Abstract
Allogeneic hematopoietic stem cell transplantation is the treatment of choice for severe primary immunodeficiencies (PIDs). For patients lacking an HLA-identical donor, gene therapy is an attractive therapeutic option. Approaches based on insertion of a functional gene using viral vectors have provided proof of concept for the ability of gene therapy to cure PIDs. However, leukemic transformation due to insertional mutagenesis has been observed, prompting development of novel approaches based on introduction of DNA double strand breaks (DSB) into the endogenous locus to achieve gene correction, or into a safe genomic location (“safe harbor”).
Homing Endonucleases (HE) and Zinc Finger Nucleases (ZFN) are target-specific endonucleases that induce site-specific DSB, hence facilitating homologous recombination around their target sites to achieve gene correction or gene insertion into safe harbors. An alternative approach to achieve site-specific insertion of functional genes is based on transposons, DNA elements that spontaneously translocate from a specific chromosomal location to another. These novel tools may lead to efficient and safer strategies to achieve gene therapy for PIDs and other disorders.
Keywords: Primary Immunodeficiencies, Severe Combined Immunodeficiency, Gene Therapy, Gene Correction, Locus-specific Targeting, Homing Endonucleases, Meganucleases, Zinc Finger Nucleases, Safe Harbors, Transposons
Introduction
Primary immunodeficiencies (PIDs) comprise over 150 different disorders resulting from abnormal development and/or function of the immune system1. Most of these disorders are the consequence of a monogenic defect and hence follow a simple mendelian inheritance. Severe combined immunodeficiency (SCID) includes a group of disorders characterized by impaired development of T cells and functional B cell deficiency. Patients with SCID are highly prone to infections and typically die within the first two years of life, unless immune reconstitution is achieved. Allogeneic hematopoietic stem cell transplantation (HSCT) is the treatment of choice for SCID and for other severe forms of PIDs. HSCT from an HLA-identical donor can cure more than 95% of SCID infants, with no need for chemotherapy because of the patient's inability to reject donor-derived cells 2. However this option is available only to 15% of patients with SCID. T-cell depleted HSCT from a mismatched related donor without chemotherapy results in >95% survival if the transplant is performed within the first 3.5 months of life, and 70% survival at later ages 3. However, the majority of the patients fail to achieve functional B cell reconstitution and require life-long administration of intravenous immunoglobulins 3. If pre-transplant chemotherapy is used to favor the engraftment of donor-derived hematopoietic stem cells (HSCs), robust and complete immune reconstitution is often achieved, however treatment-related toxicity and GvHD cause significant mortality2. Furthermore, use of chemotherapy and immunosuppressive drugs to prevent GvHD cause a delay in immune reconstitution, and hence pose an increased risk of infections early after HSCT. Similar considerations apply also to other forms of severe PID, such as Wiskott-Aldrich syndrome (WAS), chronic granulomatous disease (CGD), and hemophagocytic lymphohistiocytosis (HLH) 2. Furthermore, in spite of continuous progress, survival after HSCT for non-SCID PIDs remains lower as compared to what is observed for SCID, even when an HLA-identical donor is available2. For these reasons, gene therapy is an attractive therapeutic option for SCID patients lacking an HLA-identical donor and for patients affected with severe non-SCID PIDs.
