Abstract
For more than 30 years the only genetic factor associated with susceptibility to multiple sclerosis (MS) was the HLA region. Recent advancements in genotyping platforms and the development of more effective statistical methods, resulted in the identification of 16 more genes by genome-wide association analyses (GWAS) in the last 3 years alone. While the effect of each of these genes is modest compared to that of HLA, this list is expected to grow significantly in the near future, thus defining a complex landscape in which susceptibility may be determined by a combination of allelic variants in different pathways according to ethnic background, disease sub-type, and specific environmental triggers. A considerable overlap of susceptibility genes among multiple autoimmune diseases is becoming evident and integration of these genetic variants with our current knowledge of affected biological pathways will greatly improve our understanding of mechanisms of general autoimmunity and of tissue specificity.
Introduction
Autoimmune disorders arise when physiological tolerance to “self” antigens is lost. Although several mechanisms may be involved in this pathogenic process, dysregulation of T-cell and B-cell activation and of pathways leading to inflammation are logical candidates. Multiple sclerosis (MS) is a common inflammatory disorder of the central nervous system (CNS) characterized by myelin loss, gliosis, varying degrees of axonal pathology, and progressive neurological dysfunction. A large body of research supports a multifactorial etiology, with an underlying genetic susceptibility likely acting in concert with undefined environmental exposures [1–3]. Although the exact pathogenic mechanisms underlying MS remains unknown, it has been proposed that lymphocytes activated in the periphery by a microbial mimic home to the CNS, become attached to receptors on endothelial cells, and then proceed to cross the blood-brain barrier (BBB) directly into the interstitial matrix. T cells are then reactivated in-situ by fragments of myelin antigens exposed in the context of human leukocyte antigen (HLA) molecules on the surface of antigen presenting cells (macrophages, microglia, and perhaps, astrocytes). Reactivation induces the release of proinflammatory cytokines that open further the BBB and stimulate chemotaxis, resulting in a second, larger wave of inflammatory cell recruitment and leakage of pathogenic antibodies and other plasma proteins into the nervous system.
Susceptibility to MS has been associated with multiple factors including genetics, epigenetics, and the environment. While the modest concordance rate in monozygotic twins (30–35%) suggests that environmental factors are major players in MS, increased heritability within families and the decrease in risk with degree of relatedness all argue in favor of genetic factors. Modern genomics developments have made available miniaturization and automation of genotyping platforms, and more than 200 genome-wide association studies (GWAS) have been performed in different diseases to date [4,5] including 31 studies in 7 common AID (7 in MS alone). In this chapter the findings on these studies will be summarized, and hypotheses about the possible pathogenic mechanisms implicated in MS will be elaborated.
The role of the HLA system in MS
The HLA region has been associated with hundreds of human diseases, including most autoimmune diseases (e.g. B27 with Ankylosing spondylitis, DR3 with Grave’s disease, Myasthenia Gravis, systemic lupus erythematosus (SLE), and Type 1 Diabetes (T1D); DR15 with MS) (for a review, see [6]). For most of these diseases, however, it has not yet been possible to show the molecular mechanisms underlying disease association with a particular HLA molecule(s). One possible explanation for this shortcoming is that it has often been difficult to unequivocally ascertain the primary disease-risk HLA gene(s) due to the remarkably strong linkage disequilibrium across the HLA [7]. There is still debate, for example, as to whether the HLA-DRB1*15:01 association explains the entire HLA–class II genetic linkage signal in MS [8–12] and whether susceptibility genes also exist within the class III region[13] and/or are telomeric to the class I region[14–16]. In addition, MS could result from the combination of different HLA molecules expressed at various loci (class-I and/or class-II) rather than the result of one HLA variant only. The situation has been complicated by the fact that susceptibility to MS is clearly polygenic and particular HLA allele(s), in combination with other genetic variants and environmental factors, may be required to develop the disease. Moreover, disease-relevant autoantigen(s) are largely unknown, which prevents a thorough three-dimensional analysis of HLA–peptide interactions. Last, but not least, MS is phenotypically very heterogeneous, in terms of clinical presentation, age at onset, co-morbidities with other autoimmune disorders, and severity, and it is likely that different alleles, or allelic combinations at different loci, will predispose to different forms of the disease [17].
Gene discovery in MS
Linkage studies
The strategy for a genetic linkage study requires the collection of family pedigrees with more than one affected member to track the inheritance and use highly polymorphic genetic markers to identify discrete chromosomal segments that deviate from independent segregation and co-segregate with the trait. Linkage screens with different levels of resolution and genome coverage have been completed in more than 30 datasets of familial MS cases [18]. Each of these studies suggested multiple chromosomal regions with potential involvement in disease susceptibility, consistent with the long-held view that MS is a polygenic disorder. However, only the HLA-class II locus on chromosome 6p21.3 has ever exceeded the threshold for formal statistical significance (a LOD>3 is considered proof of linkage) and no other region of statistically confident linkage has ever been observed. Using the values of HLA allele sharing by descent in sibships, it has been estimated that the HLA locus accounts for 20–60% of the genetic susceptibility in MS [19]. Even at the higher end of this estimate, much of the heritability of MS remains to be explained.
