Abstract
This paper presents a new approach to inverting (fitting) models of coupled dynamical systems based on state-of-the-art (cubature) Kalman filtering. Crucially, this inversion furnishes posterior estimates of both the hidden states and parameters of a system, including any unknown exogenous input. Because the underlying generative model is formulated in continuous time (with a discrete observation process) it can be applied to a wide variety of models specified with either ordinary or stochastic differential equations. These are an important class of models that are particularly appropriate for biological time-series, where the underlying system is specified in terms of kinetics or dynamics (i.e., dynamic causal models). We provide comparative evaluations with generalized Bayesian filtering (dynamic expectation maximization) and demonstrate marked improvements in accuracy and computational efficiency. We compare the schemes using a series of difficult (nonlinear) toy examples and conclude with a special focus on hemodynamic models of evoked brain responses in fMRI. Our scheme promises to provide a significant advance in characterizing the functional architectures of distributed neuronal systems, even in the absence of known exogenous (experimental) input; e.g., resting state fMRI studies and spontaneous fluctuations in electrophysiological studies. Importantly, unlike current Bayesian filters (e.g. DEM), our scheme provides estimates of time-varying parameters, which we will exploit in future work on the adaptation and enabling of connections in the brain.
Keywords: Neuronal, fMRI, Blind deconvolution, Cubature Kalman filter, Smoother, Stochastic, Hemodynamic modeling, Dynamic Expectation Maximization, Nonlinear
Introduction
The propagation of neuronal activity in the brain is a dynamic process, which mediates the communication among functional brain areas. Although, recent advances in neuroimaging allow for greater insights into brain function, all available noninvasive brain mapping techniques provide only indirect measures of the underlying electrophysiology. For example, we cannot observe the time-varying neuronal activation in the brain but we can measure the electrical field it generates on the scalp using electroencephalography (EEG). Similarly, in functional magnetic resonance imaging (fMRI) we measure hemodynamic responses, which represent changes in blood flow and blood oxygenation that follow neuronal activation. Crucially, the form of this hemodynamic response can vary across subjects and different brain regions (Aguirre et al., 1998; Handwerker et al., 2004). This complicates the estimation of hidden neuronal states and identification of the effective connectivity (i.e. directed influence) between different brain regions (David, 2009; David et al., 2008; Friston, 2009; Roebroeck et al., 2009a, b).
In general, the relationship between initial neuronal activation and our observations rests on a complex electro/bio-physiological process. If this process is known and well described, it can be approximated by mathematical modeling. Inversion of the ensuing model allows us to estimate hidden states of neuronal systems (e.g., the neuronal activation) from observations. The resulting estimate will be affected by the accuracy of the inversion (formulated as an optimization problem) and by the precision of the observation itself (temporal resolution, signal to noise ratio (SNR), etc.). In signal processing theory, this problem is called blind deconvolution and is described as estimating the unknown input to a dynamic system, given output data, when the model of the system contains unknown parameters. A note on terminology is needed here: although convolution is usually defined as a linear operation, the term deconvolution is generally used in reference to the inversion of nonlinear (generalized) convolution models (i.e. restoration); we adhere to this convention.
In fMRI, the physiological mechanisms mediating the relationship between neuronal activation and vascular/metabolic systems have been studied extensively (Attwell et al., 2010; Iadecola, 2002; Magistretti and Pellerin, 1999) and models of hemodynamic responses have been described at macroscopic level by systems of differential equations. The hemodynamic model (Friston et al., 2000) links neuronal activity to flow and subsumes the balloon-windkessel model (Buxton et al., 1998; Mandeville et al., 1999a), linking flow to observed fMRI signals. The hemodynamic model includes model of neurovascular coupling (i.e., how changes in neuronal activity cause a flow-inducing signal) and hemodynamic processes (i.e. changes in cerebral blood flow (CBF), cerebral blood volume (CBV), and total de-oxyhemoglobin (dHb)). In this paper, we will focus on a hemodynamic model of a single region in fMRI, where experimental studies suggest that the neuronal activity that drives hemodynamic responses corresponds more to afferent synaptic activity (as opposed to efferent spiking activity (Lauritzen, 2001; Logothetis, 2002)). In the future work, we will use exactly the same scheme to model distributed neuronal activity as observed in multiple regions.
The hemodynamic model is nonlinear in nature (Berns et al., 1999; Mechelli et al., 2001). Therefore, to infer the hidden states and parameters of the underlying system, we require methods that can handle these nonlinearities. In Friston et al. (2000), the parameters of a hemodynamic model were estimated using a Volterra kernel expansion to characterize the hemodynamic response. Later, Friston et al. (2002) introduced a Bayesian estimation framework to invert (i.e., fit) the hemodynamic model explicitly. This approach accommodated prior constraints on parameters and avoided the need for Volterra kernels. Subsequently, the approach was generalized to cover networks of coupled regions and to include parameters controlling the neuronal coupling (effective connectivity) among brain regions (Friston et al., 2003). The Bayesian inversion of these models is known as dynamic causal modeling (DCM) and is now used widely to analyses effective connectivity in fMRI and electrophysiological studies. However, current approaches to hemodynamic and causal models only account for noise at the level of the measurement; where this noise includes thermally generated random noise and physiological fluctuations. This is important because physiological noise represents stochastic fluctuations due to metabolic and vascular responses, which affect the hidden states of the system; furthermore, neuronal activity can show pronounced endogenous fluctuations (Biswal et al., 1995; Krüger and Glover, 2001). Motivated by this observation, Riera et al. (2004) proposed a technique based on a fully stochastic model (i.e. including physiological noise) that used the local linearization filter (LLF) (Jimenez and Ozaki, 2003), which can be considered a form of extended Kalman filtering (EKF) (Haykin, 2001) for continuous dynamic systems. Besides estimating hemodynamic states and parameters, this approach allows one to estimate the system’s input, i.e. neuronal activity; by its parameterization via radial basis functions (RBFs). In Riera et al. (2004), the number of RBFs was considered fixed a priori, which means that the solution has to lie inside a regularly distributed but sparse space (otherwise, the problem is underdetermined). Recently, the LLF technique was applied by Sotero at al. (2009) to identify the states and parameters of a metabolic/hemodynamic model.
The hemodynamic response and hidden states of hemodynamic models possess strong nonlinear characteristics, which are prescient with respect to stimulus duration (Birn et al., 2001; Miller et al., 2001). This makes one wonder whether a linearization approach such as LLF can handle such strong nonlinearities. Johnston et al. (2008) proposed particle filtering, a sequential Monte Carlo method, that accommodates true nonlinearities in the model. The approach of Johnston et al. was shown to be both accurate and robust, when used to estimate hidden physiologic and hemodynamic states; and was superior to the LLF. Similarly, two-pass particle filtering, including a smoothing (backwards pass) procedure, was introduced by Murray et al. (2008). Another attempt to infer model parameters and hidden states used the unscented Kalman filter (UKF), which is more suitable for highly nonlinear problems (Hu et al., 2009). Finally, Jacobson et al. (2008) addressed inference on model parameters, using a Metropolis–Hastings algorithm for sampling their posterior distribution.
None of the methods mentioned above, except (Riera et al., 2004) with its restricted parameterization of the input, can perform a complete deconvolution of fMRI signals and estimate both hidden states and input; i.e. the neuronal activation, without knowing the input (stimulation function). Here, an important exception is the methodology introduced by Friston et al. (2008) called dynamic expectation maximization (DEM) and its generalizations: variational filtering (Friston, 2008a) and generalized filtering (Friston et al., 2010). DEM represents a variational Bayesian technique (Hinton and van Camp, 1993; MacKay, 1995), that is applied to models formulated in terms of generalized coordinates of motion. This scheme allows one to estimate not only the states and parameters but also the input and hyperparameters of the system generating those states. Friston et al. (2008) demonstrated the robustness of DEM compared to standard Bayesian filtering methods, particularly the extended Kalman filter and particle filter, on a selection of difficult nonlinear/linear dynamic systems. They concluded that standard methods are unable to perform joint estimation of the system input and states, while inferring the model parameters.
In this paper, we propose an estimation scheme that is based on nonlinear Kalman filtering, using the recently introduced cubature Kalman filter (CKF) (Arasaratnam and Haykin, 2009), which is recognized as the closest known approximation to Bayesian filtering. Our procedure applies a forward pass using the CKF that is finessed by a backward pass of the cubature Rauch-Tung-Striebel smoother. Moreover, we utilize the efficient square-root formulation of these algorithms. Crucially, we augment the hidden states with both parameters and inputs, enabling us to identify hidden states, model parameters and estimate the system input. We will show that we can obtain accurate estimates of hidden hemodynamic and neuronal states, well beyond the temporal resolution of fMRI.
The paper is structured as follows: First, we review the general concept of nonlinear continuous-discrete state-space models for simultaneous estimation of the system hidden states, its input and parameters. We then introduce the forward-backward cubature Kalman estimation procedure in its stable square-root form, as a suitable method for solving this complex inversion problem. Second, we provide a comprehensive evaluation of our proposed scheme and compare it with DEM. For this purpose, we use the same nonlinear/linear dynamic systems that were used to compare DEM with the EKF and particle filter algorithms (Friston et al., 2008). Third, we devote special attention to the deconvolution problem, given observed hemodynamic responses; i.e. to the estimation of neuronal activity and parameter identification of a hemodynamic model. Again, we provide comparative evaluations with DEM and discuss the advantages and limitations of each approach, when applied to fMRI data.
Nonlinear continuous-discrete state-space models
Nonlinear filtering problems are typically described by state-space models comprising a process and measurement equations. In many practical problems, the process equation is derived from the underlying physics of a continuous dynamic system, and is expressed in the form of a set of differential equations. Since the measurements y are acquired by digital devices; i.e. they are available at discrete time points (t = 1,2, …, T), we have a model with a continuous process equation and a discrete measurement equation. The stochastic representation of this state-space model, with additive noise, can be formulated as:
| (1) |
where θt represents unknown parameters of the equation of motion h and the measurement function g, respectively; ut is the exogenous input (the cause) that drives hidden states or the response; rt is a vector of random Gaussian measurement noise, rt~ (0, Rt); I(xt, t) can be a function of the state and time; and βt denotes a Wiener process or state noise that is assumed to be independent of states and measurement noise.
(0, Rt); I(xt, t) can be a function of the state and time; and βt denotes a Wiener process or state noise that is assumed to be independent of states and measurement noise.
The continuous time formulation of the stochastic differential equations (SDE) in (1) can also be expressed using Riemann and Ito integrals (Kloeden and Platen, 1999):
| (2) |
where the second integral is stochastic. This equation can be further converted into a discrete-time analogue using numerical integration such as Euler-Maruyama method or the local linearization (LL) scheme (Biscay et al., 1996; Ozaki, 1992). This leads to the standard form of a first order autoregressive process (AR(1)) of nonlinear state-space models:
| (3) |
where qt is a zero-mean Gaussian state noise vector; qt~ (0, Qt). Our preference is to use LL-scheme, which has been demonstrated to improve the order of convergence and stability properties of conventional numerical integrators (Jimenez et al., 1999). In this case, the function f is evaluated through:
(0, Qt). Our preference is to use LL-scheme, which has been demonstrated to improve the order of convergence and stability properties of conventional numerical integrators (Jimenez et al., 1999). In this case, the function f is evaluated through:
| (4) |
where fxt is a Jacobian of h and Δt is the time interval between samples (up to the sampling interval). The LL method allows integration of a SDE near discretely and regularly distributed time instants, assuming local piecewise linearity. This permits the conversion of a SDE system into a state-space equation with Gaussian noise. A stable reconstruction of the trajectories of the state-space variables is obtained by a one step prediction. Note that expression in (4) is not always the most practical; it assumes the Jacobian has full rank. See (Jimenez, 2002) for alternative forms.
Probabilistic inference
The problem of estimating the hidden states (causing data), parameters (causing the dynamics of hidden states) and any non-controlled exogenous input to the system, in a situation when only observations are given, requires probabilistic inference. In a Markovian setting, the optimal solution to this problem is given by the recursive Bayesian estimation algorithm which recursively updates the posterior density of the system state as new observations arrive. This posterior density constitutes the complete solution to the probabilistic inference problem, and allows us to calculate an “optimal” estimate of the state. In particular, the hidden state xt, with initial probability p (x0), evolves over time as an indirect or partially observed first-order Markov process, according to the conditional probability density p (xt|xt−1). The observations yt are conditionally independent, given the state, and are generated according to the conditional posterior probability density p (yt|xt). In this sense, the discrete-time variant of state-space model presented in Eq. (3) can also be written in terms of transition densities and a Gaussian likelihood:
| (5) |
The state transition density p (xt|xt−1) is fully specified by f and the state noise distribution p (qt), whereas g and the measurement noise distribution p(rt) fully specify the observation likelihood p (yt|xt). The dynamic state-space model, together with the known statistics of the noise (and the prior distribution of the system states), defines a probabilistic generative model of how system evolves over time and of how we (partially or inaccurately) observe this hidden state (Van der Merwe, 2004).
Unfortunately, the optimal Bayesian recursion is usually tractable only for linear, Gaussian systems, in which case the closed-form recursive solution is given by the classical Kalman filter (Kalman, 1960) that yields the optimal solution in the minimum-mean-square-error (MMSE) sense, the maximum likelihood (ML) sense, and the maximum a posteriori (MAP) sense. For more general real-world (nonlinear, non-Gaussian) systems the optimal Bayesian recursion is intractable and an approximate solution must be used.
