrogels was dependent on the degree of hydrogel crosslinking, and was optimized to result in > 90% release of the equilibrium value within 6 hours of culture. Total release rate and total release was decreased with increasing UV exposure during crosslinking (Supplemental Material, SFigure 1). In the devices used for this study we employed PEGDA as an encapsulation material; however, the system is potentially amenable to other co-polymers and hydrogels for controlled chemical release.23 These concepts represents an important advancement over previous devices, as controlled release and the potential for different hydrogel chemistry creates a device that can be used to explore experimental parameters of drug screening including, controlled release, dose profile, and a variety of chemical properties of relevant drugs.
Figure 3.
Microarray device alignment and characterization. A) Photograph of a microarray device with arrayed chemical-laden hydrogels (red arrow) sandwiched on to arrayed microwells (yellow arrow). Alignment features are indicated with black arrows. B, C) Phase contrast images with overlaid fluorescent images of an aligned device (scale bar = 100 μm). D, E, F) Phase contrast and fluorescent images, and quantification of fluorescence of released chemicals (green is FITC-labeled dextran and red is Rhodamine B). G–K) MCF-7 viability in sealed microwells at 0, 6, 12 and 24 hour time points (Live/Dead staining with calcein-AM, green, and ethidium homodimer, red).
Previously we employed a peripheral device to align and sandwich the two parts of the device.19 In this paper the alignment was accomplished with the aid of 2× magnification and integrated alignment features. The alignment tolerances in the current system are relaxed in comparison with our previous design in which arrayed microscale posts were used to deliver chemicals to the arrayed microwells. The diameter of the arrayed posts in our previous design were only slightly less than the diameter of the arrayed microwells, thus necessitating highly accurate alignment prior to sandwiching. In the current design, each chemical-laden hydrogel was approximately one-quarter the diameter of the corresponding microwell, thus successful sandwiching, where each hydrogel addresses a single microwell, can be accomplished despite imperfect alignment. In three separate trials, an average alignment of 97% was achieved (677±23 out of 700 successfully aligned microwells, n = 3), where a microwell and hydrogel were considered aligned when the distance from the center of a microwell and the center of an arrayed hydrogel was less than 160 μm (SFigure 2).
PDMS patterned with arrayed chemical-laden hydrogels was used to create arrays of sealed microscale cultures. As PDMS is permeable to oxygen, gas exchange was permitted and cell viability in sealed cultures was unaffected up to 24 hours (Figure 3G–K). Similar results were previously reported19, and were also observed with human hepatocellular carcinoma cells (HEPG2) (cell viability greater than 90% at 24 hours; SFigure 3).
Screening of cellular apoptosis and necrosis
It has previously been demonstrated that chemical cytotoxicity can be measured on live cell arrays and in arrayed microwell cultures in a high throughput manner using microscale devices.4, 6, 19 Here, we used a previously developed assay to determine the extent of chemical-induced apoptosis and necrosis in a microarray device for HTS at the bench top. Fluorescently labeled annexin V was used as an indicator of apoptosis, as annexin V binds to phophatidylserine translocated to the outer membrane surface during the early stages of apoptosis.24 A nucleic acid stain, SYTOX-Orange, that penetrates cells with compromised membranes was used as an indicator of necrotic, or dead, cells.25 The SYTOX family of DNA binding dyes do not permeate the membranes of early apoptotic cells, and as such we anticipate that apoptotic cells in our system will show high annexin V-APC staining relative to SYTOX-Orange staining as indicated by the manufacture’s protocols. The use of fluorescent indicators enabled rapid analysis with a microarray scanner.