An International Multiple Sclerosis Genetics Consortium (IMSGC) reported in late 2005 the results of a linkage screen in 730 multi-case MS families using more than 4500 SNPs [20]. The peak logarithm of the odds (LOD) score of 11.7 found in the HLA illustrates the substantially greater power achieved by this high-density screen compared with earlier efforts. Strikingly, however, although numerous regions on multiple chromosomes revealed possible linkage signals of interest, no other locus reached genome-wide significance. These data indicate that any susceptibility allele common in the population and outside the HLA region is likely to increase MS risk by a factor of less than of 2. Similar findings have been reported for other autoimmune diseases.
Genome-wide association studies (GWAS)
The aim of a GWAS is to characterize the genetic architecture of a complex genetic trait through the identification of disease variants against the background of random variation seen in a population as a whole. In a typical study, hundreds of thousands of markers covering a significant portion of the common variation in the population are tested simultaneously in cases and controls and the allelic frequencies of each marker are compared between the two groups. Compared to linkage, association studies have greater statistical power to detect common genetic variants that confer a modest risk for a disease [21•]. Although population stratification and the introduction of phenocopies due to poor ascertainment of study participants are important confounding factors in association studies, inadequate sample size is undoubtedly the main reason why most published claims of association are suspected type I errors [22]. Hence, the identification of genes influencing the development of MS needs to rely primarily on association-based methods and must involve very large patient cohorts.
A total of seven positive GWAS have been reported to date in MS. The classic HLA-DRB1 risk locus stood out in all studies with remarkably strong statistical significance (e.g. P < 1×10−32 in [23]). These studies were followed by extensive replications efforts of the top hits [24–28] that together with a comprehensive replicated meta-analysis [29] provided robust evidence for approximately a dozen non-HLA loci affecting disease susceptibility (Table 1).
Table 1.
GWAS in MS
| Study | Year | Platform [SNPs passing QC] | Initial Sample Size | Replication Sample Size | Region | Gene(s) | SNPs | p-Value | OR | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Nischwitz et al. | 2010 | Illumina [~300,000] | 590 cases 825 controls |
NR | 10q11.21 19p13.2 9q34.2 6p21.32 |
Intergenic ZNF433 VAV2 DQA1 |
rs2503875 rs3745672 rs3780792 rs9271366 |
2.00E-07 1.00E-06 1.00E-06 4.00E-17 |
1.66 7.39 1.6 2.62 |
[61] |
| Sanna et al. | 2010 | Affymetrix [6,607,266] (imputed) | 882 cases 872 controls |
1,775 cases 2,005 controls |
6p21.32 3q13.11 |
HLA-DRB, HLA-DQB1 CBLB |
rs2040406 rs9657904 |
1.00E-20 2.00E-10 |
2.05 1.4 |
[62] |
| Jakkula et al. | 2010 | Illumina [297,343] | 68 cases 136 controls |
4,570 cases 10,143 controls |
17q21.2 6p21.32 |
STAT3 HLA |
rs744166 rs3135338 |
3.00E-10 2.00E-25 |
1.15 3.43 |
[63] |
| Bahlo et al. | 2009 | Illumina [302,098] | 1,618 cases 3,413 controls |
2,256 cases 2,310 controls |
10p15.1 20q13.12 16p12.1 1p22.1 1p13.1 6p21.32 8q24.21 12q14.1 |
IL2RA CD40 NR EVI5, RPL5 CD58 HLA-DRB1 ASAP1, DDEF1 METTL1, CYP27B1 |
rs2104286 rs6074022 rs8049603 rs6604026 rs1335532 rs9271366 rs6984045 rs703842 |
7.00E-06 1.00E-07 1.00E-06 3.00E-06 1.00E-07 7.00E-184 2.00E-06 5.00E-11 |
1.16 1.2 1.19 1.17 1.28 2.78 1.59 1.23 |
[64] |
| De Jager et al.* | 2009 | Affymetrix & Illumina [~2.56 million] (imputed) | 2,624 cases 7,220 controls |
2,215 cases 2,116 controls |
6p21.32 1p13.1 6p22.1 3q25.33 10p15.1 12p13.31 2q22.1 12p13.31 16p13.13 5p13.2 16q24.1 11q12.2 5p13.1 12q24.31 10q22.3 |
HLA-DRB1 CD58 HLA-B IL12A IL2RA TNFRSF1A CXCR4 TNFRSF1A CLEC16A IL7R IRF8 CD6 PTGER4 MPHOSPH9 ZMIZ1 |
rs3135388 rs2300747 rs2523393 rs4680534 rs2104286 rs4149584 rs882300 rs1800693 rs11865121 rs6897932 rs17445836 rs17824933 rs6896969 rs1790100 rs1250540 |
4.00E-225 3.00E-10 1.00E-17 6.00E-06 9.00E-08 5.00E-06 1.00E-07 2.00E-11 2.00E-07 2.00E-06 4.00E-09 4.00E-09 2.00E-07 7.00E-07 2.00E-06 |
2.75 1.3 1.28 1.12 1.15 1.58 1.19 1.2 1.15 1.12 1.25 1.18 1.1 1.11 1.12 |
[29] |
| Baranzini et al. | 2008 | Illumina [551,642] | 978 cases 883 controls |
NR | 13q31.3 9p22.2 8p23.2 12q12 3q23 4q35.1 20p13 2p25.1 3q24 2q14.2 |
GPC5 SH3GL2 CSMD1 PDZRN4 SLC25A36 MGC45800 C20orf46 DDEF2 ZIC1 EN1 |
rs9523762 rs1755289 rs1529316 rs1458175 rs908821 rs7672826 rs397020 rs1109670 rs1841770 rs651477 |
1.00E-06 3.00E-06 2.00E-06 2.00E-06 3.00E-06 8.00E-06 8.00E-07 9.00E-06 8.00E-06 7.00E-06 |
1.36 1.35 1.36 1.34 1.37 1.37 1.41 1.38 1.34 1.38 |