Numerous approximation solutions to the recursive Bayesian estimation problem have been proposed over the last couple of decades, in a variety of fields. These methods can be loosely grouped into the following four main categories:
Gaussian approximate methods: These methods model the pertinent densities by Gaussian distributions, under assumption that a consistent minimum variance estimator (of the posterior state density) can be realized through the recursive propagation and updating of the first and second order moments of the true densities. Nonlinear filters that fall under this category are (in chronological order): a) the extended Kalman filter (EKF), which linearizes both the nonlinear process and measurement dynamics with a first-order Taylor expansion about current state estimate; b) the local linearization filter (LLF) is similar to EKF, but the approximate discrete time model is obtained from piecewise linear discretization of nonlinear state equation; c) the unscented Kalman filter (UKF) (Julier et al., 2002) chooses deterministic sample (sigma) points that capture the mean and covariance of a Gaussian density. When propagated through the nonlinear function, these points capture the true mean and covariance up to a second-order of the nonlinear function; d) the divided difference filter (DDF) (Norgaard et al., 2000) uses Stirling’s interpolation formula. As with the UKF, DDF uses a deterministic sampling approach to propagate Gaussian statistics through the nonlinear function; e) the Gaussian sum filters (GSF) approximates both the predicted and posterior densities as sum of Gaussian densities, where the mean and covariance of each Gaussian density is calculated using separate and parallel instances of EKF or UKF; f) the quadrature Kalman filter (QKF) (Ito and Xiong, 2002) uses the Gauss-Hermite numerical integration rule to calculate the recursive Bayesian estimation integrals, under a Gaussian assumption; g) the cubature Kalman filter (CKF) is similar to UKF, but uses the spherical-radial integration rule.
Direct numerical integration methods: these methods, also known as grid-based filters (GBF) or point-mass method, approximate the optimal Bayesian recursion integrals with large but finite sums over a uniform N-dimensional grid that covers the complete state-space in the area of interest. For even moderately high dimensional state-spaces, the computational complexity can become untenably large, which precludes any practical use of these filters (Bucy and Senne, 1971).
Sequential Monte-Carlo (SMC) methods: these methods (called particle filters) use a set of randomly chosen samples with associated weights to approximate the density (Doucet et al., 2001). Since the basic sampling dynamics (importance sampling) degenerates over time, the SMC method includes a re-sampling step. As the number of samples (particles) becomes larger, the Monte Carlo characterization of the posterior density becomes more accurate. However, the large number of samples often makes the use of SMC methods computationally prohibitive.
Variational Bayesian methods: Variational Bayesian methods approximate the true posterior distribution with a tractable approximate form. A lower bound on the marginal likelihood (evidence) of the posterior is then maximized with respect to the free parameters of this approximation (Jaakkola, 2000).
The selection of suitable sub-optimal approximate solutions to the recursive Bayesian estimation problem represents a trade-off between global optimality on one hand and computational tractability (and robustness) on the other hand. In our case, the best criterion for sub-optimality is formulated as: “Do as best as you can, and not more”. Under this criterion, the natural choice is to apply the cubature Kalman filter (Arasaratnam and Haykin, 2009). The CKF is the closest known direct approximation to the Bayesian filter, which outperforms all other nonlinear filters in any Gaussian setting, including particle filters (Arasaratnam and Haykin, 2009; Fernandez-Prades and Vila-Valls, 2010; Li et al., 2009). The CKF is numerically accurate, can capture true nonlinearity even in highly nonlinear systems, and it is easily extendable to high dimensional problems (the number of sample points grows linearly with the dimension of the state vector).
Cubature Kalman filter
The cubature Kalman filter is a recursive, nonlinear and derivative free filtering algorithm, developed under the Kalman filtering framework. It computes the first two moments (i.e. mean and covariance) of all conditional densities using a highly efficient numerical integration method (cubature rules). Specifically, it utilizes the third-degree spherical-radial cubature rule to approximate the integrals of the form (nonlinear function × Gaussian density) numerically using a set of m equally weighted symmetric cubature points :
| (6) |
| (7) |
where ξi is the i-th column of the cubature points matrix ξ with weights ωi and N is dimension of the state vector.
In order to evaluate the dynamic state-space model described by (3), the CKF includes two steps: a) a time update, after which the predicted density p (xt|y1:t−1) =  (x̂t|t−1, Pt|t−1) is computed; and b) a measurement update, after which the posterior density p (xt|y1:t) =
(x̂t|t−1, Pt|t−1) is computed; and b) a measurement update, after which the posterior density p (xt|y1:t) =  (x̂t|t, Pt|t) is computed. For a detailed derivation of the CKF algorithm, the reader is referred to (Arasaratnam and Haykin, 2009). We should note that even though CKF represents a derivative-free nonlinear filter, our formulation of the continuous-discrete dynamic system requires first order partial derivatives implicit in the Jacobian, which is necessary for implementation of LL scheme. Although, one could use simple Euler’s methods to approximate the numerical solution of the system (Sitz et al., 2002), local linearization generally provides more accurate solutions (Valdes Sosa et al., 2009). Note that since the Jacobian is only needed to discretise continuous state variables in the LL approach (but for each cubature point), the main CKF algorithm remains discrete and derivative-free.
(x̂t|t, Pt|t) is computed. For a detailed derivation of the CKF algorithm, the reader is referred to (Arasaratnam and Haykin, 2009). We should note that even though CKF represents a derivative-free nonlinear filter, our formulation of the continuous-discrete dynamic system requires first order partial derivatives implicit in the Jacobian, which is necessary for implementation of LL scheme. Although, one could use simple Euler’s methods to approximate the numerical solution of the system (Sitz et al., 2002), local linearization generally provides more accurate solutions (Valdes Sosa et al., 2009). Note that since the Jacobian is only needed to discretise continuous state variables in the LL approach (but for each cubature point), the main CKF algorithm remains discrete and derivative-free.
Parameters and input estimation
Parameter estimation sometimes referred to as system identification, can be regarded as a special case of general state estimation in which the parameters are absorbed into the state vector. Parameter estimation involves determining the nonlinear mapping:
| (8) |
where the nonlinear map 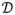 (.) is, in our case, the dynamic model f (.) parameterized by the vector θt. The parameters θt correspond to a stationary process with an identity state-transition matrix, driven by an “artificial” process noise wt~
(.) is, in our case, the dynamic model f (.) parameterized by the vector θt. The parameters θt correspond to a stationary process with an identity state-transition matrix, driven by an “artificial” process noise wt~ (0, Wt) (the choice of variance Wt determines convergence and tracking performance and is generally small). The input or cause of motion on hidden states ut can also be treated in this way, with input noise vt~
(0, Wt) (the choice of variance Wt determines convergence and tracking performance and is generally small). The input or cause of motion on hidden states ut can also be treated in this way, with input noise vt~ (0, Vt). This is possible because of the so-called natural condition of control (Arasaratnam and Haykin, 2009), which says that the input ut can be generated using the state prediction x̂t|t−1.
(0, Vt). This is possible because of the so-called natural condition of control (Arasaratnam and Haykin, 2009), which says that the input ut can be generated using the state prediction x̂t|t−1.
A special case of system identification arises when the input to the nonlinear mapping function 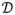 (.), i.e. our hidden states xt, cannot be observed. This then requires both state estimation and parameter estimation. For this dual estimation problem, we consider a discrete-time nonlinear dynamic system, where the system state xt, the parameters θt and the input ut, must be estimated simultaneously from the observed noisy signal yt. A general theoretical and algorithmic framework for dual Kalman filter based estimation was presented by Nelson (2000) and Van der Merwe (2004). This framework encompasses two main approaches, namely joint estimation and dual estimation. In the dual filtering approach, two Kalman filters are run simultaneously (in an iterative fashion) for state and parameter estimation. At every time step, the current estimate of the parameters θt is used in the state filter as a given (known) input and likewise, the current estimate of the state x̂t is used in the parameter filter. This results in a stepwise optimization within the joint state-parameter space. On the other hand, in the joint filtering approach, the unknown system state and parameters are concatenated into a single higher-dimensional joint state vector, x̃t.= [xt, ut, θt]T It was shown in (Van der Merwe, 2004) that parameter estimation based on nonlinear Kalman filtering represents an efficient online 2nd order optimization method that can be also interpreted as a recursive Newton-Gauss optimization method. They also showed that nonlinear filters like UKF and CKF are robust in obtaining globally optimal estimates, whereas EKF is very likely to get stuck in a non-optimal local minimum.
(.), i.e. our hidden states xt, cannot be observed. This then requires both state estimation and parameter estimation. For this dual estimation problem, we consider a discrete-time nonlinear dynamic system, where the system state xt, the parameters θt and the input ut, must be estimated simultaneously from the observed noisy signal yt. A general theoretical and algorithmic framework for dual Kalman filter based estimation was presented by Nelson (2000) and Van der Merwe (2004). This framework encompasses two main approaches, namely joint estimation and dual estimation. In the dual filtering approach, two Kalman filters are run simultaneously (in an iterative fashion) for state and parameter estimation. At every time step, the current estimate of the parameters θt is used in the state filter as a given (known) input and likewise, the current estimate of the state x̂t is used in the parameter filter. This results in a stepwise optimization within the joint state-parameter space. On the other hand, in the joint filtering approach, the unknown system state and parameters are concatenated into a single higher-dimensional joint state vector, x̃t.= [xt, ut, θt]T It was shown in (Van der Merwe, 2004) that parameter estimation based on nonlinear Kalman filtering represents an efficient online 2nd order optimization method that can be also interpreted as a recursive Newton-Gauss optimization method. They also showed that nonlinear filters like UKF and CKF are robust in obtaining globally optimal estimates, whereas EKF is very likely to get stuck in a non-optimal local minimum.
There is a prevalent opinion that the performance of joint estimation scheme is superior to dual estimation scheme (Ji and Brown, 2009; Nelson, 2000; Van der Merwe, 2004). Therefore, the joint CKF is used below to estimate states, input, and parameters. Note that since the parameters are estimated online with the states, the convergence of parameter estimates depends also on the length of the time series.
The state-space model for joint estimation scheme is then formulated as:
| (9) |
Since the joint filter concatenates the state and parameter variables into a single state vector, it effectively models the cross-covariances between the state, input and parameters estimates:
| (10) |
This full covariance structure allows the joint estimation framework not only to deal with uncertainty about parameter and state estimates (through the cubature-point approach), but also to model the interaction (conditional dependences) between the states and parameters, which generally provides better estimates.
Finally, the accuracy of the CKF can be further improved by augmenting the state vector with all the noise components (Li et al., 2009; Wu et al., 2005), so that the effects of process noise, measurement noise and parameter noise are explicitly available to the scheme. By augmenting the state vector with the noise variables (Eqs. 11 and 12), we account for uncertainty in the noise variables in the same manner as we do for the states during the propagation of cubature-points. This allows for the effect of the noise on the system dynamics and observations to be treated with the same level of accuracy as the state variables (Van der Merwe, 2004). It also means that we can model noise that is not purely additive. Because this augmentation increases the number of cubature points (by the number of noise components), it may also capture high-order moment information (like skew and kurtosis). However, if our problem does not require more than the first two moments; augmented CKF furnishes the same results as non-augmented CKF.
Square-root cubature Kalman filter
In practice, Kalman filters are known to be susceptible to numerical errors due to limited word-length arithmetic. Numerical errors can lead to propagation of an asymmetric, non-positive-definite covariance, causing the filter to diverge (Kaminski et al., 1971). As a robust solution to this, a square-root Kalman filter is recommended. This avoids the matrix square-rooting operations P = SST that are necessary in the regular CKF algorithm by propagating the square-root covariance matrix S directly. This has important benefits: preservation of symmetry and positive (semi) definiteness of the covariance matrix, improved numerical accuracy, double order precision, and reduced computational load. Therefore, we will consider the square-root version of CKF (SCKF), where the square-root factors of the predictive posterior covariance matrix are propagated (Arasaratnam and Haykin, 2009).
Bellow, we summarize the steps of SCKF algorithm. First, we describe the forward pass of a joint SCKF for the simultaneous estimation of states, parameters, and of the input, where we consider the state-space model in (9). Second, we describe the backward pass of the Rauch-Tung-Striebel (RTS) smoother. This can be derived easily for SCKF due to its similarity with the RTS smoother for square-root UKF (Simandl and Dunik, 2006). Finally, we will use the abbreviation SCKS to refer to the combination of SCKF and our RTS square-root cubature Kalman smoother. In other words, SCKF refers to the forward pass, which is supplemented with a backward pass in SCKS.
Forward filtering pass
Filter initialization
During initialization step of the filter we build the augmented form of state variable:
| (11) |
The effective dimension of this augmented state is N = nx + nu + nθ + nq + nv + nw + nr, where nx is the original state dimension, nu is dimension of the input, nθ is dimension of the parameter vector, {nq, nv, nw} are dimensions of the noise components (equal to nx, nu, nθ, respectively), and nr is the observation noise dimension (equal to the number of observed variables). In a similar manner, the augmented state square-root covariance matrix is assembled from the individual (square-root) covariance matrices of x, u, θ, q, v, w, and r:
| (12) |
| (13) |
where Px, Pu, Pθ are process error covariance matrices for states, input and parameters. Q, V, W are their corresponding process noise covariances, respectively and R is the observation noise covariance. The square-root representations of these matrices are calculated (13), where the “chol” operator represents a Cholesky factorization for efficient matrix square-rooting and “diag” forms block diagonal matrix.
Time update step
We evaluate the cubature points (i = 1,2, …, m = 2N):
| (14) |
where the set of sigma points ξ is pre-calculated at the beginning of algorithm (Eq. 7). Next, we propagate the cubature points through the nonlinear dynamic system of process equations and add noise components:
| (15) |
where F comprises [f(xt−1, θt−1, ut−1), ut−1, θt−1]T as expressed in process equation (9). The superscripts distinguish among the components of cubature points, which correspond to the states x, input u, parameters θ and their corresponding noise variables (q, v, w) that are all included in the augmented matrix 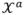 a. Note that the size of new matrix
is only (nx + nu + nθ) × m.
a. Note that the size of new matrix
is only (nx + nu + nθ) × m.