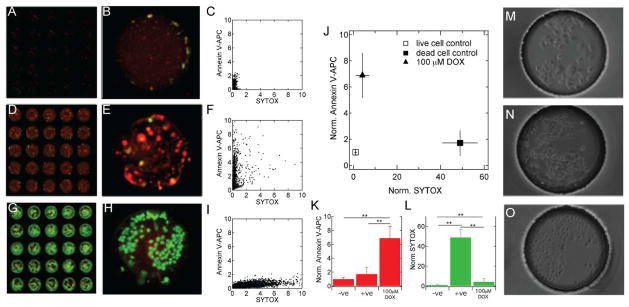
Figure 4 shows the results of control compounds for apoptotic cells, dead cells (membrane compromised cells), and lives cells (no chemical addition). Doxorubicin, a known chemotherapeutic, that has previously been shown to be cytotoxic and apoptotic to MCF-7 cells was used as a positive control for chemical-induced apoptosis.7,19,13 Addition of the surfactant Triton X-100 to culture media, which results in compromised cell membranes, was used here as a positive control for inducing necrosis. Finally, a live cell control where no chemicals were added during culture was used as a negative control. PEGDA hydrogels containing doxorubicin, Triton X-100, and no additives were printed in array format on PDMS device tops, and aligned and sandwiched to arrays of microwells containing MCF-7 cells. Microscale cultures containing, separately, 100 μM doxorubicin as a positive apoptosis controls, 0.01% Triton X-100 as a positive necrosis controls, and negative controls were cultured for 12 hours at 37 °C in a humidified incubator with 5% CO2. Fluorescent scanning images of the arrayed microwells after incubating with allophycocyanain (APC)-conjugated annexin V and SYTOX-Orange clearly shows annexin V positive cells that were treated with doxorubicin and membrane compromised cells that were treated with the surfactant (Figure 4D,E). Negative control microwell cultures showed only minimal fluorescence due to either stain (Figure 4A,B). These results are also shown in scatter plots of pixel intensity. In the negative control cultures the pixel intensity of both annexin V-APC and SYTOX-orange channels was low (Figure 4C). A population of pixels with high annexin V-APC fluorescence, but low SYTOX-Orange intensity was observed with doxorubicin treatment (Figure 4F). In contrast, the treatment with 0.01% Triton X-100 resulted in low annexin V-APC intensity and high SYTOX-Orange intensity (Figure 4I).
Figure 4.
Measuring chemical-induced apoptosis. Microwell MCF-7 cell cultures exposed to (A,B) no chemicals (negative control; −ve), (D,E) 100 μM doxorubicin (DOX), and (G,H) 0.01% Triton X-100 (positive control; +ve) for 12 hours. Fluorescent images are of microwells stained with annexin V-APC (red) and SYTOX-Orange (green) (Ex:Em, 632:695/15 and 532:575/25, respectively). Pixel intensity (×10−3) due to each stain is shown in (C,F,I). The average fluorescence intensity of each control condition normalized to the negative control (−ve) is presented in (J). ANOVA analysis of the average annexin V-APC and SYTOX-Orange fluorescence for each condition are presented in (K) and (L), respectively (n ≥ 30, ** p<0.01, * p<0.05). Phase contrast images of −ve, 100 μM DOX, and +ve are shown in (M,N,O), respectively.
The extent of apoptosis and necrosis resultant from each control condition is shown in Figure 4J, where the mean fluorescence due to annexin V binding is plotted as a function of the mean fluorescence due to SYTOX-Orange staining. Early apoptotic cultures appear in the upper left quadrant, while necrotic cultures appear in the lower right quadrant. Negative control cultures were used to normalize each signal, and as such appear in the lower left quadrant of the plot. In comparison with the negative control, MCF-7 microscale cultures treated with 100 μM doxorubicin showed a 7-fold (6.8) increase in annexin V fluorescence (p < 0.05, ANOVA), and showed no statistical different in SYTOX-Orange staining (Figure 4K,L). Conversely, the necrotic control showed no significant difference in annexin V fluorescence with the live cell control, but showed a 50-fold increase (48.9) SYTOX-Orange staining (p < 0.01 ANOVA). Qualitative differences between treatments were also observed in phase contrast micrographs. Cells in the negative control cultures were large in size and adhered to the bottom of the microwells (Figure 4M), while doxorubicin treated cells showed morphological characteristics of early apoptosis as they were slightly granular and smaller (Figure 4N).13 The cultures treated with surfactant were small and round and did not adhere to the microwell bottom and were not packed in a confluent layer (Figure 4O). Taken together, these results demonstrate the ability of the device to quantitatively distinguish between apoptotic and necrotic cells.