[23] |
| Comabella et al. | 2008 | Affymetrix[428,867] (pooled) | 242 cases 242 controls |
375 cases 375 controls |
6p21.32 | HLA-DRB1 | rs3129934 | 9.00E-11 | 3.3 | [65] |
| Hafler et al.. | 2007 | Affymetrix [334,923] | 931 trios 2,431 controls |
609 trios 2,322 cases 2,987 controls |
9q33 6p21.32 16p13.13 10p15.1 5p13.2 1p22.1 |
DBC1 HLA-DRA KIAA0350 IL2RA IL7RA RPL5 |
rs10984447 rs3135388 rs6498169 rs12722489 rs6897932 rs6604026 |
8.00E-06 9.00E-81 4.00E-06 3.00E-08 3.00E-07 8.00E-06 |
1.17 1.99 1.14 1.25 1.18 1.15 |
[66] |
Meta-analysis
It is important to note that these markers may not represent necessarily the causal disease variant themselves, explaining in part the very modest independent odds values for each allele. Additional follow-up experiments refined some of the association signals and revealed early mechanistic insights into the functional role of the identified genes, most notable a change in the soluble vs. membrane-bound ratio for the IL2 and IL7 receptors [30–32••] and diminished expression of CD58 mRNA [31]. It is noteworthy that some of the MS allelic variants were also proposed to be involved in other autoimmune diseases, suggesting common mechanisms underlying different autoimmune conditions [28,33–35••]. For example, the IL2RA-mediated susceptibility effect is shared among MS, T1D, Graves’ disease, and rheumatoid arthritis (RA) [32]. Interestingly, the direction of the association is not consistent across diseases as the IL2RA allelic variant associated with susceptibility to MS appears to confer resistance to T1D. A second allele confers susceptibility to both diseases, whereas yet a third allele is associated with susceptibility to T1D only [32,36]. Similarly, while the minor allele of the R620W polymorphism in PTPN22 has been associated with susceptibility to T1D, RA [37–40], and SLE [41,42], it appears to confer protection to CD [43]. Functional studies aimed at detecting tissue (or cell) specific variation in the expression or function of the target gene may contribute to elucidate the pathogenic mechanisms operating in each AID [36].
Altogether, the GWAS data seems to support the long-held view that MS susceptibility is conferred by the action of common (i.e. those with a risk allele frequency of >5%) sequence allelic variants in multiple genes [44]. Even with this expanding roster of risk loci, much of the heritability of MS remains unexplained. For example, the sorting and classification of duplications or deletions of genomic segments generating copy number variants (CNVs) lag behind. CNVs are a major source of human genetic diversity and have been shown to influence rare genomic disorders as well as complex traits and diseases [45–47]. Recent studies have reported associations between CNV and several autoimmune conditions such as SLE, psoriasis, CD, RA and T1D [48].
The second phase of the Wellcome Trust Case Control Consortium (WTCCC2), a massive collaborative project that is genotyping a total of 120,000 samples in MS and 12 other diseases (including ankylosing spondylitis, ulcerative colitis, and psoriasis) is now near completion [49]. The MS component of this project includes 10,000 cases genotyped with a high-SNP/CNV density platform. Such study will be adequately powered to identify common risk alleles with odds ratio as modest as 1.2 [50,51] thus practically guaranteeing identification of the complete set of common variants involved in MS.
Comparative analysis of susceptibility genes for MS and other autoimmune diseases
In most GWAS the number of markers in which the evidence for association exceeds the genome-wide significance threshold is small, and markers that do not exceed this threshold are generally neglected. A plausible strategy to increase the prior odds of finding a true significant marker is to incorporate prior biological knowledge into the GWAS data in the form of gene ontologies or pathways [52–56•]. The advantage of these methods is that even if markers in individual genes do not reach genome-wide significance, several modest associations in genes from the same biological pathway may highlight collectively its involvement in a disease process. Building on this rationale we merged statistical evidence from GWA analyses with experimental evidence of protein interaction from yeast two hybrid or chromatin immunoprecipitation (ChIP) studies to discover sub-networks (or modules) of interacting proteins associated with MS susceptibility [57]. Through this approach we were able to identify novel susceptibility pathways in MS, such as axon guidance and glutamate metabolism. We also uncovered genetic overlaps between MS and Alzheimer’s disease and bipolar disorder. In addition, the presence of common variants in the MHC region between MS, RA, and T1D, but not T2D or CD with MHC alleles was highlighted.