We then compute the predicted mean x̂t|t−1 and estimate the square-root factor of predicted error covariance St|t−1 by using weighted and centered (by subtracting the prior mean x̂t|t−1) matrix Xt|t−1:
| (16) |
| (17) |
| (18) |
The expression S = qr (X) denotes triangularization, in the sense of the QR decomposition1, where resulting S is a lower triangular matrix.
Measurement update step
During the measurement update step we propagate the cubature points through the measurement equation and estimate the predicted measurement:
| (19) |
| (20) |
Subsequently, the square-root of the innovation covariance matrix Syy,t|t−1 is estimated by using weighted and centered matrix Yt|t−1:
| (21) |
| (22) |
This is followed by estimation of the cross-covariance Pxy,t|t−1 matrix and Kalman gain Kt:
| (23) |
| (24) |
The use symbol/represents the matrix right divide operator; i.e. the operation A/B, applies the back substitution algorithm for an upper triangular matrix B and the forward substitution algorithm for lower triangular matrix A.
Finally, we estimate the updated state x̂t|t and the square-root factor of the corresponding error covariance:
| (25) |
| (26) |
The difference yt − ŷt|t−1 in Eq. (25) is called the innovation or the residual. It basically reflects the difference between the actual measurement and predicted measurement (prediction error). Further, this innovation is weighted by Kalman gain, which minimizes the posterior error covariance St|t.
In order to improve convergence rates and tracking performance, during parameter estimation, a Robbins-Monro stochastic approximation scheme for estimating the innovations (Ljung and Söderström, 1983; Robbins and Monro, 1951) is employed. In our case, this involves approximation of square-root matrix of parameter noise covariance SWt by:
| (27) |
where K̃t is the partition of Kalman gain matrix corresponding to the parameter variables, and λW ∈ (0,1] is scaling parameter usually chosen to be a small number (e.g. 10−3). Moreover, we constrain SWt to be diagonal, which implies an independence assumption on the parameters. Van der Merwe (2004) showed that the Robbins-Monro method provides the fastest rate of convergence and lowest final MMSE values. Additionally, we inject process noise artificially by annealing the square-root covariance of process noise with , using λq = 0.9995, λq ∈ (0,1] (Arasaratnam and Haykin, 2008).
Backward smoothing pass
The following procedure is a backward pass, which can be used for computing the smoothed estimates of time step t from estimates of time step t + 1. In other words, a separate backward pass is used for computing suitable corrections to the forward filtering results to obtain the smoothing solution . Because the smoothing and filtering estimates of the last time step T are the same, we make . This means the recursion can be used for computing the smoothing estimates of all time steps by starting from the last step t = T and proceeding backward to the initial step t = 0. To accomplish this, all estimates of x̂0:T and S0:T from the forward pass have to be stored and are then called at the beginning of each time step of backward pass (28, 29).
Square-root cubature RTS smoother
Each time step of the smoother is initialized by forming an augmented state vector and square-root covariance , using estimates from the SCKF forward pass, x̂t|T, St|T, and square-roots covariance matrices of the noise components:
| (28) |
| (29) |
We then evaluate and propagate cubature points through nonlinear dynamic system (SDEs are integrated in forward fashion):
| (30) |
| (31) |
We compute the predicted mean and corresponding square-root error covariance matrix:
| (32) |
| (33) |
| (34) |
Next, we compute the predicted cross-covariance matrix, where the weighted and centered matrix is obtained by using the partition (x, u, θ) of augmented cubature point matrix and the estimated mean before it propagates through nonlinear dynamic system (i.e. the estimate from forward pass):
| (35) |
| (36) |
Finally, we estimate the smoother gain At, the smoothed mean and the square-root covariance :
| (37) |
| (38) |
| (39) |
Note that resulting error covariance will be smaller than St|t from the forward run, as the uncertainty over the state prediction is smaller when conditioned on all observations, than when conditioned only on past observations.
This concludes our description of the estimation procedure, which can be summarized in the following steps:
- Evaluate the forward pass of the SCKF, where the continuous dynamic system of process equations is discretized by an LL-scheme for all cubature points. Note that both time update and measurement update steps are evaluated with an integration step Δt, and we linearly interpolate between available observation values. In this case, we weight all noise covariances by . In each time step of the filter evaluation we obtain predicted {x̂t|t−1, ût|t−1, θ̂t|t−1} and filtered {x̂t|t, ût|t, θ̂t|t} estimates of the states, parameters and the inputs. These predicted estimates are used to estimate prediction errors et = yt − ŷt, which allows us to calculate the log-likelihood of the model given the data as:
(40) Evaluate the backward pass of the SCKS to obtain smoothed estimates of the states , the input , and the parameters . Again, this operation involves discretization of the process equations by the LL-scheme for all cubature points.
Iterate until the stopping condition is met. We evaluate log-likelihood (40) at each iteration and terminate the optimization when the increase of the (negative) log-likelihood is less than a tolerance value of e.g. 10−3.
Before we turn to the simulations, we provide with a brief description of DEM, which is used for comparative evaluations.
Dynamic expectation maximization
DEM is based on variational Bayes, which is a generic approach to model inversion (Friston et al., 2008). Briefly, it approximates the conditional density p (ϑ|y, m) on some model parameters, ϑ = {x, u, θ, η}, given a model m, and data y, and it also provides lower-bound on the evidence p (y|m) of the model itself. In addition, DEM assumes a continuous dynamic system formulated in generalized coordinates of motion, where some parameters change with time, i.e. hidden states x and input u, and rest of the parameters are time-invariant. The state-space model has the form:
| (41) |
where
| (42) |
Here, g̃ and f̃ are the predicted response and motion of the hidden states, respectively. D is derivative operator whose first leading diagonal contains identity matrices, and which links successive temporal derivatives (x′, x″, …; u′, u″, …). These temporal derivatives are directly related to the embedding orders2 that one can specify separately for input (d) and for states (n) a priori. We will use embedding orders d = 3 and n = 6.
DEM is formulated for the inversion of hierarchical dynamic causal models with (empirical) Gaussian prior densities on the unknown parameters of generative model m. These parameters are {θ, η}, where θ represents set of model parameters and η = {α, β, σ} are hyperparameters, which specify the amplitude of random fluctuations in the generative process. These hyperparameters correspond to (log) precisions (inverse variances) on the state noise (α), the input noise (β), and the measurement noise (σ), respectively. In contrast to standard Bayesian filters, DEM also allows for temporal correlations among innovations, which is parameterized by additional hyperparameter γ called temporal precision.
DEM comprises three steps that optimize states, parameters and hyperparameters receptively: The first is the D-step, which evaluates Eq. (41), for the posterior mean, using the LL-scheme for integration of SDEs. Crucially, DEM (and its generalizations) does not use a recursive Bayesian scheme but tries to optimize the posterior moments of hidden states (and inputs) through an generalized (“instantaneous”) gradient ascent on (free-energy bound on) the marginal likelihood. This generalized ascent rests on using the generalized motion (time derivatives to high order) of variables as part of the model generating or predicting discrete data. This means that DEM is a formally simpler (although numerically more demanding) than recursive schemes and only requires a single pass though the time-series to estimate the states.
DEM comprises additional E (expectation) and M (maximization) steps that optimize the conditional density on parameters and hyperparameters (precisions) after the D (deconvolution) step. Iteration of these steps proceeds until convergence. For an exhaustive description of DEM, see (Friston et al., 2008). A key difference between DEM (variational and generalized filtering) and SCKS is that the states and parameters are optimized with respect to (a free-energy bound on) the log-evidence or marginal likelihood, having integrated out dependency on the parameters. In contrast, SCKS optimizes the parameters with respect to the log-likelihood in Equation (40), to provide maximum likelihood estimates of the parameters, as opposed to maximum a posteriori (MAP) estimators. This reflects the fact that DEM uses shrinkage priors on the parameters and hyperparameters, whereas SCKS does not. SCKS places priors on the parameter noise that encodes our prior belief that they do not change (substantially) over time. This is effectively a constraint on the volatility of the parameters (not their values per se), which allows the parameters to ‘drift’ slowly to their maximum likelihood value. This difference becomes important when evaluating one scheme in relation to the other, because we would expect some shrinkage in the DEM estimates to the prior mean, which we would not expect in the SCKS estimates (see next section).
DEM rests on a mean-field assumption used in variational Bayes; in other words, it assumes that the states, parameters and hyperparameters are conditionally independent. This assumption can be relaxed by absorbing the parameters and hyperparameters into the states as in SCKS. The resulting scheme is called generalized filtering (Friston et al., 2010). Although generalized filtering is formally more similar to SCKS than DEM (and is generally more accurate), we have chosen to use DEM in our comparative evaluations because of DEM has been validated against EKF and particle filtering (whereas generalized filtering has not). Furthermore, generalized filtering uses prior constraints on both the parameters and how fast they can change. In contrast, SCKS and DEM only use one set of constraints on the change and value of the parameters respectively. However, we hope to perform this comparative evaluation in a subsequent paper; where we will consider Bayesian formulations of cubature smoothing in greater detail and relate its constraints on changes in parameters to the priors used in generalized filtering.
Finally, for simplicity, we assume that the schemes have access to all the noise (precision) hyperparameters, meaning that they are not estimated. In fact, for SCKS we assume only the precision of measurement noise to be known and update the assumed values of the hyperparameters for fluctuations in hidden states and input during the inversion (see Eq. (27)). We can do this because we have an explicit representation of the errors on the hidden states and input.
Inversion of dynamic models by SCKF and SCKS
In this section, we establish the validity and accuracy of the SCKF and SCKS schemes in relation to DEM. For this purpose, we analyze several nonlinear and linear continuous stochastic systems that were previously used for validating of DEM, where its better performance was demonstrated in relation to the EKF and particle filtering. In particular, we consider the well known Lorenz attractor, a model of a double well potential, a linear convolution model and, finally, we devote special attention to the inversion of a hemodynamic model. Even though some of these models might seem irrelevant for hemodynamic and neuronal modeling, they are popular for testing the effectiveness of inversion schemes and also (maybe surprisingly) exhibit behaviors that can be seen in models used in neuroimaging.
To assess the performance of the various schemes, we perform Monte Carlo simulations, separately for each of these models; where the performance metric for the statistical efficiency of the estimators was the squared error loss function (SEL). For example, we define the SEL for states as:
| (43) |
Similarly, we evaluate SEL for the input and parameters (when appropriate). Since the SEL is sensitive to outliers; i.e. when summing over a set of (xt − x̂t)2, the final sum tends to be biased by a few large values. We consider this a convenient property when comparing the accuracy of our cubature schemes and DEM. Furthermore, this measure of accuracy accommodates the different constraints on the parameters in DEM (shrinkage priors on the parameters) and SCKS (shrinkage priors on changes in the parameters). We report the SEL values in natural logarithmic space; i.e. log(SEL).
Note that all data based on the above models were simulated through the generation function in the DEM toolbox (spm_DEM_generate.m) that is available as part of SPM8 (http://www.fil.ion.ucl.ac.uk/spm/).
Lorenz attractor
The model of the Lorenz attractor exhibits deterministic chaos, where the path of the hidden states diverges exponentially on a butterfly-shaped strange attractor in a three dimensional state-space. There are no inputs in this system; the dynamics are autonomous, being generated by nonlinear interactions among the states and their motion. The path begins by spiraling onto one wing and then jumps to the other and back in chaotic way. We consider the output to be the simple sum of all three states at any time point, with innovations of unit precision σ = 1 and γ = 8. We further specified a small amount of the state noise (α = e16). We generated 120 time samples using this model, with initial state conditions x0 = [0.9, 0.8, 30]T, parameters θ = [18, −4, 46.92]T and an LL-integration step Δt = 1.
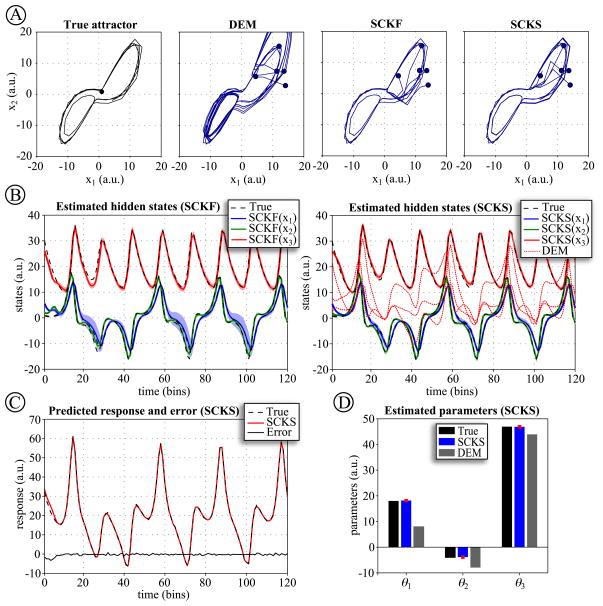
This sort of chaotic system shows sensitivity to initial conditions; which, in the case of unknown initial conditions, is a challenge for any inversion scheme. Therefore, we first compare SCKF, SCKS and DEM when the initial conditions x1 differ from the true starting values, with known model parameters. This simulation was repeated five times with random initializations and different innovations. Since we do not estimate any parameters, only a single iteration of the optimization process is required. We summarized the resulting estimates in terms of the first two hidden states and plotted their trajectories against each other in their corresponding state-space (Fig. 1A). It can be seen that all three inversion schemes converge quickly to the true trajectories. DEM provides the least accurate estimate (but still exhibits high performance when compared to EKF and particle filters (Friston, 2008a; Friston et al., 2008)). The SCKF was able to track the true trajectories more closely. This accuracy is even more improved by SCKS, where the initial residuals are significantly smaller, hence providing the fastest convergence.