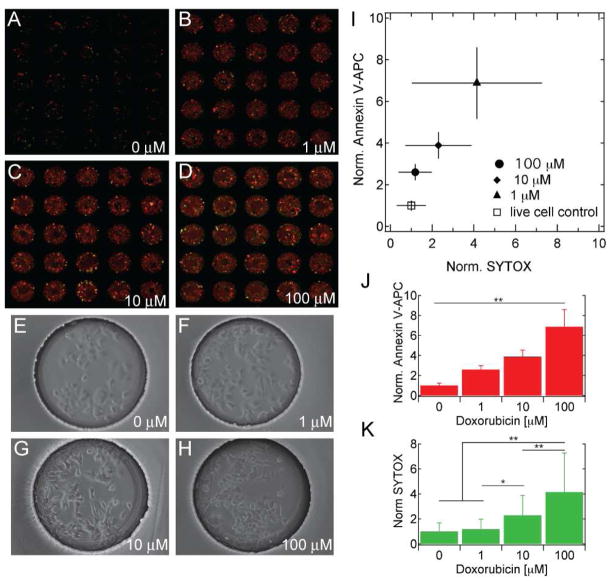
The results shown in Figure 5 suggest that not only does doxorubicin induce apoptosis, but also that doxorubicin concentration has a significant effect on the extent of apoptosis. After 12 hours of exposure, 100 μM doxorubicin induced an average apoptosis of 2.6 fold higher than 1 μM (p < 0.05 ANOVA) and 1.5 fold higher than 10 μM (p < 0.05 ANOVA), while 10 μM induced the average apoptosis of 1.8 fold higher than 1 μM (p < 0.05 ANOVA; Figure 5A–D,I–J). These observations suggest that not only does doxorubicin induces significantly higher apoptosis in comparison with the negative control, but there is also, significant difference in the extent of apoptosis related to the concentration of the doxorubicin. Concomitant with the increase in apoptosis was a small, relative to the positive necrotic controls, but significant increase in necrosis (Figure 5K). In support of the annexin V binding data, high magnification phase contrast micrographs of representative microwell cultures (Figure 5E–H) show decreasing cell size and increasing granularity that coincides with increased doxorubicin concentration. The data presented in Figure 5 demonstrates the sensitivity of the device to measure the extent of apoptosis based on the concentration of the chemical. The concentration dependent apoptotic response of MCF-7 to doxorubicin was confirmed in 96-well plate assays (SFigure 4).
Figure 5.
Increasing doxorubicin concentrations result in increased apoptosis. A–H) Fluorescent scanner and phase contrast images of microwell MCF-7 cell cultures exposed to 0, 1, 10 and 100 mM of doxorubicin. Fluorescent images show annexin V-APC (red) and SYTOX-Orange (green). I–K) Normalized annexin V-APC and SYTOX-Orange fluorescence for each doxorubicin condition, with associated ANOVA analyses (n ≥ 30, ** p<0.01, * p<0.05).
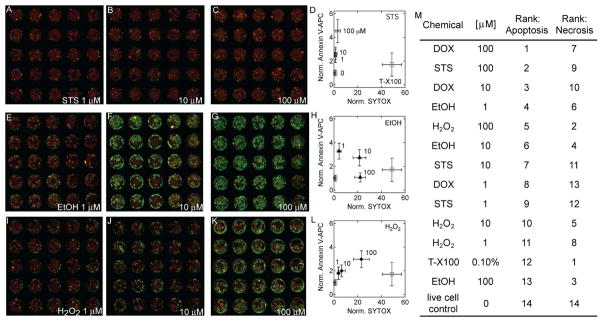
To further demonstrate the use of the device as a bench top cell-based chemical screening technology, arrayed hydrogels were laden with staurosporine (STS), ethanol (EtOH), and hydrogen peroxide (H2O2) to simultaneously evaluate the concentration dependent cytotoxicity and differences in apoptotic-induction. These chemicals were selected as each has been shown to induce apoptosis. At high concentrations EtOH is known to be cytotoxic, but at low concentrations it has been shown to preferentially cause apoptosis.26, 27 STS is a well known inducer of apoptosis, and shows potency at low concentrations.28–30 Reactive oxygen species, including H2O2, have also been shown to induce apoptosis.31, 32
Analysis of the small chemical library in the microarray sandwich device revealed that doxorubicin induced significantly higher apoptosis in MCF-7 cells in comparison with STS, EtOH, H2O2, and the positive necrotic control (p < 0.05 ANOVA). Each chemical treatment was found to be concentration dependent, and with the exception of EtOH, higher concentrations resulted in increased apoptosis. In the case of STS, 100 μM induced significantly higher apoptosis in comparison with 10 μM and 1 μM (p < 0.05 ANOVA), but a 10 μM treatment did not induce a significant increase in apoptosis above a 1 μM treatment. A similar trend was observed with H2O2 treatment. However, for EtOH the induction of apoptosis reached a maximum at 1 μM, and decreased at 10 and 100 μM.