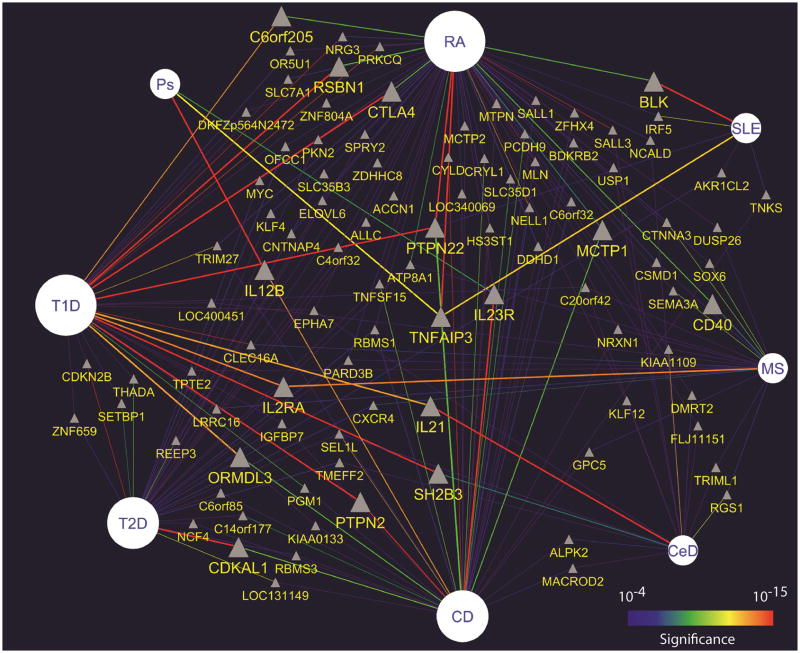
In order to systematically study genetic commonalities among autoimmune disorders we used publicly available information [5,58] to extract all moderately significant (p<10−4) associations from studies in celiac disease (CeD), Crohn’s disease (CD,) MS, psoriasis, RA, SLE, and T1D (plus those in T2D). Altogether, 1,201 genes with modest evidence for association in at least one of these autoimmune disorders were identified. We then used a network-based approach to visualize all the reported associations at once and identified 71 non-MHC genes shared by at least two diseases (Figure 1), 7 by three diseases, and only 2 by four diseases (Table 2) [33••]. This select list of potentially “general” autoimmunity genes includes PTPN22, a tyrosine phosphatase strongly associated with T1D (aggregate p<10−226), RA (aggregate p<10−90), and to a lesser extent with CD (aggregate p<10−8). TNFAIP3 is highly associated with RA (aggregate p<10−20), SLE (aggregate p<10−11), Ps (aggregate p<10−11), and moderately associated with CD (aggregate p<10−5). Other general autoimmunity genes are IL23R, KIAA1109 and CTLA4. Altogether, the data presented here suggests that genes involved in activation, proliferation, and homeostasis of cells involved in adaptive immune responses are more likely to represent general autoimmunity genes. This is further supported by the observation that a large proportion of these genes physically interact among each other (S. Baranzini, unpublished observation), thus possibly taking part in the same or highly overlapping biological pathways. A corollary to this observation indicates that associations unique to each disease would be responsible for attracting immune responses to specific tissues, although more functional studies in animal models will be needed to confirm this.
Figure 1. Autoimmune disease-gene network.
Top genetic associations in 7 autoimmune diseases and T2D. The most significant SNP per gene was selected. Only associations with significance of at least p<10−4 are visualized. If a given gene was identified in more than one disease, multiple lines connecting it with each disease were drawn. Lines are colored using a “heat” scheme according to the evidence for association. Thus “hot” edges (e.g. red, orange) represent more significant associations than “cold” edges (e.g. purple, blue). Diseases are depicted by circles of size proportional to the number of associated genes, non-MHC genes by grey triangles. To facilitate visualization, only genes shared by at least two diseases are shown.
Table 2.
Genes shared by at least three diseases at (aggregate) p< 10−4
| Gene | Chr | Description | Crohn’s | RA | T1D | Celiac | MS | SLE | Ps |
|---|---|---|---|---|---|---|---|---|---|
| PTPN22* | 1 | Protein tyrosine phosphatase, non-receptor type | 10−8 | 10−90 | 10−226 | 10−5 | |||
| IL23R | 1 | Interleukin 23 receptor | 10−102 | 10−4 | 10−7 | ||||
| NRXN1 | 2 | Neurexin 1 isoform beta precursor | 10−4 | 10−4 | 10−4 | ||||
| KIAA1109 | 4 | Hypothetical protein LOC84162 | 10−5 | 10−12 | 10−5 | ||||
| EPHA7 | 6 | Ephrin receptor EphA7 | 10−4 | 10−4 | 10−5 | ||||
| TRIM27 | 6 | Tripartite motif-containing 27 | 10−5 | 10−6 | 10−11 | ||||
| TNFAIP3* | 6 | Tumor necrosis factor, alpha-induced protein 3 | 10−5 | 10−20 | 10−11 | 10−11 | |||
| TNKS | 8 | Tankyrase | 10−4 | 10−4 | 10−6 | ||||
| C20orf42 | 20 | Fermitin family homolog 1 | 10−5 | 10−4 | 10−4 |
Genes involved in 4 diseases.