Figure 1.
(A) The Lorenz attractor simulations were repeated five times, using different starting conditions (dots) and different random innovations. The hidden states of this model were estimated using DEM, SCKF and SCKS. Here, we summarize the resulting trajectories in terms of the first two hidden states, plotted against each other in their corresponding state-space. The true trajectories are shown on the upper left. (B) The inversion of Lorenz system by SCKF, SCKS and DEM. The true trajectories are shown as dashed lines, DEM estimates with dotted lines, and SCKF and SCKS estimates with solid lines including the 90% posterior confidence intervals (shaded areas). (C) Given the close similarity between the responses predicted by DEM and SCKS, we show only the result for SCKS. (D) The parameters estimates are summarized in lower left in terms of their expectation and 90% confidence intervals (red lines). Here we can see that DEM is unable to estimate the model parameters.
Next, we turned to testing the inversion schemes when both initial conditions and model parameters are unknown. We used initial state conditions x0 = [2, 8, 22]T and parameters θ0 = [10, −8, 43]T, where their true values were the same as above. We further assumed an initial prior precision on parameter noise p(θ) =  (0,0.1), and allowed the algorithm to iterate until the convergence. The SCKF and SCKS converged in 6 iteration steps, providing very accurate estimates of both states and parameters (Fig. 1B,D). This was not the case for DEM, which did not converge, exceeding the maximum allowed number of iteration, 50.
(0,0.1), and allowed the algorithm to iterate until the convergence. The SCKF and SCKS converged in 6 iteration steps, providing very accurate estimates of both states and parameters (Fig. 1B,D). This was not the case for DEM, which did not converge, exceeding the maximum allowed number of iteration, 50.
The reason for DEM’s failure is that the updates to the parameters are not properly regularized in relation to their highly nonlinear impact on the trajectories of hidden states. In other words, DEM makes poor updates, which are insensitive to the highly nonlinear form of this model. Critically, SCKF and SCKS outperformed DEM because it uses an online parameter update scheme and were able to accommodate nonlinearities much more gracefully, through its cubature-point sampling. Heuristically, cubature filtering (smoothing) can be thought of as accommodating nonlinearities by relaxing the strong assumptions about the form of the likelihood functions used in optimizing estimates. DEM assumes this form is Gaussian and therefore estimates its local curvature with second derivatives. A Gaussian form will be exact for linear models but not non-linear models. Conversely, cubature filtering samples this function over greater distances in state or parameter space and relies less on linear approximations.
MC simulations
To verify this result, we conducted a series of 100 Monte Carlo simulations under three different estimation scenarios. In the 1st scenario, we considered unknown initial conditions of hidden states but known model parameters. The initial conditions were sampled randomly from uniform distribution x0 ~ 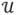 (0,20), and the true values were the same as in all previous cases. In the 2nd scenario, the initial states were known but the model parameters unknown, being sampled from the normal distribution around the true values θ0 ~
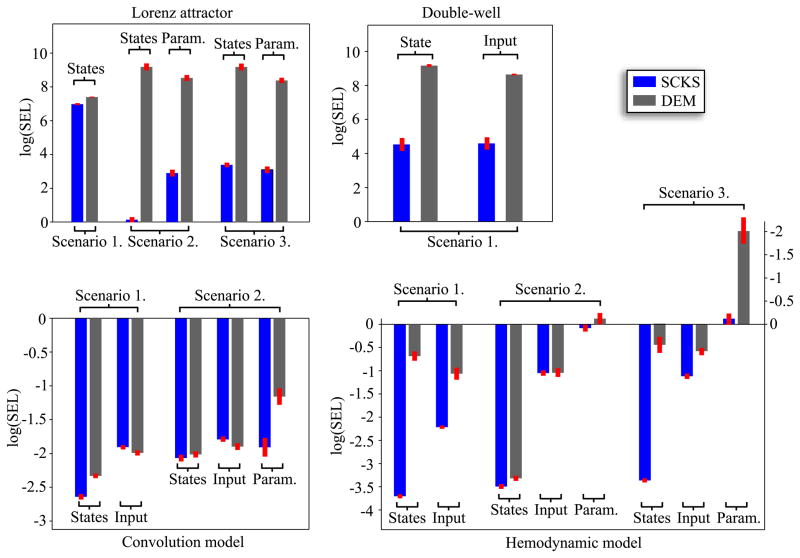
(0,20), and the true values were the same as in all previous cases. In the 2nd scenario, the initial states were known but the model parameters unknown, being sampled from the normal distribution around the true values θ0 ~  (θtrue, 10). Finally, the 3rd scenario was combination of the first two; with both initial conditions and parameters unknown. In this case, the states were always initialized with x0 = [2, 8, 22]T and parameters sampled from the normal distribution. Results, in terms of average log(SEL), comparing the performance of SCKS and DEM are shown in Fig. 4.
(θtrue, 10). Finally, the 3rd scenario was combination of the first two; with both initial conditions and parameters unknown. In this case, the states were always initialized with x0 = [2, 8, 22]T and parameters sampled from the normal distribution. Results, in terms of average log(SEL), comparing the performance of SCKS and DEM are shown in Fig. 4.
Figure 4.
The Monte Carlo evaluation of estimation accuracy using an average log(SEL) measure for all models under different scenarios. The SEL measure is sensitive to outliers, which enables convenient comparison between different algorithms tested on the same system. However, it cannot be used to compare performance among different systems. A smaller log(SEL) value reflects a more accurate estimate. For quantitative intuition, a value of log(SEL) = −2 is equivalent to mean square error (MSE) of about 2 · 10−3 and a log(SEL) = 7 is a MSE of about 7 · 101.
Double-well
The double-well model represents a dissipative system with bimodal variability. What makes this system particularly difficult to invert for many schemes is the quadratic form of the observation function, which renders inference on the hidden states and their causes ambiguous. The hidden state is deployed symmetrically about zero in a double-well potential, which makes the inversion problem even more difficult. Transitions from one well to other can be then caused either by input or high amplitude fluctuations. We drove this system with slow sinusoidal input
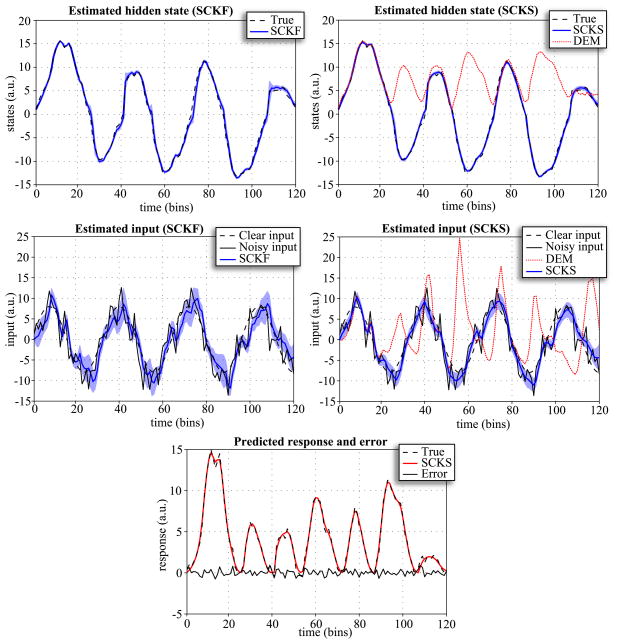
and generated 120 time points response with noise precision σ = e2, a small amount of state noise α = e16, and with a reasonable level of input noise β = 1/8. The temporal precision was γ = 2 and LL-integration step again Δt = 1, with initial condition x0 = 1, and mildly informative (initial) prior on the input precision p(u) =  (0.1). We tried to invert this model using only observed responses by applying SCKF, SCKS and DEM. Fig. 2 shows that DEM failed to estimate the true trajectory of the hidden state, in the sense that the state is always positive. This had an adverse effect on the estimated input and is largely because of the ambiguity induced by the observation function. Critically, the accuracy of the input estimate will be always lower than that of the state, because the input is expressed in measurement space vicariously through the hidden states. Nevertheless, SCKF and SCKS were able to identify this model correctly, furnishing accurate estimates for both the state and the input, even though this model represents a non-Gaussian (bimodal) problem (Fig. 2).
(0.1). We tried to invert this model using only observed responses by applying SCKF, SCKS and DEM. Fig. 2 shows that DEM failed to estimate the true trajectory of the hidden state, in the sense that the state is always positive. This had an adverse effect on the estimated input and is largely because of the ambiguity induced by the observation function. Critically, the accuracy of the input estimate will be always lower than that of the state, because the input is expressed in measurement space vicariously through the hidden states. Nevertheless, SCKF and SCKS were able to identify this model correctly, furnishing accurate estimates for both the state and the input, even though this model represents a non-Gaussian (bimodal) problem (Fig. 2).
Figure 2.
Inversion of the double-well model, comparing estimates of the hidden state and input from SCKF, SCKS and DEM. This figure uses the same format as Fig. 1B,C. Again, the true trajectories are depicted with dashed lines and the shaded area represents 90% posterior confidence intervals. Given the close similarity between the responses predicted by DEM and SCKS, we show only the result for SCKS.
MC simulations
To evaluate the stability of SCKS estimates in this context, we repeated the simulations 100 times, using different innovations. It can be seen from the results in Fig. 4 that the SCKS estimates of the state and input are about twice as close to the true trajectories than the DEM estimates. Nevertheless, the SCKS was only able to track the true trajectories of the state and input completely (as shown in Fig 2.) in about 70% of all simulations. This is still an excellent result for this difficult nonlinear and non-Gaussian model. In remaining 30% SCKS provided results where some half-periods of hidden state trajectories had the wrong sign; i.e. were flipped around zero. At the present time, we have no real insight into why DEM fails consistently to cross from positive to negative conditional estimates, while the SCKS scheme appears to be able to do this. One might presume this is a reflection of cubature filtering’s ability to handle the nonlinearities manifest at zero crossings. The reason this is a difficult problem is that the true posterior density over the hidden state is bimodal (with peaks at positive and negative values of the hidden state). However, the inversion schemes assume the posterior is a unimodal Gaussian density, which is clearly inappropriate. DEM was not able to recover the true trajectory of the input for any simulation, which suggests that the cubature-point sampling in SCKS was able to partly compensate for the divergence between the true (bimodal) and assumed unimodal posterior.
Convolution model
The linear convolution model represents another example that was used in (Friston, 2008a; Friston et al., 2008) to compare DEM, EKF, particle filtering and variational filtering. In this model (see Tab. 1), the input perturbs hidden states, which decay exponentially to produce an output that is a linear mixture of hidden states. Specifically, we used the input specified by Gaussian bump function of the form , two hidden states and four output responses. This is a single input-multiple output system with the following parameters:
Table 1.
State and observation equations for dynamic systems
| f(x,u, θ) | g(x, θ) | |||
|---|---|---|---|---|
| Lorenz attractor |
|
x1 + x2 + x3 | ||
| Double-well |
|
|
||
| Convolution model | θ2x + θ3u | θ1x | ||
| Hemodynamic model |
|
V0[k1(1 − x4) + k2(1 − x4/x3) + k3(1 − x3)] |
We generated data over 32 time points, using innovations sampled from Gaussian densities with precision σ = e8, a small amount of state noise α = e12 and minimal input noise β = e16. The LL-integration step was Δt = 1 and temporal precision γ = 4. During model inversion, the input and four model parameters are unknown and are subject to mildly informative prior precisions, p(u) =  (0,0.1), and p(θ) =
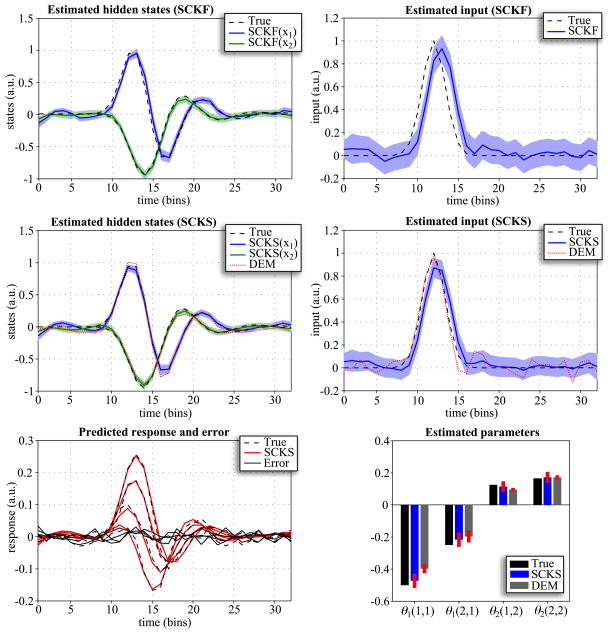
(0,0.1), and p(θ) =  (0, 10−4), respectively. Before initializing the inversion process, we set parameters θ1(1,1); θ1(2,1); θ2(1,2); and θ2(2,2); to zero. Fig. 3, shows that applying only a forward pass with SCKF does not recover the first hidden state and especially the input correctly. The situation is improved with the smoothed estimates from SCKS, when both hidden states match the true trajectories. Nevertheless, the input estimate is still slightly delayed in relation to the true input. We have observed this delay repeatedly, when inverting this particular convolution model with SCKS. The input estimate provided by DEM is, in this case, correct, although there are more perturbations around the baseline compared to the input estimated by SCKS. The reason that DEM was able to track the input more accurately is that is has access to generalized motion. Effectively this means it sees the future data in a way that recursive update schemes (like SCKF) do not. This becomes important when dealing with systems based on high-order differential equations, where changes in a hidden state or input are expressed in terms of high-order temporal derivatives in data space (we will return to this issue later). Having said this, the SCKS identified the unknown parameters more accurately than DEM, resulting in better estimates of hidden states.