At higher concentrations of EtOH and H2O2 substantially more necrosis was observed. In fact, 100 μM H2O2 and 100 and 10 μM EtOH induced the highest levels of necrosis behind the positive necrotic control (Figure 6M). When taking into consideration the rank order of apoptosis and necrosis, 100 and 10 μM doxorubicin and 100 μM STS exhibited the highest preferential induction of apoptosis. Comparatively, 100 μM H2O2 and 100 μM EtOH induced high levels of necrosis and low levels of apoptosis in MCF-7 cells. Combined, the data here demonstrates that the microarray sandwich device can be used for screening chemical and drug libraries. The extent of apoptosis and necrosis can effectively be measured and comparisons between chemicals and across a wide range of concentrations can be easily made.
Figure 6.
Microarrays for cell-based screening of chemical-induced apoptosis. Concentration dependent apoptosis and necrosis (as judged by annexin V-APC and SYTOX-Orange, respectively) for (A–D) staurosporine (STS), (E–H) ethanol (EtOH), and (I–L) hydrogen peroxide (H2O2). M) The rank order of all chemicals for apoptosis and necrosis as measured in the microarray device.
Recapitulation of traditional plated-based HTS technologies requires systems and devices that can screen thousands of individual assays simultaneously and that are amenable to different cell cultures and cell-based assays. Additionally, read-outs and assays results must be generated in a rapid and quantifiable way. The microarray sandwich device presented here creates an array of 2100 assays. Each sealed microwell contains less than 40 nL of culture media and approximately 70 cells. The low MW PEGDA microwell array prevents diffusion of solutes between microwells, thus isolating each microscale culture, and gas exchange is permitted through the PDMS top seal. We quantified the extent of apoptosis and necrosis in MCF-7 cells, but the device is amenable to the study of different cells types, and can potentially be use for cell aggregates and embryoid bodies19, 33. Additionally, microwells can be modified with extra-cellular matrix proteins to evaluate the potential contributions of microenvironment on cell viability and apoptosis in drug screens, as well as potential contributions of microenvironmental factors to drug resistance. Using fluorescent based assays allowed for the rapid analysis of quantifiable outcomes. Here, we use a microarray scanner traditionally used for analysis of nucleic acid microarray, but it is also possible to use fluorescent microscopy with automated staging.
Replication of HTS at the bench top requires devices that are easy and inexpensive to operate and are easy to manufacture. As soft lithography techniques are becoming widely accessible the equipment required to fabricate our microarray sandwich device is becoming commonplace. In addition to soft lithography equipment, fabrication of our device requires only minimal robotic equipment: a contact spotter or inkjet printer. Both of these printing technologies are rapidly becoming more accessible and new lower cost equipments are now becoming available. Importantly, the chemical-laden hydrogel arrays can be prepared beforehand, or can be prepared offsite, and stored until use. Additionally, operational costs are kept to a minimum as high screening concentration (100 μM) requires less than 4 nanomoles per assays. Finally, the operation of the device was simple, such that we were able to consistently align and sandwich a single device in less than 5 minutes.
Conclusions
We have developed a microarray sandwich device for cell-based screening of chemical libraries at the bench top. The device uses a microarray of chemical-laden PEGDA hydrogels to individually address microscale cell cultures, thus creating a microarray of individual assays. With the device we screened a small library of chemical compounds known to induce apoptosis and screened each over a range of concentrations. The device is simple to use and can simultaneously perform 2100 individual assays, thus creating an accessible bench top-screening device. Furthermore, the device is potentially amenable to many different fluorescent-based assays and can be used for a variety of screening applications. We anticipate broad application of this platform as it is simple, scalable, and robust, and can be applied to many different screening experiments.