The next steps in MS genetics research
Despite the success of GWA studies in identifying novel susceptibility loci that withstood the challenge of independent replication, many questions remain concerning the genetic architecture of MS [59]. Noteworthy, all of the reported associations are derived from microarray-based studies, where only common variants (minor allele frequency > 10%) are interrogated in GWAS. Thus, the almost certain influence of evolutionarily younger (rare) variants has not been adequately evaluated. With the advent of next generation sequencing, more data is expected to be gathered in the near future to address this important question. The 1000 genomes project (www.1000genomes.org), is an international research consortium with the goal of finding most genetic variants that have frequencies of at least 1% in several populations around the world. Data emerging from this effort will undoubtedly uncover a multitude of private variants giving rise to a new catalog of human variation, an invaluable resource in the search for novel disease associations.
In a modest, but important first step to assess this hypothesis we recently sequenced the entire genomes of a female MS-discordant MZ twin pair, generating over one billion, high quality, shot gun whole-genome reads, corresponding to approximately 22 fold coverage of each genome [60•]. These are the first female, twin, and autoimmune genome sequences reported to date. Among ~3.6 million single nucleotide polymorphisms (SNPs), ~200,000 insertion-deletion polymorphisms (indels), 27 copy number variations (CNVs) and 1.1 million mCpG dinucleotides detected, no SNPs, indels or CNVs and methylation of cytosine residues differed between the twins. We also analyzed the full mRNA transcriptome and epigenome sequences of CD4+ lymphocytes from 3 pair MZ, discordant twins for MS. While 19,000 genes were expressed in each of the CD4+ T cell preparations, no reproducible transcriptional differences were identified between MS-affected and unaffected twins. Surprisingly, only 2–176 differences in methylation of ~2 million CpG dinucleotides were detected between siblings in three twin pairs, in contrast to ~800 methylation differences between T cells of unrelated individuals and several thousand differences between tissues or normal and cancerous tissues. The sequence of many more discordant twin pairs, patients and control genomes is necessary to realize the power of this approach. Given the impressive rate of advances in the field, the technology for whole genome sequencing will be accurate and inexpensive enough for large-scale application within the next few years.
Conclusions
The genetic bases of autoimmune diseases are just starting to be uncovered. While several bona-fide susceptibility genes have been identified in most common traits, technological advances in high throughput genotyping platforms and more affordable second generation sequencing methods will contribute to significantly expand these lists. In addition, structural variants (insertion/deletion polymorphisms, copy number variations, etc) are also likely to play a significant role in determining susceptibility to AID. Taking into account the known susceptibility loci for each trait, disease-specific custom genotyping chips will be designed so as to cover a wide spectrum of variants in larger cohorts of individuals. At the same time, deep sequencing of candidate regions will be carried out to identify rare (private) mutations and structural variants that affect only a few individuals. Together, these approaches will eventually discover most if not all of the genetic contribution to these diseases and allow for the systematic search of similarity and differences among them.
While the identification of the precise pathways involved in susceptibility to AID will clearly require additional time and effort, integration of data from multiple diseases represents the logical next step in discovering similarities and differences among them. While similarities will shed light into the general mechanism behind autoimmunity, genetic features private to a given disease will help pinpoint the basis for tissue specificity. One obvious benefit of this new knowledge would be the cross-utilization of drugs for diseases with similar genetic fingerprint. Ultimately, this high resolution genetic disease landscape will contribute to more accurate models of pathogenesis setting the bases for the development of more rational therapeutic approaches.
Acknowledgments
SEB is a Harry Weaver Neuroscience Scholar of the National Multiple Sclerosis Society (NMSS). This work was also supported by grants from the NMSS (RG4051A2, RG2901D9), and from the National Institute for Neurological Disorders and Stroke (RO1NS049477).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hauser SL, Goodin DS. Multiple Sclerosis and other demyelinating diseases. In: Fauci AS, editor. Harrison’s principles of internal medicine. 17. McGraw-Hill Medical; 2008. pp. 405–422. [Google Scholar]
- 2.Compston A. McAlpine’s multiple sclerosis. 4. Philadelphia: Churchill Livingstone Elsevier; 2005. [Google Scholar]
- 3.Oksenberg JR, Baranzini SE, Barcellos LF, Hauser SL. Multiple sclerosis: genomic rewards. J Neuroimmunol. 2001;113:171–184. doi: 10.1016/s0165-5728(00)00444-6. [DOI] [PubMed] [Google Scholar]
- 4.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.NHGRI: A Catalog of Published Genome-Wide Association Studies. Edited by. Bethesda, MD. NHGRI; 2009: The genome-wide association study (GWAS) publications listed here include only those attempting to assay at least 100,000 single nucleotide polymorphisms (SNPs) in the initial stage. Publications are organized from most to least recent date of publication, indexing from online publication if available. Studies focusing only on candidate genes are excluded from this catalog. Studies are identified through weekly PubMed literature searches, daily NIH-distributed compilations of news and media reports, and occasional comparisons with an existing database of GWAS literature (HuGE Navigator).