(0, 10−4), respectively. Before initializing the inversion process, we set parameters θ1(1,1); θ1(2,1); θ2(1,2); and θ2(2,2); to zero. Fig. 3, shows that applying only a forward pass with SCKF does not recover the first hidden state and especially the input correctly. The situation is improved with the smoothed estimates from SCKS, when both hidden states match the true trajectories. Nevertheless, the input estimate is still slightly delayed in relation to the true input. We have observed this delay repeatedly, when inverting this particular convolution model with SCKS. The input estimate provided by DEM is, in this case, correct, although there are more perturbations around the baseline compared to the input estimated by SCKS. The reason that DEM was able to track the input more accurately is that is has access to generalized motion. Effectively this means it sees the future data in a way that recursive update schemes (like SCKF) do not. This becomes important when dealing with systems based on high-order differential equations, where changes in a hidden state or input are expressed in terms of high-order temporal derivatives in data space (we will return to this issue later). Having said this, the SCKS identified the unknown parameters more accurately than DEM, resulting in better estimates of hidden states.
Figure 3.
Results of inverting the linear convolution model using SCKF, SCKS and DEM; summarizing estimates of hidden states, input, four model parameters and the response. This figure uses the same format as Fig. 1B,C,D.
MC simulations
For Monte Carlo simulation we looked at two different scenarios. First, we inverted the model when treating only the input as unknown, and repeated the simulations 100 times with different innovations. In the second scenario, which was also repeated 100 times with different innovations, both input and the four model parameters were treated as unknown. The values of these parameters were sampled from the normal distribution θ0 =  (0,1). Fig. 4, shows that DEM provides slightly more accurate estimates of the input than SCKS. This is mainly because of the delay issue above. However, SCKS again furnishes more accurate estimates, with a higher precision on inverted states and markedly higher accuracy on the identified model parameters.
(0,1). Fig. 4, shows that DEM provides slightly more accurate estimates of the input than SCKS. This is mainly because of the delay issue above. However, SCKS again furnishes more accurate estimates, with a higher precision on inverted states and markedly higher accuracy on the identified model parameters.
Hemodynamic model
The hemodynamic model represents a nonlinear “convolution” model that was described extensively in (Buxton et al., 1998; Friston et al., 2000). The basic kinetics can be summarized as follows: Neural activity u causes an increase in vasodilatory signal h1 that is subject to auto-regulatory feedback. Blood flow h2 responds in proportion to this signal and causes changes in blood volume h3 and deoxyhemoglobin content, h4. These dynamics are modeled by a set of differential equations and the observed response is expressed as a nonlinear function of blood volume and deoxyhemoglobin content (see Tab. 3). In this model, the outflow is related to the blood volume through Grubb’s exponent α. The relative oxygen extraction is a function of flow, where ϕ is a resting oxygen extraction fraction. The description of model parameters, including the prior noise precisions is provided in Tab. 3.
Table 3.
Hemodynamic model parameters.
| Biophysical parameters of the state equations | |||
|---|---|---|---|
| Description | Value | Prior on noise variance | |
| κ | Rate of signal decay | 0.65 s−1 |
p (θκ) =  (0, 10−4) (0, 10−4) |
| χ | Rate of flow-dependent elimination | 0.38 s−1 |
p (θχ) =  (0, 10−4) (0, 10−4) |
| τ | Hemodynamic transit time | 0.98 s |
p (θτ) =  (0, 10−4) (0, 10−4) |
| α | Grubb’s exponent | 0.34 |
p (θα) =  (0, 10−8) (0, 10−8) |
| ϕ | Resting oxygen extraction fraction | 0.32 |
p (θϕ) =  (0, 10−8) (0, 10−8) |
| ε | Neuronal efficiency | 0.54 |
p (θε) =  (0, 10−8) (0, 10−8) |
| Fixed biophysical parameters of the observation equation | ||
|---|---|---|
| Description | Value | |
| V0 | Blood volume fraction | 0.04 |
| k1 | Intravascular coefficient | 7ϕ |
| k2 | Concentration coefficient | 2 |
| k3 | Extravascular coefficient | 2ϕ-0.2 |
In order to ensure positive values of the hemodynamic states and improve numerical stability of the parameter estimation, the hidden states are transformed xi = log(hi) ⇔ hi = exp (xi). However, before evaluating the observation equation, the log-hemodynamic states are exponentiated. The reader is referred to (Friston et al., 2008; Stephan et al., 2008) for a more detailed explanation.
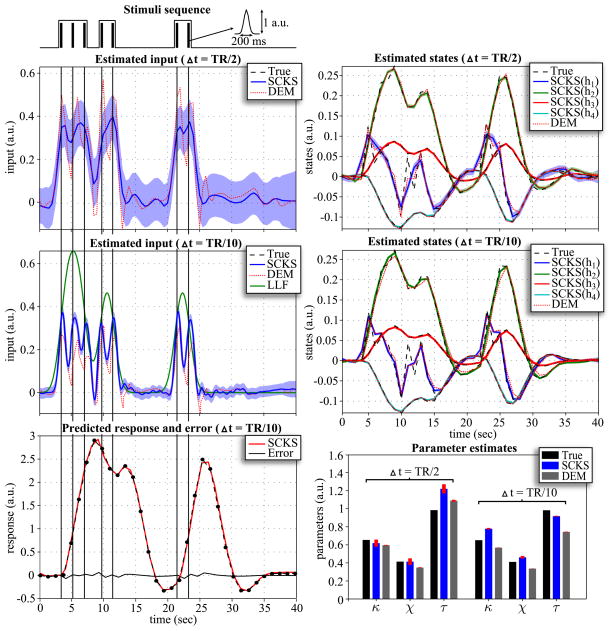
Although there are many practical ways to use the hemodynamic model with fMRI data, we will focus here on its simplest instance; a single-input, single-output variant. We will try to estimate the hidden states and input though model inversion, and simultaneously identify model parameters from the observed response. For this purpose, we generated data over 60 time points using the hemodynamic model, with an input in the form of a Gaussian bump functions with different amplitudes centered at positions (10; 15; 39; and 48), and model parameters as reported in Tab. 3. The sampling interval or repeat time (TR) was equal to TR = 1 sec. We added innovations to the output with a precision σ = e6. This corresponds to a noise variance of about 0.0025, i.e. in range of observation noise previously estimated in real fMRI data (Johnston et al., 2008; Riera et al., 2004), with a temporal precision γ = 1. The precision of state noise was α =e8 and precision of the input noise β = e8. At the beginning of the model inversion, the true initial states were x0 = [0,0,0,0]T. Three of the six model parameters, specifically θ = {κ,χ,τ}, were initialized randomly, sampling from the normal distribution centered on the mean of the true values . The remaining parameters were based on their true values. The reasons for omitting other parameters will be discussed later in the context of parameter identifiability. The prior precision of parameter noise are given in Tab. 3, where we allowed a small noise variance (10−8) in the parameters that we considered to be known {α,ϕ,ε}; i.e. these parameters can only experience very small changes during estimation. The parameter priors for DEM were as reported in (Friston et al., 2010) with the exception of {α,ϕ}, which we fixed to their true values.
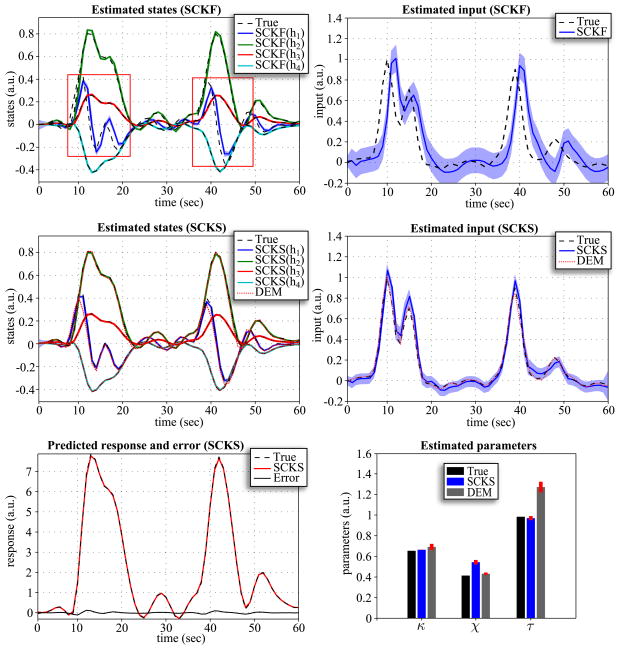
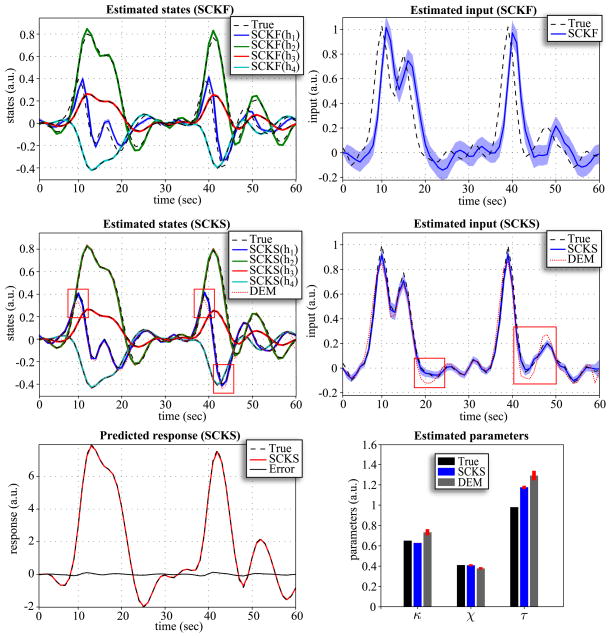
For model inversion we considered two scenarios that differed in the size of the integration step. First, we applied an LL-integration step of Δt = 0.5; in the second scenario, we decreased the step to Δt = 0.2. Note that all noise precisions are scaled by before estimation begins. The same integration steps were also used for DEM, where we additionally increased the embedding orders (n = d = 8) to avoid numerical instabilities. The results are depicted in Fig. 5 and 6. It is noticeable that in both scenarios neither the hidden states nor input can be estimated correctly by SCKF. For Δt = 0.5, SCKS estimates the input less accurately than DEM, with inaccuracies in amplitude and in the decaying part of the Gaussian input function, compared to the true trajectory. This occurred even though the hidden states were tracked correctly. The situation is very different for Δt = 0.2: Here the results obtained by SCKS are very precise for both the states and input. This means that a finer integration step had beneficial effects on both SCKF and SCKS estimators. In contrast, the DEM results did not improve. Here, including more integration steps between observation samples decreased the estimation accuracy for the input and the states. This means that DEM, which models high order motion, does not require the small integration steps necessary for SCKF and SCKS. Another interesting point can be made regarding parameter estimation. As we mentioned above, SCKS estimated the hidden states in both scenarios accurately, which might lead to the conclusion that the model parameters were also indentified correctly. However, although some parameters were indeed identified optimally (otherwise we would not obtain correct states) they were not equal to the true values. This is due to the fact that the effects of some parameters (on the output) are redundant, which means different sets of parameter values can provide veridical estimates of the states. For example, the effects of increasing the first parameter can be compensated by decreasing the second, to produce exactly the same output. This feature of the hemodynamic model has been discussed before in (Deneux and Faugeras, 2006) and is closely related to identifiably issues and conditional dependence among parameters estimates.
Figure 5.
Results of the hemodynamic model inversion by SCKF, SCKS and DEM, with an integration step of Δt = 0.5 and the first three model parameters were identified. This figure uses the same format as Fig 1B,C,D.
Figure 6.
Results of the hemodynamic model inversion by SCKF, SCKS and DEM, with an integration step of Δt = 0.2 and the first three model parameters were identified. This figure uses the same format as Fig 1B,C,D.
MC simulations
We examined three different scenarios for the hemodynamic model inversion. The simulations were inverted using an integration step Δt = 0.2 for SCKF and SCKS and Δt = 0.5 for DEM. First, we focus on performance when the input is unknown, we have access to the true (fixed) parameters and the initial states are unknown. These were sampled randomly from the uniform distribution x0 ~  (0,0.5). In the second scenario, the input is again unknown, and instead of unknown initial conditions we treated three model parameters θ = {κ,χ,τ} as unknown. Finally in the last scenario, all three variables (i.e. the initial conditions, input, and three parameters) are unknown. All three simulations were repeated 100 times with different initializations of x0, θ0, innovations, and state and input noise. From the MC simulation results, the following interesting behaviors were observed. Since the DEM estimates are calculated only in a forward manner, if the initial states are incorrect, it takes a finite amount of time before they converge to their true trajectories. This error persists over subsequent iterations of the scheme (E-steps) because they are initialized with the same incorrect state. This problem is finessed with SCKS: Although the error will be present in the SCKF estimates of the first iteration, it is efficiently corrected during the smoothing by SCKS, which brings the initial conditions closer to their true values. This enables an effective minimization of the initial error over iterations. This feature is very apparent from MC results in terms of log(SEL) for all three scenarios. When the true initial state conditions are known (2nd scenario), the accuracy of the input estimate is the same for SCKS and DEM, SCKS has only attained slightly better estimates of the states, hence also better parameter estimates. However, in the case of unknown initial conditions, SCKS is superior (see Fig. 4).