Current experience with gene therapy for PIDs: success and limitations
SCID is a particularly attractive disease candidate for gene therapy, because of the severity of the condition, the proven efficacy of HSCT, the easiness to isolate and manipulate target cells (HSCs), and the strong selective advantage of gene-targeted cells, as indicated by experience in animal models and by the emergence of mature and diversified T cells in vivo in SCID patients with gene reversion in lymphoid progenitor cells. Initial attempts to gene therapy for SCID used retroviruses (RV) as vectors, and expression of the therapeutic gene was driven by the RV long terminal repeat (LTR). RV-mediated integration of the transgene into the host genome allows stable transmission to progeny cells, thus maintaining efficacy during lymphoid development. Adenosine deaminase (ADA) deficiency was the first PID in which gene therapy was performed 20 years ago. Initially, gene targeting was addressed to peripheral blood lymphocytes as was shown for 2 patients with ADA-SCID that had normalized T cell counts and improved cellular and humoral responses following reinfusion of peripheral T cells that were treated by retroviral mediated transfer of the normal ADA gene4. However CD34+ cells rapidly became the target of interest. Efficient targeting of CD34+ cells has been achieved using a RV vector to deliver the ADA gene. In combination with a submyeloablative conditioning regimen to favor engraftment of gene-corrected cells, this protocol has resulted in long-term immune reconstitution in 13 out of 15 patients treated in Milan, with no need for concurrent use of enzyme replacement therapy5. Gene therapy trials with use of RV vectors and reduced intensity conditioning are also carried out in London (NCT01279720), Los Angeles (NCT00794508) and at the NIH (NCT00018018). Two trials have formally demonstrated the efficacy of gene therapy for X-linked SCID, leading to full normalization of T lymphocyte development and function without use of chemotherapy6. Ten patients each were treated in Paris or London between 1999 and 2006. These two trials made use of RV vectors that contained an amphotropic envelope or the gibbon ape envelope, respectively; in both vectors, the IL2RG cDNA was placed under the control of viral LTR. Seventeen out of these 20 patients are alive with T cell reconstitution. However, 5 patients have developed leukemic proliferation between 2.5 and 5 years after treatment; four of these patients have been successfully treated and now remain leukemia-free with robust T cell function, but one patient had died from treatment-refractory leukemia. These leukemic events were associated with integration of the RV within oncogenes loci; the LMO-2 oncogene was targeted in 4 of these 5 cases. The RV-LTR enhancer led to increased and deregulated expression of the oncogene; secondary mutations contributed to clonal proliferation.
The risk of insertional mutagenesis associated with use of RV vectors was confirmed in two other gene therapy trials for CGD and WAS. Two adult patients with CGD were treated by Grez and colleagues with gene therapy following chemotherapy; in this trial, expression of the CYBB cDNA was driven by the LTR of the spleen focus forming virus, to allow for robust expression of the transgene in myeloid cells. Clinical benefit was associated with restoration of NADPH oxidase activity. However, clonal myelopoiesis was observed, with expansion of cells carrying retroviral integration near the Evi-1, PRD1M16 and STBP1 oncogenes. This may have contributed to the short term clinical benefit by allowing expansion of cells with normal NADPH oxidase activity, however one of the two patients succumbed to myelodysplasia as the result of insertional mutagenesis7. More recently, Klein and colleagues have reported on successful use of gene therapy in patients with WAS8. In this trial, patients received submyeloablative chemotherapy followed by re-infusion of autologous CD34+ cells that had been transduced with a RV in which expression of the WAS cDNA was under control of the myeloproliferative sarcoma virus LTR. Functional reconstitution in T, NK and myeloid cells and a significant increase in platelet number have been observed in the two patients treated with this protocol for whom extensive follow-up data are available, and preliminary data indicate that beneficial effects were observed 9 out of 10 patients treated. However, one patient developed acute T cell leukemia due to insertional mutagenesis and activation of the LMO-2 proto-oncogene (Cristoph Klein, personal communication).
Development of novel viral vectors for gene therapy of PIDs
In order to reduce the risk of insertional mutagenesis, self-inactivating (SIN)-LTR RV vectors have been developed, with use of internal promoters and deletion of LTR enhancer activity, so-called Self-inactivating (SIN)-LTR RV vectors9. A multi-site (London, Paris, Boston, Los Angeles and Cincinnati) clinical protocol for treatment of X-linked SCID using SIN-LTR RV vector has recently been initiated (NCT01129544, NCT01175239). Additional modifications proposed to increase safety of RV vectors include use of insulators (to prevent transactivation of neighboring genes) and of suicide genes in the construct. In spite of these technical advances, there is still concern that RV vectors may pose risks, because of the preferential integration within 5′ regulatory regions and coding regions of genes that are actively expressed in target cells, and especially of proto-oncogenes.