- 6.Shiina T, Inoko H, Kulski JK. An update of the HLA genomic region, locus information and disease associations: 2004. Tissue Antigens. 2004;64:631–649. doi: 10.1111/j.1399-0039.2004.00327.x. [DOI] [PubMed] [Google Scholar]
- 7.Miretti MM, Walsh EC, Ke X, Delgado M, Griffiths M, Hunt S, Morrison J, Whittaker P, Lander ES, Cardon LR, et al. A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:634–646. doi: 10.1086/429393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukazawa T, Sasaki H, Kikuchi S, Hamada T, Tashiro K. Genetics of multiple sclerosis. Biomed Pharmacother. 2000;54:103–106. doi: 10.1016/S0753-3322(00)88860-5. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez O, Fernandez V, Alonso A, Caballero A, Luque G, Bravo M, Leon A, Mayorga C, Leyva L, de Ramon E. DQB1*0602 allele shows a strong association with multiple sclerosis in patients in Malaga, Spain. J Neurol. 2004;251:440–444. doi: 10.1007/s00415-004-0350-2. [DOI] [PubMed] [Google Scholar]
- 10.Khare M, Mangalam A, Rodriguez M, David CS. HLA DR and DQ interaction in myelin oligodendrocyte glycoprotein-induced experimental autoimmune encephalomyelitis in HLA class II transgenic mice. J Neuroimmunol. 2005;169:1–12. doi: 10.1016/j.jneuroim.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 11.Prat E, Tomaru U, Sabater L, Park DM, Granger R, Kruse N, Ohayon JM, Bettinotti MP, Martin R. HLA-DRB5*0101 and -DRB1*1501 expression in the multiple sclerosis-associated HLA-DR15 haplotype. J Neuroimmunol. 2005;167:108–119. doi: 10.1016/j.jneuroim.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 12.Sospedra M, Muraro PA, Stefanova I, Zhao Y, Chung K, Li Y, Giulianotti M, Simon R, Mariuzza R, Pinilla C, et al. Redundancy in antigen-presenting function of the HLA-DR and -DQ molecules in the multiple sclerosis-associated HLA-DR2 haplotype. J Immunol. 2006;176:1951–1961. doi: 10.4049/jimmunol.176.3.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Jong BA, Huizinga TWJ, Zanelli E, Giphart MJ, Bollen ELEM, Uitdehaag BMJ, Polman CH, Westendorp RGJ. Evidence for additional genetic risk indicators of relapse-onset MS within the HLA region. Neurology. 2002;59:549–555. doi: 10.1212/wnl.59.4.549. [DOI] [PubMed] [Google Scholar]
- 14.Marrosu MG, Murru R, Murru MR, Costa G, Zavattari P, Whalen M, Cocco E, Mancosu C, Schirru L, Solla E, et al. Dissection of the HLA association with multiple sclerosis in the founder isolated population of Sardinia. Hum Mol Genet. 2001;10:2907–2916. doi: 10.1093/hmg/10.25.2907. [DOI] [PubMed] [Google Scholar]
- 15.Rubio JP, Bahlo M, Butzkueven H, van Der Mei IA, Sale MM, Dickinson JL, Groom P, Johnson LJ, Simmons RD, Tait B, et al. Genetic dissection of the human leukocyte antigen region by use of haplotypes of Tasmanians with multiple sclerosis. Am J Hum Genet. 2002;70:1125–1137. doi: 10.1086/339932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeo TW, De Jager PL, Gregory SG, Barcellos LF, Walton A, Goris A, Fenoglio C, Ban M, Taylor CJ, Goodman RS, et al. A second major histocompatibility complex susceptibility locus for multiple sclerosis. Ann Neurol. 2007;61:228–236. doi: 10.1002/ana.21063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caillat-Zucman S. Molecular mechanisms of HLA association with autoimmune diseases. Tissue Antigens. 2009;73:1–8. doi: 10.1111/j.1399-0039.2008.01167.x. [DOI] [PubMed] [Google Scholar]
- 18.Fernald GH, Yeh RF, Hauser SL, Oksenberg JR, Baranzini SE. Mapping gene activity in complex disorders: Integration of expression and genomic scans for multiple sclerosis. J Neuroimmunol. 2005;167:157–169. doi: 10.1016/j.jneuroim.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 19.Haines JL, Terwedow HA, Burgess K, Pericak-Vance MA, Rimmler JB, Martin ER, Oksenberg JR, Lincoln R, Zhang DY, Banatao DR, et al. Linkage of the MHC to familial multiple sclerosis suggests genetic heterogeneity. The Multiple Sclerosis Genetics Group. Hum Mol Genet. 1998;7:1229–1234. doi: 10.1093/hmg/7.8.1229. [DOI] [PubMed] [Google Scholar]
- 20.Sawcer S, Ban M, Maranian M, Yeo TW, Compston A, Kirby A, Daly MJ, De Jager PL, Walsh E, Lander ES, et al. A high-density screen for linkage in multiple sclerosis. Am J Hum Genet. 2005;77:454–467. doi: 10.1086/444547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. In this seminal article the authors describe the oportunities and limitations of genetics analyses and set parameters for adequately powered studies. [DOI] [PubMed] [Google Scholar]
- 22.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 23.Baranzini SE, Wang J, Gibson RA, Galwey N, Naegelin Y, Barkhof F, Radue EW, Lindberg RL, Uitdehaag BM, Johnson MR, et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet. 2009;18:767–778. doi: 10.1093/hmg/ddn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubio JP, Stankovich J, Field J, Tubridy N, Marriott M, Chapman C, Bahlo M, Perera D, Johnson LJ, Tait BD, et al. Replication of KIAA0350, IL2RA, RPL5 and CD58 as multiple sclerosis susceptibility genes in Australians. Genes Immun. 2008;9:624–630. doi: 10.1038/gene.2008.59. [DOI] [PubMed] [Google Scholar]