(0,0.5). In the second scenario, the input is again unknown, and instead of unknown initial conditions we treated three model parameters θ = {κ,χ,τ} as unknown. Finally in the last scenario, all three variables (i.e. the initial conditions, input, and three parameters) are unknown. All three simulations were repeated 100 times with different initializations of x0, θ0, innovations, and state and input noise. From the MC simulation results, the following interesting behaviors were observed. Since the DEM estimates are calculated only in a forward manner, if the initial states are incorrect, it takes a finite amount of time before they converge to their true trajectories. This error persists over subsequent iterations of the scheme (E-steps) because they are initialized with the same incorrect state. This problem is finessed with SCKS: Although the error will be present in the SCKF estimates of the first iteration, it is efficiently corrected during the smoothing by SCKS, which brings the initial conditions closer to their true values. This enables an effective minimization of the initial error over iterations. This feature is very apparent from MC results in terms of log(SEL) for all three scenarios. When the true initial state conditions are known (2nd scenario), the accuracy of the input estimate is the same for SCKS and DEM, SCKS has only attained slightly better estimates of the states, hence also better parameter estimates. However, in the case of unknown initial conditions, SCKS is superior (see Fig. 4).
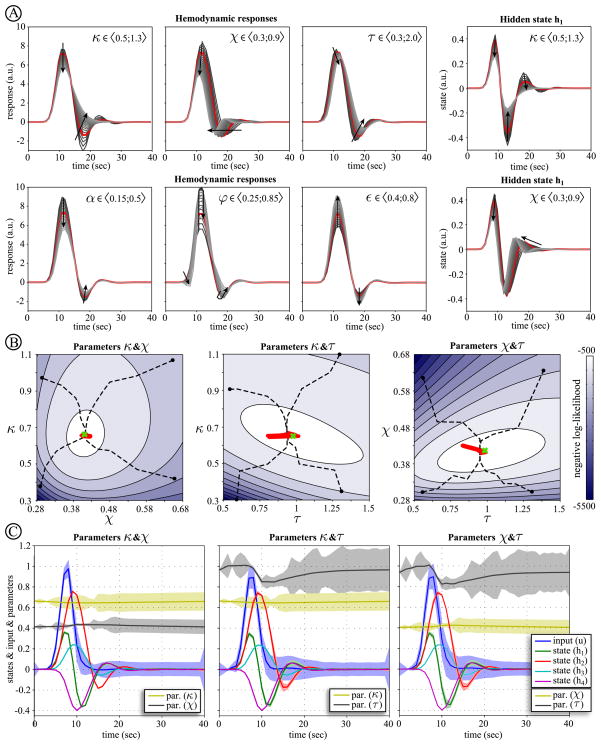
Effect of model parameters on hemodynamic response and their estimation
Although the biophysical properties of hemodynamic states and their parameters were described extensively in (Buxton et al., 1998; Friston et al., 2000), we will revisit the contribution of parameters to the final shape of hemodynamic response function (see Fig. 7A). In particular, our interest is in the parameters θ = {κ,χ,τ,α,ϕ,ε}, which play a role in the hemodynamic state equations. We evaluated changes in hemodynamic responses over a wide range of parameters values (21 regularly spaced values for each parameter). In Fig. 7A, the red lines represent biologically plausible mean parameter values that were estimated empirically in (Friston et al., 2000), and which are considered to be the true values here (Tab. 3). The arrows show change in response when these parameters are increased. The first parameter is κ = 1/τs, where τs is the time constant of signal decay. Increasing this parameter dampens the hemodynamic response to any input and suppresses its undershoot. The second parameter χ = 1/τf is defined by the time constant of the auto-regulatory mechanism τf. The effect of increasing parameter χ (decreasing the feedback time constant τf) is to increase the frequency of the response and lower its amplitude, with small change of the undershoot (see also the effect on the first hemodynamic state h1). The parameter τ is the mean transit time at rest, which determines the dynamics of the signal. Increasing this parameter slows down the hemodynamic response, with respect to flow changes. It also slightly reduces response amplitude and more markedly suppresses the undershoot. The next parameter is the stiffness or Grub’s exponent α, which is closely related to the flow-volume relationship. Increasing this parameter increases the degree of nonlinearity of the hemodynamic response, resulting in decreases of the amplitude and weaker suppression of undershoot. Another parameter of hemodynamic model is resting oxygen extraction fraction ϕ. Increasing this parameter can have quite profound effects on the shape of the hemodynamic response that bias it towards an early dip. This parameter has an interesting effect on the shape of the response: During the increase of ϕ, we first see an increase of the response peak amplitude together with deepening of undershoot, whereas after the value passes ϕ = 0.51, the undershoot is suppressed. Response amplitude continues to grow until ϕ = 0.64 and falls rapidly after that. Additionally, the early dip starts to appear with ϕ = 0.68 and higher values. The last parameter is the neuronal efficacy ε, which simply modulates the hemodynamic response. Increasing this parameter scales the amplitude of the response.
Figure 7.
(A)The tops row depicts the effect of changing the hemodynamic model parameters on the response and on the first hidden state. For each parameter, the range of values considered is reported, comprising 21 values. (B) The middle row shows the optimization surfaces (manifolds) of negative log-likelihood obtained via SCKS for combinations of the first three hemodynamic model parameters {κ,χ,τ}. The trajectories of convergence (dashed lines) for four different parameter initializations (dots) are superimposed. The true values (at the global optima) are depicted by the green crosshair and the dynamics of the parameters over the final iteration correspond to the thick red line. (C) The bottom row shows the estimates of hidden states and input for the corresponding pairs of parameters obtained during the last iteration, where we also show the trajectory of the parameters estimates over time.
In terms of system identification, it has been shown (Deneux and Faugeras, 2006) that very little accuracy is lost when the values of Grub’s exponent and resting oxygen extraction fraction are fixed to physiologically plausible values. This is in accordance with (Riera et al., 2004), where these parameters were also fixed. Grub’s exponent is supposed to be stable during steady-state stimulation (Mandeville et al., 1999b); α = 0.38 ± 0.1 with almost negligible effects on the response within this range. The resting oxygen extraction fraction parameter is responsible for the early dip that is rarely observed in fMRI data. Its other effects can be approximated by combining the parameters {κ, τ}. In our case, where the input is unknown, the neuronal efficiency parameter ε is fixed as well. This is necessary, because a change in this parameter is degenerate with respect to the amplitude of neuronal input.
To pursue this issue of identifiably we examined the three remaining parameters θ = {κ, χ, τ} in terms of the (negative) log-likelihood for pairs of these three parameters; as estimated by the SCKS scheme (Fig. 7B). The curvature (Hessian) of this log-likelihood function is, in fact, the conditional precision (inverse covariance) used in variational schemes like DEM and is formally related to the Fisher Information matrix for the parameters in question. A slow curvature (shallow) basin means that we are conditionally uncertain about the precise value and that large changes in parameters will have relatively small effects on the observed response or output variables. The global optimum (true values) is marked by the green crosslet. To compute these log-likelihoods we ran SCKS for all combinations of parameters within their selected ranges, assuming the same noise precisions as in the hemodynamic simulations above (Tab. 3). Note that we did not perform any parameter estimation, but only evaluated log-likelihood for different parameter values, having optimized the states. Looking at the ensuing (color-coded) optimization manifolds, particularly at the white area bounded by the most inner contour, we can see how much these parameters can vary around the global optimum and still provide reasonably accurate predictions (of output, hidden states and input). This range is especially wide for the mean transit time τ. One can see from the plot at the top of Fig. 7A that changing τ = 〈0.3; 2.0〉 over a wide range has little effect on the response. The region around the global maximum also discloses conditional dependencies and redundancy among the parameters. These dependencies make parameter estimation a generally more difficult task.
Nevertheless, we were curious if, at least under certain circumstances, the true parameter values could be estimated. Therefore, we allowed for faster dynamics on the parameters {κ, χ, τ} by using higher noise variances (4 · 10−4, 2 · 10−4, 10−2, respectively) and evaluated all three possible parameter combinations using SCKS. In other words, we optimized two parameters with the third fixed, over all combinations. These noise parameters were chosen after intensive testing, to establish the values that gave the best estimates. We repeated these inversions four times, with different initial parameter estimates selected within the manifolds shown in Fig. 7A. In Fig. 7B, we can see how the parameters moved on the optimization surface, where the black dashed line depicts the trajectory of the parameter estimates over successive iterations, starting from the initial conditions (black dot) and terminating around the global optimum (maximum). The red thick line represents the dynamic behavior of parameters over time during the last iteration. The last iteration estimate for all states, input and parameters is depicted in Fig. 7C. Here the dynamics of transit time (τ) is especially interesting; it drops with the arrival of the neuronal activation and is consequently restored during the resting period. This behavior is remarkably similar to that observed by Mandeville et al. in rat brains, where mean transit time falls during activation. Clearly, we are not suggesting that the transit time actually decreased during activation in our simulations (it was constant during the generation of data). However, these results speak to the interesting application of SCKS to identify time-dependent changes in parameters. This could be important when applied to dynamic causal models of adaptation or learning studies that entail changes in effective connectivity between neuronal populations. The key message here is that if one can (experimentally) separate the time scale of true changes in parameters from the (fast) fluctuations inherent in recursive Bayesian filtering (or generalized filtering), it might be possible to estimate (slow) changes in parameters that are of great experimental interest.
In general, enforcing slow dynamics on the parameters (with a small noise variance) will ensure more accurate results for both states and input, provided the true parameters also change slowly. Moreover, we prefer to consider all the parameters of the hemodynamic state equations as unknown and limit their variations with high prior precisions. This allows us to treat all the unknown parameters uniformly; were certain (assumed) parameters can be fixed to their prior mean using an infinitely high prior precision.
Beyond the limits of fMRI signal
One of the challenges in fMRI research is to increase a speed of brain volume sampling; i.e. to obtain data with a higher temporal resolution. Higher temporal resolution allows one to characterize changes in the brain more accurately, which is important in many aspects of fMRI. In this section, we will show that estimating unobserved (hidden) hemodynamic states and, more importantly, the underlying neuronal drives solely from observed data by blind deconvolution can significantly improve the temporal resolution and provide estimates of the underlying neuronal dynamics at a finer temporal scale. This may have useful applications in the formation of things like psychophysiological interactions (Gitelman et al., 2003).
In the hemodynamic model inversions above we did not use very realistic neuronal input, which was a Gaussian bump function and the data were generated with a temporal resolution of 1 s. This was sufficient for our comparative evaluations; however in real data, the changes in underlying neuronal activation are much faster (possibly in the order of milliseconds) and may comprise a rapid succession of events. The hemodynamic changes induced by this neuronal activation manifest as a rather slow response, which peaks at about 4–6 s.
To make our simulations more realistic, we considered the following generation process, which is very similar to the simulation and real data used previously in Riera et al. (2004). First, we generated our data with a time step of 50 ms using the sequence of neuronal events depicted at the top of Fig. 8. These Gaussian-shaped neuronal events (inputs) had a FWTM (full-width at tenth of maximum) of less than 200 ms. Otherwise, the precisions on innovations, states noise, and input noise were identical to the hemodynamic simulations above. Next we down-sampled the synthetic response with a realistic TR = 1.2 s, obtaining data of 34 time points from the original 800. For estimation, we used the same priors on the input p(u) =  (0,0.1) and parameters as summarized in Tab. 3.
(0,0.1) and parameters as summarized in Tab. 3.
Figure 8.
Inversion of the hemodynamic model for more realistic neuronal inputs (top left) and fMRI observations sampled with a TR = 1.2 s (bottom left – dotted line). The input and hidden states estimates obtained by SCKS and DEM are shown for an integration step Δt = TR/2 (top row) and Δt = TR/10 (middle row). The parameter estimates are shown on the bottom right. The best estimate of the input that could be provided by the local linearization filter is depicted on the middle left panel by the solid green line.
Our main motivation was the question: How much of the true underlying neuronal signal can we recover from this simulated sparse observation, when applying either SCKS or DEM? To answer this, two different scenarios were considered. The first used an integration step Δt = TR/2 = 0.6 s which had provided quite favorable results above. The top row of Fig. 8 shows the estimated input and states provided by SCKS and DEM. It can be seen that the states are traced very nicely by both approaches. For the input estimates, SCKS captures the true detailed neuronal structure deficiently, although the main envelope is correct. For DEM, the input estimate is much closer to the true structure of the neuronal signal, distinguishing all seven events. However, one can not overlook sharp undershoots that appear after the inputs. The reason for these artifacts rests on the use of generalized coordinates of motion, where the optimization of high order temporal derivatives does not always produce the optimal low order derivatives (as shown in the Fig. 8).
In the second scenario, where we decreased the integration step to Δt = TR/10 = 0.12 s, we see that the SCKS estimate of the input has improved markedly. For DEM the input estimate is actually slightly worse than in the previous case. Recalling the results from previous simulations (Fig. 5 and 6) it appears that the optimal integration step for DEM is Δt = TR/2, and decreasing this parameter does not improve estimation (as it does for SCKS). Conversely, an excessive decrease of Δt can downgrade accuracy (without an appropriate adjustment of the temporal precision).
Here we can also compare our results with the results obtained in (Riera et al., 2004), where the LL-innovation technique was used with a constrained nonlinear optimization algorithm (Matlab’s fmincon.m function) to estimate the neuronal activation. In our simulations the neuronal input was parameterized by a set of RBFs, regularly spaced with an inter-distance interval equal to TR, where the amplitudes of RBFs together with the first three hemodynamic model parameters, including noise variances, were subject to estimation. The resulting estimate is depicted by the solid green line at the bottom of Fig. 8. It is obvious that this only captures the outer envelope of the neuronal activation. Although this approach represented the most advanced technique at the time of its introduction (2004), its use is limited to relatively short time-series that ensures the number of parameters to be estimated is tractable.
We conclude that inversion schemes like DEM and especially SCKS can efficiently reconstruct the dynamics of neuronal signals from fMRI signal, affording a considerable improvement in effective temporal resolution.