Supplementary Material
Acknowledgments
This paper was supported by the National Institutes of Health (EB009196; DE019024; EB007249; HK092836), the National Science Foundation CAREER award (DMR0847287) and the Office of Naval Research Young Investigator award.
Footnotes
Author contributions
C.H.K, I.W., and A.K., conceived the idea and designed experiments. N.N.K., H.B., S.S., and J.K. helped design some experiments and interpret data. C.H.K., S.H.L. J.F. conducted experiments. C.H.K, I.W., N.N.K., analyzed the data. I.W., C.H.K, and N.N.K wrote the manuscript. C.H.K, I.W., J.W.K, and A.K. revised the manuscript.
Supporting Information Available
Results of doxorubicin, staurosporine, and Triton X-100 release for PEGDA hydrogels with different crosslinking conditions. Device alignment accuracy data. HEPG2 viability in sealed microwell cultures from 6 to 24 hours. Apoptotic assays for doxorubicin and staurosporine in 96-well plate format. Fluorescence data for the library screen for all conditions. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Schuster D, Laggner C, Langer T. Curr Pharm Design. 2005;11(27):3545–3559. doi: 10.2174/138161205774414510. [DOI] [PubMed] [Google Scholar]
- 2.Bleicher KH, Bohm HJ, Muller K, Alanine AI. Nat Rev Drug Discov. 2003;2(5):369–378. doi: 10.1038/nrd1086. [DOI] [PubMed] [Google Scholar]
- 3.Kang LF, Chung BG, Langer R, Khademhosseini A. Drug Discov Today. 2008;13(1–2):1–13. doi: 10.1016/j.drudis.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey SN, Sabatini DM, Stockwell BR. Proc Natl Acad Sci U S A. 2004;101(46):16144–16149. doi: 10.1073/pnas.0404425101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tavana H, Jovic A, Mosadegh B, Lee QY, Liu X, Luker KE, Luker GD, Weiss SJ, Takayama S. Nat Mater. 2009;8(9):736–41. doi: 10.1038/nmat2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee MY, Park CB, Dordick JS, Clark DS. Proc Natl Acad Sci U S A. 2005;102(4):983–987. doi: 10.1073/pnas.0406755102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee MY, Kumar RA, Sukumaran SM, Hogg MG, Clark DS, Dordick JS. Proc Natl Acad Sci U S A. 2008;105(1):59–63. doi: 10.1073/pnas.0708756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lii J, Hsu WJ, Parsa H, Das A, Rouse R, Sia SK. Anal Chem. 2008;80(10):3640–3647. doi: 10.1021/ac8000034. [DOI] [PubMed] [Google Scholar]
- 9.Toh YC, Lim TC, Tai D, Xiao GF, van Noort D, Yu HR. Lab Chip. 2009;9(14):2026–2035. doi: 10.1039/b900912d. [DOI] [PubMed] [Google Scholar]
- 10.Ma B, Zhang GH, Qin JH, Lin BC. Lab Chip. 2009;9(2):232–238. doi: 10.1039/b809117j. [DOI] [PubMed] [Google Scholar]
- 11.Khademhosseini A, Yeh J, Eng G, Karp J, Kaji H, Borenstein J, Farokhzad OC, Langer R. Lab Chip. 2005;5(12):1380–1386. doi: 10.1039/b508096g. [DOI] [PubMed] [Google Scholar]
- 12.King KR, Wang SH, Irimia D, Jayaraman A, Toner M, Yarmush ML. Lab Chip. 2007;7(1):77–85. doi: 10.1039/b612516f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye NN, Qin JH, Shi WW, Lin BC. Electrophoresis. 2007;28(7):1146–1153. doi: 10.1002/elps.200600450. [DOI] [PubMed] [Google Scholar]