- 25.Ban M, Goris A, Lorentzen AR, Baker A, Mihalova T, Ingram G, Booth DR, Heard RN, Stewart GJ, Bogaert E, et al. Replication analysis identifies TYK2 as a multiple sclerosis susceptibility factor. Eur J Hum Genet. 2009;17:1309–1313. doi: 10.1038/ejhg.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D’Netto MJ, Ward H, Morrison KM, Ramagopalan SV, Dyment DA, DeLuca GC, Handunnetthi L, Sadovnick AD, Ebers GC. Risk alleles for multiple sclerosis in multiplex families. Neurology. 2009;72:1984–1988. doi: 10.1212/WNL.0b013e3181a92c25. [DOI] [PubMed] [Google Scholar]
- 27.Hoppenbrouwers IA, Aulchenko YS, Ebers GC, Ramagopalan SV, Oostra BA, van Duijn CM, Hintzen RQ. EVI5 is a risk gene for multiple sclerosis. Genes Immun. 2008;9:334–337. doi: 10.1038/gene.2008.22. [DOI] [PubMed] [Google Scholar]
- 28.Hafler JP, Maier LM, Cooper JD, Plagnol V, Hinks A, Simmonds MJ, Stevens HE, Walker NM, Healy B, Howson JM, et al. CD226 Gly307Ser association with multiple autoimmune diseases. Genes Immun. 2009;10:5–10. doi: 10.1038/gene.2008.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Jager PL, Jia X, Wang J, de Bakker PI, Ottoboni L, Aggarwal NT, Piccio L, Raychaudhuri S, Tran D, Aubin C, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Gregory SG, Schmidt S, Seth P, Oksenberg JR, Hart J, Prokop A, Caillier SJ, Ban M, Goris A, Barcellos LF, et al. Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat Genet. 2007;39:1083–1091. doi: 10.1038/ng2103. The association with a variant in IL7Ra was reported. Authors demostrate that this SNP affects the amount of soluble vs. membrane bound receptor in the surface of immune cells. This was one of the first two non-HLA genes involved in MS susceptibility. [DOI] [PubMed] [Google Scholar]
- 31••.De Jager PL, Baecher-Allan C, Maier LM, Arthur AT, Ottoboni L, Barcellos L, McCauley JL, Sawcer S, Goris A, Saarela J, et al. The role of the CD58 locus in multiple sclerosis. Proc Natl Acad Sci U S A. 2009;106:5264–5269. doi: 10.1073/pnas.0813310106. Authors identify a variant in the CD58 gene that is associated with MS and has a protective effect. Functional studies demostrate a dose dependent effect in the expression of CD58 conditioned to the genotype and a higher expression in patietns during remission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Maier LM, Lowe CE, Cooper J, Downes K, Anderson DE, Severson C, Clark PM, Healy B, Walker N, Aubin C, et al. IL2RA genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS Genet. 2009;5:e1000322. doi: 10.1371/journal.pgen.1000322. This work report functional analysis and the presence of allelic heterogeneity in the IL2Ra gene, the second confirmed non-HLA signal in MS. A variant that confers susceptibility to MS is associated with protection in T1D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Baranzini SE. The genetics of autoimmune diseases: a networked perspective. Curr Opin Immunol. 2009;21:596–605. doi: 10.1016/j.coi.2009.09.014. A systematic comparison of all available genetic susceptibility information is perfomed using a network-based visualization system. Unbiased, quantitative estimates of disease similarities and differences are provided. [DOI] [PubMed] [Google Scholar]
- 34••.IMSGC. The expanding genetic overlap between multiple sclerosis and type I diabetes. Genes Immun. 2009;10:11–14. doi: 10.1038/gene.2008.83. Two polymorphisms associated with T1D (CD226 and CLEC16A) were also found to be associated with MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sirota M, Schaub MA, Batzoglou S, Robinson WH, Butte AJ. Autoimmune disease classification by inverse association with SNP alleles. PLoS Genet. 2009;5:e1000792. doi: 10.1371/journal.pgen.1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dendrou CA, Plagnol V, Fung E, Yang JH, Downes K, Cooper JD, Nutland S, Coleman G, Himsworth M, Hardy M, et al. Cell-specific protein phenotypes for the autoimmune locus IL2RA using a genotype-selectable human bioresource. Nat Genet. 2009;41:1011–1015. doi: 10.1038/ng.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gregersen PK, Amos CI, Lee AT, Lu Y, Remmers EF, Kastner DL, Seldin MF, Criswell LA, Plenge RM, Holers VM, et al. REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat Genet. 2009;41:820–823. doi: 10.1038/ng.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raychaudhuri S, Remmers EF, Lee AT, Hackett R, Guiducci C, Burtt NP, Gianniny L, Korman BD, Padyukov L, Kurreeman FA, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40:1216–1223. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, Liew A, Khalili H, Chandrasekaran A, Davies LR, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.WTCCC. Genome-wide association study of 14, 000 cases of seven common diseases and 3, 000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, Lee AT, Chung SA, Ferreira RC, Pant PV, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 43.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.IMSGC. Comprehensive follow-up of the first genome-wide association study of multiple sclerosis identifies KIF21B and TMEM39A as susceptibility loci. Hum Mol Genet. 2010;19:953–962. doi: 10.1093/hmg/ddp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wain LV, Armour JA, Tobin MD. Genomic copy number variation, human health, and disease. Lancet. 2009;374:340–350. doi: 10.1016/S0140-6736(09)60249-X. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, Martens JW, Yu JX, Jiang J, Sieuwerts AM, Smid M, Klijn JG, Wang Y, Foekens JA. Copy number alterations that predict metastatic capability of human breast cancer. Cancer Res. 2009;69:3795–3801. doi: 10.1158/0008-5472.CAN-08-4596. [DOI] [PubMed] [Google Scholar]