Lentiviral (LV) vectors may be safer, because they do not show any preference for integration within 5′ regulatory regions. A gene therapy trial based on use of LV vectors for WAS, in which the WAS cDNA is under control of the endogenous promoter, has been recently started in Milan, and should open soon in London, Paris and Boston. It is expected that additional protocols based on use of LV vectors will be started soon for various forms of PID. However, it should be noted that even use of LV vectors does not eliminate the risk of clonal dominance, as indicated by recent experience in a trial for β-thalassemia, although no leukemic outgrowth was observed in this case10. To further increase safety, non integrating lentiviral (NILV) vectors have been proposed. Following reverse transcription, the LV DNA does not integrate into the host genome, but tends to form episomal circles, that are diluted with cell division. Therefore, use of NILV as such is not attractive for the treatment of disorders (such as PIDs) that involve proliferating cells. However, NILV vectors may become of interest to deliver enzymes (homing endonucleases, zinc-finger nucleases) that may induce true gene correction or targeting of the transgene to a suitable chromosomal location (“safe harbor”).
Novel strategies to achieve gene correction and locus-specific targeting
In parallel with the development of safer viral vectors, scientists have focused on alternative approaches to achieve disease correction by gene therapy using strategies that would avoid deregulated expression of other genes. True “in situ” gene correction and targeting of safe harbors are receiving particular interest.
Gene correction relies upon replacing the region of the gene containing the mutation with a fragment of DNA containing the wild-type sequencing, while leaving all neighboring sequences intact. If successful, this strategy will “repair” the mutation and allow for normal protein expression. This strategy also has the inherent advantage of using the endogenous promoter and enhancer regulatory elements, thus maintaining physiological expression of the gene. This approach would be particularly attractive for correction of PIDs due to mutations of tightly regulated genes (such as CD40 ligand, CD40LG), and would also be of special interest to correct disorders that have proven refractory to conventional approaches of gene therapy. One group of such disorders is RAG1 deficiency (associated with SCID, Omenn syndrome and various forms of leaky SCID in humans). Attempts to cure Rag1−/− mice by RV-mediated gene transfer have failed, unless stem cells are transduced at high multiplicity of infection to integrate a high copy number of the transgene, but this approach exposes to the risk of insertional mutagenesis and lymphoma development11. Furthermore, even the addition of known endogenous Rag1 regulatory elements in the RV construct does not permit correction of the leaky SCID phenotype of Rag1S723C/S723C mice (Notarangelo LD and Mostoslavsky G, unpublished observations). Finally, “in situ” gene repair should be considered for rare forms of PID resulting from dominant-negative mutations.
An alternative approach to gene therapy, that would minimize risks of insertional mutagenesis, is based on targeting of specific and safe loci in the genome (“safe harbors”), that are devoid of oncogenes and whose disruption does not lead to deleterious consequences. This approach may be particularly interesting for PIDs, because in most cases PID-causing mutations are scattered through the diseased gene, making gene repair by use of specific HEs or ZFNs unpractical. Potential candidates for locus-specific “safe harbor” targeting include the human Rosa26 locus on chromosome 3 and the Adeno-Associated Virus integration Site 1 (AAVS1) on chromosome 19q13, that encodes for the ubiquitously expressed PPP1R12C gene.
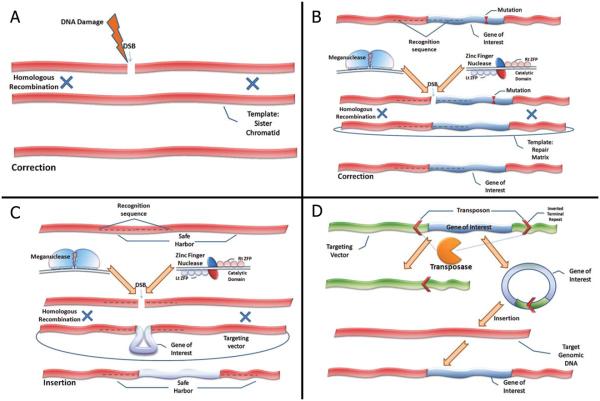
Both gene repair and specific targeting into “safe harbors” require insertion of exogenous DNA sequences into the host genome. Most of the strategies that are used to achieve this goal utilize the endogenous DNA repair mechanisms that are based on homologous recombination (HR). This is one of the critical mechanisms responsible for repairing DNA double-stranded breaks (DSB) that result from exposure to alkylating agents, radiation, but that can also occur spontaneously, especially during chromosome replication. By harnessing the second copy of the affected gene that is coded on the sister chromatid as a template; the HR mechanism facilitates the replacement of the DNA surrounding the DSB by recombination (Fig 1). If extra-sequences flanked by 5′ and 3′ homology arms are included in the template, HR may promote insertion of these sequences into the repaired site. This mechanism has been historically used for the creation of knock-out and knock-in animal models. Similarly, spontaneous HR may induce gene correction when a template carrying the normal sequence, but otherwise homologous to the targeted mutated area, is introduced into the cell. However the rate of spontaneous HR is extremely low (< 1/106 cells), and is even lower in HSCs than in embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs). To circumvent this problem, novel strategies have been designed to introduce locus-specific DNA breaks and promote introduction of exogenous DNA templates into the genome.