Discussion
We have proposed a nonlinear Kalman filtering based on an efficient square-root cubature Kalman filter (SCKF) and RTS smoother (SCKS) for the inversion of nonlinear stochastic dynamic causal models. We have illustrated its application by estimating neuronal activity by (so called) blind deconvolution from fMRI data. Using simulations of different stochastic dynamic systems, including validation via Monte Carlo simulations, we have demonstrated its estimation and identification capabilities. Additionally, we have compared its performance with an established (DEM) scheme, previously validated in relation to EKF and particle filtering (Friston et al., 2008).
In particular, using a nonlinear model based on the Lorenz attractor, we have shown that SCKF and SCKS outperform DEM when the initial conditions and model parameters are unknown. The double-well model turned out (as anticipated) to be difficult to invert. In this case, both SCKF and SCKS could invert both states and input correctly; i.e. to track their true trajectories in about 70% of the simulations (unlike DEM). Both the Lorenz attractor and double-well system are frequently used for testing the robustness of new nonlinear filtering methods and provide a suitable forum to conclude that SCKF and SCKS show a higher performance in nonlinear and non-Gaussian setting than DEM. The third system we considered was a linear convolution model, were the performance of both inversion schemes was comparable. In contrast to the previous models, the SCKF alone was not sufficient for successful estimation of the states and input. Although DEM provided a better estimate of the input, the SCKS was more precise in tracking hidden states and inferring unknown model parameters.
We then turned to the hemodynamic model proposed by Buxton et al. (1998) and completed by Friston et al. (2000), which comprises nonlinear state and observation equations. The complexity of this model, inherent in a series of nonlinear differential equations (i.e. higher order ODEs) makes the inversion problem fairly difficult. If the input is unknown, it cannot be easily solved by a forward pass of the SCKF or any other standard nonlinear recursive filter. It was precisely this difficulty that motivated Friston et al. (2008) to develop DEM by formulating the deconvolution problem in generalized coordinates of motion. The same problem motivated us to derive a square-root formulation of the Rauch-Tung-Striebel smoother and solve the same problem with a recursive scheme.
Both DEM and SCKS (SCKF) use an efficient LL-scheme for the numerical integration of non-autonomous multidimensional stochastic differential equations (Jimenez, 2002). Using simulations, we have demonstrated that for a successful inversion of the hemodynamic model, SCKS requires an integration step of at least Δt = TR/2 for the accurate estimation of hidden states, and preferably a smaller integration step for an accurate inference on the neuronal input. Unlike SCKS, DEM provides the best estimates of the input when the integration step is Δt = TR/2. This is because it uses future and past observations to optimize a path or trajectory of hidden states, in contrast to recursive schemes that update in a discrete fashion. Nevertheless, with smaller integration steps, SCKS affords more precise estimates of the underlying neuronal signal than DEM under any integration step. Additionally, in the case of more realistic hemodynamic simulations we have shown that with the smaller integration step of about Δt = TR/10 we were able to recover the true dynamics of neuronal activity that cannot be observed (or estimated) at the temporal resolution of the measured signal. This takes us beyond the limits of the temporal resolution of hemodynamics underlying the fMRI signal.
An interesting aspect of inversion schemes is their computational cost. Efficient implementations of SCKS with the integration step of Δt = TR/10 (including parameter estimation) are about 1.3 times faster than DEM (with an integration step of Δt = TR/2 and a temporal embedding n = 6 and d = 3). If the integration step is the same, then SCKS is about 5 times faster, which might have been anticipated, given that DEM is effectively dealing with six times the number of (generalized) hidden states.
We have also examined the properties of parameter identification of hemodynamic model under the SCKS framework. Based on the previous experience (Deneux and Faugeras, 2006; Riera et al., 2004), we constrained the hemodynamic model by allowing three parameters to vary; i.e. rate of signal decay, rate of flow-dependent elimination, and mean transit time. The remaining parameters were kept (nearly) constant, because they had only minor effects on the hemodynamic response function.
Our procedure for parameter identification uses a joint estimation scheme, where both hidden states and parameters are concatenated into a single state vector and inferred simultaneously in dynamic fashion. The SCKS is iterated until the parameters converge. Moreover, the convergence is enhanced by a stochastic Robbins-Monro approximation of the parameter noise covariance matrix. This enabled very efficient parameter identification in all of the stochastic models we considered, including the hemodynamic model. However, specifically in the case of the hemodynamic model, we witnessed a particular phenomenon, which was also reported by Deneux et al. (2006). Put simply, the effects of some parameters on the hemodynamic response are degenerate, in that different combinations can still provide accurate predictions of observed responses. In this context, we have shown in Fig. 7A that different sets of parameters can produce a very similar hemodynamic response function. This degeneracy or redundancy is a ubiquitous aspect of model inversion and is usually manifest as conditional dependency among the parameter estimates. The problem of conditional dependencies is usually finessed by optimizing the model in terms of its evidence. Model evidence ensures that the conditional dependences are suppressed by minimizing complexity (which removes redundant parameters). In our setting, we are estimating both states and parameters and have to contend with possible conditional dependences between the states and parameters. In principle, this can be resolved by comparing the evidence for different models and optimizing the parameterization to provide the most parsimonious model. We will pursue this in a subsequent paper, in which we examine the behavior of model evidence, as estimated under cubature smoothing. It should be noted, that this work uses models that have already been optimized over the past few years, so that they provide the right balance of accuracy and complexity, when trying to explain typical fMRI data. However, we may have to revisit this issue when trying to estimate the hidden neuronal states as well as parameters.
There are further advantages of SCKS compared to DEM. Since DEM performs inference on states and input in a forward manner only, it is sensitive to misspecification of initial conditions. Critically, recent implementations of DEM (Friston et al., 2008) start each iteration with the same initial values of the states and the input, resulting in significant error at the initial phase of deconvolution. This is not the case for SCKS, which, by applying smoothing backward step, minimizes the initial error and converges to the true initial value over iterations. Next, DEM can produce sharp undershoots in the input estimate when the hidden states or their causes change too quickly. The SCKS does not have this problem. However, the use of generalized motion enables DEM to be applied online. Additionally, this framework also allows DEM to model temporal dependencies in the innovations or fluctuations of hidden states, which might be more plausible for biological systems. In Kalman filtering, these fluctuations are generally assumed to be Markovian. Having said this, it is possible to cast dynamical models in generalized coordinates of motion as classical Markovian models, where the innovations are successively colored before entering the state equation (see Eq. 3 in Friston et al. (2008b)).
Based on our MC simulations, we conclude that in general SCKS provided a more accurate inversion of nonlinear dynamic models, including estimation of the states, input and parameters, than DEM. Since DEM has been shown to outperform EKF and particle filtering, it makes the SCKS the most efficient blind nonlinear deconvolution schemes for dynamic state-space models.
Finally, all evaluations of the proposed approach, including the comparison with DEM, were performed under the assumption that SCKS algorithm had access to the true precision parameter on the measurement noise and DEM had access to precisions on all noise components. However, for application to the real data we have to be able to estimate these precision parameters as well. DEM is formulated as a hierarchical dynamic model, which allows for an elegant triple inference on hidden states, input, parameters and hyperparameters. In the case of SCKS we have introduced dynamic approximation techniques for the efficient estimation of the parameter state noise covariance matrices. We also observed that the input noise variance can be considered time-invariant, with a reasonable value (for the hemodynamic model) of about V = 0.1. This value seemed to be consistent over different levels of noise and different input. The last outstanding unknown quantity is the measurement noise covariance. We have found a robust solution (Särkkä and Hartikainen, Under revision; Särkkä and Nummenmaa, 2009) that combines the variational Bayesian method with the nonlinear Kalman filtering algorithm for the joint estimation of states and time-varying measurement noise covariance in a nonlinear state-space model. We have implemented this approach for our SCKS scheme with a minimal increase in computational cost. Although this variational Bayesian extension was not utilized in our proposal (for simplicity), it is now part of SCKS algorithm for future application to the real data.
There are several application domains we hope to explore within our framework: Since SCKF-SCKS can recover the underlying time course of synaptic activation, we can model effective connectivity at synaptic (neuronal) level. Because no knowledge about the input is necessary, one can use this scheme to invert the dynamic causal models on the resting state data, or pursue connectivity analyses in the brain regions that are dominated by endogenous activity fluctuations, irrespective of task-related responses. We will also consider conventional approaches to causal inference that try to identify the direction of the information flow between different brain regions (e.g. Granger causality, dynamic Bayesian networks, etc.). In this context, one can compare the analysis of deconvolved hidden (neuronal) states with explicit model comparison within the DCM framework. Another challenge would be to exploit the similarity among neighboring voxels in relation to their time courses. There are thousands of voxels in any volume of the human brain, and the judicious pooling of information from multiple voxels may help to improve accuracy of our deconvolution schemes. Last but not least, we hope to test variants of the hemodynamic model, starting with extension proposed by Buxton et al. (2004), which accounts for non-steady-state relationships between CBF and CBV arising due to viscoelastic effects. This is particularly interesting here, because we can, in principle, characterize these inconstant relationships in terms of time-varying parameter estimates afforded by our recursive schemes.
The Matlab code for our methods (including estimation of measurement noise covariance), which is compatible with the subroutines and variable structures used by the DEM in SPM8, is available from the authors upon request.
Conclusion
In this paper, we have introduced a robust blind deconvolution technique based on the nonlinear square-root cubature Kalman filter and Rauch-Tung-Striebel smoother, which allows an inference on hidden states, input, and model parameters. This approach is very general and can be applied to the inversion of any nonlinear continuous dynamic model that is formulated with stochastic differential equations. This first description of the technique focused on the estimation of neuronal synaptic activation by generalized deconvolution from observed fMRI data. We were able to estimate the true underlying neuronal activity with a significantly improved temporal resolution, compared to the observed fMRI signal. This speaks to new possibilities for fMRI signal analysis; especially in effective connectivity and dynamic causal modeling of unknown neuronal fluctuations (e.g. resting state data).
We validated the inversion scheme using difficult nonlinear and linear stochastic dynamic models and compared its performance with dynamic expectation maximization; one of the few methods that is capable of this sort of model inversion. Our approach afforded the same or better estimates of states, input, and model parameters, with reduced computational cost.
Table 2.
Parameters of the generative model for the simulated dynamic systems
| Lorenz | Double-well | Convolution | Hemodynamic | |||
|---|---|---|---|---|---|---|
| Observation-noise precision | Simulated | σ = 1 | σ = e2 | σ = e8 | σ = e6 | |
| Prior pdf | ~  (0.1) (0.1) |
~  (0, e−2) (0, e−2) |
~  (0, e−8) (0, e−8) |
~  (0, e−6) (0, e−6) |
||
| State-noise precision | Simulated | α = e16 | α = e16 | α = e12 | α = e8 | |
| Input-noise precision | Simulated | - |
|
β = e16 | β = e8 | |
| Prior pdf | - | ~  (0,1) (0,1) |
~  (0,0.1) (0,0.1) |
~  (0,0.1) (0,0.1) |
||
| Parameter-noise precision | Prior pdf 3 | ~  (0,0.1) (0,0.1) |
- | ~  (0, 10−4) (0, 10−4) |
Tab. 3 | |
| Initial conditions | Simulated | x0 = [0.9,0.8,30]T | x0 = 1 | x0 = [0,0]T | x0 = [0,0,0,0]T |
Acknowledgments
This work was supported by the research frame no. MSM0021630513 and no. MSM0021622404 and also sponsored by the research center DAR no. 1M0572, all funded by the Ministry of Education of the Czech Republic. Additional funding was provided by NIH grant no. R01EB000840 from the USA. KJF was funded by the Wellcome Trust. We would like to thank to Jorge Riera for providing his implementation of LL-innovation algorithm.
Footnotes
The QR decomposition is a factorization of a matrix XT into an orthogonal matrix Q and upper triangular matrix R such that XT = QR and XXT = RTQTQR = RTR = SST, where the resulting square-root (lower triangular) matrix is S = RT.
The term “embedding order” is used in analogy with lags in autoregressive modeling.
Prior precision on parameter noise is used for initialization and during CKF step the parameter noise variance is estimated by Robbins-Monro stochastic approximation (27) with scaling parameter λw = 10−2 for the Lorenz attractor and λw = 10−3 for the convolution and hemodynamic models.