- 48.Schaschl H, Aitman TJ, Vyse TJ. Copy number variation in the human genome and its implication in autoimmunity. Clin Exp Immunol. 2009;156:12–16. doi: 10.1111/j.1365-2249.2008.03865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.WTCCC2: Wellcome Trust Case Control Consortium 2. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oksenberg JR, Baranzini SE, Sawcer S, Hauser SL. The genetics of multiple sclerosis: SNPs to pathways to pathogenesis. Nat Rev Genet. 2008;9:516–526. doi: 10.1038/nrg2395. [DOI] [PubMed] [Google Scholar]
- 52•.Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat Rev Genet. 2010;11:843–854. doi: 10.1038/nrg2884. This article summarizes most of the current methods used to integrate all the genetic susceptibility information and identify biological pathways associated with a given disease. [DOI] [PubMed] [Google Scholar]
- 53.Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 2005;37:413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- 54.Lesnick TG, Papapetropoulos S, Mash DC, Ffrench-Mullen J, Shehadeh L, de Andrade M, Henley JR, Rocca WA, Ahlskog JE, Maraganore DM. A genomic pathway approach to a complex disease: axon guidance and Parkinson disease. PLoS Genet. 2007;3:e98. doi: 10.1371/journal.pgen.0030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torkamani A, Topol EJ, Schork NJ. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics. 2008;92:265–272. doi: 10.1016/j.ygeno.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang K, Li M, Bucan M. Pathway-Based Approaches for Analysis of Genomewide Association Studies. Am J Hum Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baranzini SE, Galwey NW, Wang J, Khankhanian P, Lindberg R, Pelletier D, Wu W, Uitdehaag BM, Kappos L, Polman CH, et al. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum Mol Genet. 2009;18:2078–2090. doi: 10.1093/hmg/ddp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson AD, O’Donnell CJ. An open access database of genome-wide association results. BMC Med Genet. 2009;10:6. doi: 10.1186/1471-2350-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oksenberg JR, Baranzini SE. Multiple sclerosis genetics--is the glass half full, or half empty? Nat Rev Neurol. 2010;6:429–437. doi: 10.1038/nrneurol.2010.91. [DOI] [PubMed] [Google Scholar]
- 60•.Baranzini SE, Mudge J, van Velkinburgh JC, Khankhanian P, Khrebtukova I, Miller NA, Zhang L, Farmer AD, Bell CJ, Kim RW, et al. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature. 2010;464:1351–1356. doi: 10.1038/nature08990. This is the first report of a complete genome sequencing of monozygotic twins. Although the twins are discordant for MS, no reproducible differences were found in either the genome, methylome or transcriptome of these individuals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nischwitz S, Cepok S, Kroner A, Wolf C, Knop M, Muller-Sarnowski F, Pfister H, Roeske D, Rieckmann P, Hemmer B, et al. Evidence for VAV2 and ZNF433 as susceptibility genes for multiple sclerosis. J Neuroimmunol. 2010;227:162–166. doi: 10.1016/j.jneuroim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 62.Sanna S, Pitzalis M, Zoledziewska M, Zara I, Sidore C, Murru R, Whalen MB, Busonero F, Maschio A, Costa G, et al. Variants within the immunoregulatory CBLB gene are associated with multiple sclerosis. Nat Genet. 2010;42:495–497. doi: 10.1038/ng.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jakkula E, Leppa V, Sulonen AM, Varilo T, Kallio S, Kemppinen A, Purcell S, Koivisto K, Tienari P, Sumelahti ML, et al. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene. Am J Hum Genet. 2010;86:285–291. doi: 10.1016/j.ajhg.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.ANZgene. Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet. 2009;41:824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 65.Comabella M, Craig DW, Camina-Tato M, Morcillo C, Lopez C, Navarro A, Rio J, Montalban X, Martin R. Identification of a novel risk locus for multiple sclerosis at 13q31.3 by a pooled genome-wide scan of 500,000 single nucleotide polymorphisms. PLoS ONE. 2008;3:e3490. doi: 10.1371/journal.pone.0003490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, de Bakker PI, Gabriel SB, Mirel DB, Ivinson AJ, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]