- 14.Du Y, Shim J, Vidula M, Hancock MJ, Lo E, Chung BG, Borenstein JT, Khabiry M, Cropek DM, Khademhosseini A. Lab Chip. 2009;9(6):761–7. doi: 10.1039/b815990d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mosadegh B, Kuo CH, Tung YC, Torisawa YS, Bersano-Begey T, Tavana H, Takayama S. Nat Phys. 2010;6(6):433–437. doi: 10.1038/nphys1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugiura S, Edahiro J, Kikuchi K, Sumaru K, Kanamori T. Biotechnol Bioeng. 2008;100(6):1156–1165. doi: 10.1002/bit.21836. [DOI] [PubMed] [Google Scholar]
- 17.Sugiura S, Hattori K, Kanamori T. Anal Chem. 2010;82(19):8278–8282. doi: 10.1021/ac1017666. [DOI] [PubMed] [Google Scholar]
- 18.Wu G, Irvine J, Luft C, Pressley D, Hodge CN, Janzen B. Comb Chem H Throughput Screen. 2003;6(4):303–312. doi: 10.2174/138620703106298554. [DOI] [PubMed] [Google Scholar]
- 19.Wu JH, Wheeldon I, Guo YQ, Lu TL, Du YN, Wang B, He JK, Hu YQ, Khademhosseini A. Biomat. 2011;32(3):841–848. doi: 10.1016/j.biomaterials.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karp JM, Yeh J, Eng G, Fukuda J, Blumling J, Suh KY, Cheng J, Mahdavi A, Borenstein J, Langer R, Khademhosseini A. Lab Chip. 2007;7(6):786–794. doi: 10.1039/b705085m. [DOI] [PubMed] [Google Scholar]
- 21.Khademhosseini A, Yeh J, Jon S, Eng G, Suh KY, Burdick JA, Langer R. Lab Chip. 2004;4(5):425–430. doi: 10.1039/b404842c. [DOI] [PubMed] [Google Scholar]
- 22.Kang LF, Hancock MJ, Brigham MD, Khademhosseini A. J Biomed Mater Res A. 2010;93A(2):547–557. doi: 10.1002/jbm.a.32557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Missirlis D, Kawamura R, Tirelli N, Hubbell JA. Eur J Pharm Sci. 2006;29(2):120–9. doi: 10.1016/j.ejps.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 24.van Engeland M, Nieland LJW, Ramaekers FCS, Schutte B, Reutelingsperger CPM. Cytometry. 1998;31(1):1–9. doi: 10.1002/(sici)1097-0320(19980101)31:1<1::aid-cyto1>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 25.Yan XM, Habbersett RC, Cordek JM, Nolan JP, Yoshida TM, Jett JH, Marrone BL. Anal Biochem. 2000;286(1):138–148. doi: 10.1006/abio.2000.4789. [DOI] [PubMed] [Google Scholar]
- 26.Castaneda F, Rosin-Steiner S. Int J Med Sci. 2006;3(4):160–7. doi: 10.7150/ijms.3.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castaneda F, Kinne RK. Cancer Biol Ther. 2004;3(5):430–3. doi: 10.4161/cbt.3.5.806. [DOI] [PubMed] [Google Scholar]
- 28.Bertrand R, Solary E, O’Connor P, Kohn KW, Pommier Y. Exp Cell Res. 1994;211(2):314–21. doi: 10.1006/excr.1994.1093. [DOI] [PubMed] [Google Scholar]
- 29.Feng G, Kaplowitz N. Am J Physiol Gastrointest Liver Physiol. 2002;282(5):G825–34. doi: 10.1152/ajpgi.00467.2001. [DOI] [PubMed] [Google Scholar]
- 30.Kruman I, Guo Q, Mattson MP. J Neurosci Res. 1998;51(3):293–308. doi: 10.1002/(SICI)1097-4547(19980201)51:3<293::AID-JNR3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 31.Samali A, Nordgren H, Zhivotovsky B, Peterson E, Orrenius S. Biochem Biophys Res Commun. 1999;255(1):6–11. doi: 10.1006/bbrc.1998.0139. [DOI] [PubMed] [Google Scholar]
- 32.DiPietrantonio AM, Hsieh T, Wu JM. Biochem Biophys Res Commun. 1999;255(2):477–82. doi: 10.1006/bbrc.1999.0208. [DOI] [PubMed] [Google Scholar]
- 33.Hwang YS, Chung BG, Ortmann D, Hattori N, Moeller HC, Khademhosseini A. Proc Natl Acad Sci U S A. 2009;106(40):16978–16983. doi: 10.1073/pnas.0905550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.