References
- Aguirre GK, Zarahn E, D’esposito M. The variability of human, BOLD hemodynamic responses. Neuroimage. 1998;8:360–369. doi: 10.1006/nimg.1998.0369. [DOI] [PubMed] [Google Scholar]
- Arasaratnam I, Haykin S. MICAI 2008: Advances in Artificial Intelligence. 2008. Nonlinear Bayesian Filters for Training Recurrent Neural Networks; pp. 12–33. [Google Scholar]
- Arasaratnam I, Haykin S. Cubature Kalman Filters. IEEE Transactions on Automatic Control. 2009;54:1254–1269. [Google Scholar]
- Attwell D, Buchan AM, Charpak S, Lauritzen M, MacVicar BA, Newman EA. Glial and neuronal control of brain blood flow. Nature. 2010;468:232–243. doi: 10.1038/nature09613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns GS, Song AW, Mao H. Continuous functional magnetic resonance imaging reveals dynamic nonlinearities of “dose-response” curves for finger opposition. Journal of Neuroscience. 1999;19:1–6. doi: 10.1523/JNEUROSCI.19-14-j0003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn RM, Saad ZS, Bandettini PA. Spatial heterogeneity of the nonlinear dynamics in the FMRI BOLD response. Neuroimage. 2001;14:817–826. doi: 10.1006/nimg.2001.0873. [DOI] [PubMed] [Google Scholar]
- Biscay R, Jimenez JC, Riera JJ, Valdes PA. Local linearization method for the numerical solution of stochastic differential equations. Annals of the Institute of Statistical Mathematics. 1996;48:631–644. [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magnetic Resonance in Medicine. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Bucy RS, Senne KD. Digital synthesis of non-linear filters. Automatica. 1971;7:287–298. [Google Scholar]
- Buxton RB, Uludag K, Dubowitz DJ, Liu TT. Modeling the hemodynamic response to brain activation. Neuroimage. 2004;23:S220–S233. doi: 10.1016/j.neuroimage.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Buxton RB, Wong EC, Frank LR. Dynamics of blood flow and oxygenation changes during brain activation: the balloon model. Magnetic Resonance in Medicine. 1998;39:855–864. doi: 10.1002/mrm.1910390602. [DOI] [PubMed] [Google Scholar]
- David O. fMRI connectivity, meaning and empiricism Comments on: Roebroeck et al. The identification of interacting networks in the brain using fMRI: Model selection, causality and deconvolution. NeuroImage. 2009 doi: 10.1016/j.neuroimage.2009.09.073. In Press. [DOI] [PubMed] [Google Scholar]
- David O, Guillemain I, Saillet S, Reyt S, Deransart C, Segebarth C, Depaulis A. Identifying neural drivers with functional MRI: an electrophysiological validation. PLoS Biology. 2008;6:2683–2697. doi: 10.1371/journal.pbio.0060315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneux T, Faugeras O. Using nonlinear models in fMRI data analysis: model selection and activation detection. NeuroImage. 2006;32:1669–1689. doi: 10.1016/j.neuroimage.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Doucet A, De Freitas N, Gordon N. Sequential Monte Carlo methods in practice. Springer Verlag; 2001. [Google Scholar]
- Fernandez-Prades C, Vila-Valls J. Bayesian Nonlinear Filtering Using Quadrature and Cubature Rules Applied to Sensor Data Fusion for Positioning. In proceeding of IEEE International Conference on Communications; IEEE; 2010. pp. 1–5. [Google Scholar]
- Friston K. Variational filtering. Neuroimage. 2008a;41:747–766. doi: 10.1016/j.neuroimage.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Friston K. Dynamic casual modeling and Granger causality Comments on: The identification of interacting networks in the brain using fMRI: Model selection, causality and deconvolution. Neuroimage. 2009 doi: 10.1016/j.neuroimage.2009.09.031. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ. Bayesian estimation of dynamical systems: an application to fMRI. NeuroImage. 2002;16:513–530. doi: 10.1006/nimg.2001.1044. [DOI] [PubMed] [Google Scholar]
- Friston KJ. Hierarchical models in the brain. PLoS Computional Biology. 2008b;4:e1000211. doi: 10.1371/journal.pcbi.1000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Harrison L, Penny W. Dynamic causal modelling. Neuroimage. 2003;19:1273–1302. doi: 10.1016/s1053-8119(03)00202-7. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Mechelli A, Turner R, Price CJ. Nonlinear responses in fMRI: the Balloon model, Volterra kernels, and other hemodynamics. NeuroImage. 2000;12:466–477. doi: 10.1006/nimg.2000.0630. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Stephan KE, Daunizeau J. Mathematical Problems in Engineering. 2010. Generalised Filtering; p. 2010. [Google Scholar]
- Friston KJ, Trujillo-Barreto N, Daunizeau J. DEM: a variational treatment of dynamic systems. Neuroimage. 2008;41:849–885. doi: 10.1016/j.neuroimage.2008.02.054. [DOI] [PubMed] [Google Scholar]
- Gitelman DR, Penny WD, Ashburner J, Friston KJ. Modeling regional and psychophysiologic interactions in fMRI: the importance of hemodynamic deconvolution. Neuroimage. 2003;19:200–207. doi: 10.1016/s1053-8119(03)00058-2. [DOI] [PubMed] [Google Scholar]
- Handwerker D, Ollinger J, D’Esposito M. Variation of BOLD hemodynamic responses across subjects and brain regions and their effects on statistical analyses. Neuroimage. 2004;21:1639–1651. doi: 10.1016/j.neuroimage.2003.11.029. [DOI] [PubMed] [Google Scholar]
- Haykin SS. Kalman filtering and neural networks. Wiley Online Library; 2001. [Google Scholar]
- Hinton GE, van Camp D. Keeping the neural networks simple by minimizing the description length of the weights; In proceedings of COLT-93; ACM; 1993. pp. 5–13. [Google Scholar]
- Hu Z, Zhao X, Liu H, Shi P. Nonlinear analysis of the BOLD signal. EURASIP Journal on Advances in Signal Processing. 2009;2009:1–13. [Google Scholar]
- Iadecola C. CC commentary: intrinsic signals and functional brain mapping: caution, blood vessels at work. Cerebral Cortex. 2002;12:223–224. doi: 10.1093/cercor/12.3.223. [DOI] [PubMed] [Google Scholar]
- Ito K, Xiong K. Gaussian filters for nonlinear filtering problems. Automatic Control, IEEE Transactions. 2002;45:910–927. [Google Scholar]
- Jaakkola TS. Tutorial on variational approximation methods. Advanced mean field methods: theory and practice. 2000:129–159. [Google Scholar]
- Jacobsen D, Hansen L, Madsen K. Bayesian model comparison in nonlinear BOLD fMRI hemodynamics. Neural computation. 2008;20:738–755. doi: 10.1162/neco.2007.07-06-282. [DOI] [PubMed] [Google Scholar]
- Ji Z, Brown M. Joint State and Parameter Estimation For Biochemical Dynamic Pathways With Iterative Extended Kalman Filter: Comparison With Dual State and Parameter Estimation. Open Automation and Control Systems Journal. 2009;2:69–77. [Google Scholar]
- Jimenez JC. A simple algebraic expression to evaluate the local linearization schemes for stochastic differential equations* 1. Applied Mathematics Letters. 2002;15:775–780. [Google Scholar]
- Jimenez JC, Ozaki T. Local linearization filters for non-linear continuous-discrete state space models with multiplicative noise. International Journal of Control. 2003;76:1159–1170. [Google Scholar]
- Jimenez JC, Shoji I, Ozaki T. Simulation of stochastic differential equations through the local linearization method. A comparative study. Journal of Statistical Physics. 1999;94:587–602. [Google Scholar]
- Johnston LA, Duff E, Mareels I, Egan GF. Nonlinear estimation of the BOLD signal. NeuroImage. 2008;40:504–514. doi: 10.1016/j.neuroimage.2007.11.024. [DOI] [PubMed] [Google Scholar]
- Julier S, Uhlmann J, Durrant-Whyte HF. A new method for the nonlinear transformation of means and covariances in filters and estimators. Automatic Control, IEEE Transactions. 2002;45:477–482. [Google Scholar]
- Kalman RE. A new approach to linear filtering and prediction problems. Journal of basic Engineering. 1960;82:35–45. [Google Scholar]
- Kaminski P, Bryson A, Jr, Schmidt S. Discrete square root filtering: A survey of current techniques. Automatic Control, IEEE Transactions. 1971;16:727–736. [Google Scholar]
- Kloeden PE, Platen E. Numerical Solution of Stochastic Differential Equations, Stochastic Modeling and Applied Probability. 3. Springer; 1999. [Google Scholar]
- Krüger G, Glover GH. Physiological noise in oxygenation sensitive magnetic resonance imaging. Magnetic Resonance in Medicine. 2001;46:631–637. doi: 10.1002/mrm.1240. [DOI] [PubMed] [Google Scholar]
- Lauritzen M. Relationship of spikes, synaptic activity, and local changes of cerebral blood flow. Journal of Cerebral Blood Flow & Metabolism. 2001;21:1367–1383. doi: 10.1097/00004647-200112000-00001. [DOI] [PubMed] [Google Scholar]
- Li P, Yu J, Wan M, Huang J. The augmented form of cubature Kalman filter and quadrature Kalman filter for additive noise. IEEE Youth Conference on Information, Computing and Telecommunication, YC-ICT ‘09; IEEE; 2009. pp. 295–298. [Google Scholar]
- Ljung L, Söderström T. Theory and practice of recursive identification. MIT Press; Cambridge, MA: 1983. [Google Scholar]
- Logothetis NK. The neural basis of the blood–oxygen–level–dependent functional magnetic resonance imaging signal. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences. 2002;357:1003. doi: 10.1098/rstb.2002.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay DJC. Developments in probabilistic modelling with neural networks-ensemble learning. In proceedings of 3rd Annual Symposium on Neural Networks; Nijmegen, Netherlands. 1995. pp. 191–198. [Google Scholar]
- Magistretti P, Pellerin L. Cellular Mechanisms of Brain Energy Metabolism and their Relevance to Functional Brain Imaging. Philos Trans R Soc Lond B Biol Sci. 1999;354:1155–1163. doi: 10.1098/rstb.1999.0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandeville J, Marota J, Ayata C, Zaharchuk G, Moskowitz M, Rosen B, Weisskoff R. Evidence of a cerebrovascular postarteriole windkessel with delayed compliance. Journal of Cerebral Blood Flow & Metabolism. 1999a;19:679–689. doi: 10.1097/00004647-199906000-00012. [DOI] [PubMed] [Google Scholar]
- Mandeville JB, Marota JJA, Ayata C, Zaharchuk G, Moskowitz MA, Rosen BR, Weisskoff RM. Evidence of a cerebrovascular postarteriole windkessel with delayed compliance. Journal of Cerebral Blood Flow & Metabolism. 1999b;19:679–689. doi: 10.1097/00004647-199906000-00012. [DOI] [PubMed] [Google Scholar]
- Mechelli A, Price C, Friston K. Nonlinear coupling between evoked rCBF and BOLD signals: a simulation study of hemodynamic responses. NeuroImage. 2001;14:862–872. doi: 10.1006/nimg.2001.0876. [DOI] [PubMed] [Google Scholar]
- Miller KL, Luh WM, Liu TT, Martinez A, Obata T, Wong EC, Frank LR, Buxton RB. Nonlinear temporal dynamics of the cerebral blood flow response. Human Brain Mapping. 2001;13:1–12. doi: 10.1002/hbm.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray L, Storkey A. Continuous time particle filtering for fMRI. Advances in Neural Information Processing Systems. 2008:20. [Google Scholar]
- Nelson AT. PhD thesis. Oregon Graduate Institute of Science and Technology; 2000. Nonlinear estimation and modeling of noisy time-series by dual Kalman filtering methods. [Google Scholar]
- Norgaard M, Poulsen NK, Ravn O. New developments in state estimation for nonlinear systems. Automatica. 2000;36:1627–1638. [Google Scholar]
- Ozaki T. A bridge between nonlinear time series models and nonlinear stochastic dynamical systems: a local linearization approach. Statistica Sinica. 1992;2:113–135. [Google Scholar]
- Riera J, Watanabe J, Kazuki I, Naoki M, Aubert E, Ozaki T, Kawashima R. A state-space model of the hemodynamic approach: nonlinear filtering of BOLD signals. NeuroImage. 2004;21:547–567. doi: 10.1016/j.neuroimage.2003.09.052. [DOI] [PubMed] [Google Scholar]
- Robbins H, Monro S. A stochastic approximation method. The Annals of Mathematical Statistics. 1951;22:400–407. [Google Scholar]
- Roebroeck A, Formisano E, Goebel R. Reply to Friston and David:: After comments on: The identification of interacting networks in the brain using fMRI: Model selection, causality and deconvolution. NeuroImage. 2009a doi: 10.1016/j.neuroimage.2009.09.036. In Press. [DOI] [PubMed] [Google Scholar]
- Roebroeck A, Formisano E, Goebel R. The identification of interacting networks in the brain using fMRI: Model selection, causality and deconvolution. Neuroimage. 2009b doi: 10.1016/j.neuroimage.2009.09.036. In Press. [DOI] [PubMed] [Google Scholar]
- Simandl M, Dunik J. Design of derivative-free smoothers and predictors. In proceeding of the 14th IFAC Symposium on System Identification, SYSID06; 2006. pp. 1240–1245. [Google Scholar]
- Sitz A, Schwarz U, Kurths J, Voss HU. Estimation of parameters and unobserved components for nonlinear systems from noisy time series. Physical Review E. 2002;66:1–9. doi: 10.1103/PhysRevE.66.016210. [DOI] [PubMed] [Google Scholar]
- Sotero RC, Trujillo-Barreto NJ, Jiménez JC, Carbonell F, Rodríguez-Rojas R. Identification and comparison of stochastic metabolic/hemodynamic models (sMHM) for the generation of the BOLD signal. Journal of computational neuroscience. 2009;26:251–269. doi: 10.1007/s10827-008-0109-3. [DOI] [PubMed] [Google Scholar]
- Stephan KE, Kasper L, Harrison LM, Daunizeau J, den Ouden HEM, Breakspear M, Friston KJ. Nonlinear dynamic causal models for fMRI. NeuroImage. 2008;42:649–662. doi: 10.1016/j.neuroimage.2008.04.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Särkkä S, Hartikainen J. Extension of VB-AKF to Estimation of Full Covariance and Non-Linear Systems. IEEE Transactions on Automatic Control Under revision. [Google Scholar]
- Särkkä S, Nummenmaa A. Recursive noise adaptive Kalman filtering by variational Bayesian approximations. Automatic Control, IEEE Transactions. 2009;54:596–600. [Google Scholar]
- Valdes Sosa PA, Sanchez Bornot JM, Sotero RC, Iturria Medina Y, Aleman Gomez Y, Bosch Bayard J, Carbonell F, Ozaki T. Model driven EEG/fMRI fusion of brain oscillations. Human brain mapping. 2009;30:2701–2721. doi: 10.1002/hbm.20704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Merwe R. PhD thesis. Oregon Graduate Institute of Science and Technology; 2004. Sigma-point Kalman filters for probabilistic inference in dynamic state-space models. [Google Scholar]
- Wu Y, Hu D, Wu M, Hu X. Unscented Kalman filtering for additive noise case: augmented vs. non-augmented. IEEE Signal Processing Letters. 2005;12:357–359. [Google Scholar]