Figure 1. Novel mechanisms for gene correction and insertion.
(A) Homologous recombination (HR) is an important cellular repair mechanism of DNA double-strand breaks (DSB) that occur spontaneously or following exposure to reactive oxidase species, alkylating agents or radiation. Correction is achieved by recombination with the homologous sequence coded on the sister chromatid or a homologous chromosome. (B) Both Homing Endonucleases (HEs) and Zinc Finger Nucleases (ZFN) have the ability to induce DSBs at specific DNA target sequences. HEs and ZFNs may be engineered to recognize genomic regions that are close to disease-causing mutations. ZFN are heterodimers produced by the artificial coupling of zinc finger proteins (in the figure: left ZFP, Lt ZFP; right ZFP, Rt ZFP) that bind to the specific DNA target and a DNA cleavage domain. HEs (also known as meganucleases) are heterodimers produced by modifications of naturally occurring enzymes such as I-SceI. When used in combination with a repair matrix homologous to the DNA target area (but containing the normal sequence), they may promote HR and correction of the mutation. (C) HEs and ZFNs may also be engineered to recognize locations of the genome (“safe harbors”) that do not contain oncogenes or genes encoding for micro-RNAs. When used in combination with a suitable repair matrix, containing the gene of interest flanked by 5′ and 3′ homology regions to the target DNA sequence, these HEs and ZFNs may allow for insertion of a functional gene copy into the “safe harbor”. (D) An alternative approach utilizes transposons which are DNA elements that spontaneously translocate from a specific chromosomal location to another. The transpoase enzyme recognizes inverted terminal repeats (ITR) and cleaves them, thus producing the transposon, which is later re-integrated into a different locus. Introduction of a matrix that codes for the gene of interest, flanked by the ITRs will result in excision of the gene and its re-integration into a genomic locus.
Homing endonucleases
Homing endonucleases (HEs) or Meganucleases are highly efficient, sequence-specific enzymes that induce DSB in specific loci. Originally identified in yeasts almost 25 years ago, HEs may function in various cell types derived from different organisms including plants, bacteria, yeasts, Drosophila, mice and humans, and promote HR by inducing DSBs, with a frequency that is more than 1000-fold higher than spontaneous HR12 (Fig 1).
HE-specific DNA target sites may range from 14 to 40 base pairs. Given the length of the recognition sequence, only few target sites are expected in the genome; in fact there is only one recognition site for I-SceI (the prototype of HEs) in the whole yeast genome. The LAGLIDADG family of HEs has been studied in greater detail. Its naturally occurring members are either monomeric enzymes with 2 subdomains that target nonpolyndromic DNA sequences or homodymeric proteins that target palindromic sequences. By mutagenizing in vitro the residues of I-CreI HE that are known to mediate recognition and interaction with DNA target sequences, and using high-throughput combinatorial screening, it is possible to generate and characterize a large series of engineered HEs with distinct target specificity. Using this approach, a series of HEs that specifically recognize the human RAG1 locus immediately upstream of the single coding exon 2 have been generated, and their ability to induce DNA repair has been demonstrated upon co-transfection of human cells with plasmids carrying the RAG1-specific recognition site and a repair matrix13. The efficiency of targeting is estimated to range from 0.1 to about 5% in human cells (unpublished data). While these data are encouraging, further studies are needed to determine the efficiency of targeting and the ability to induce correction of the endogenous RAG1 locus. Furthermore since the target sequence of the RAG1-specific HE is not conserved among species and is unique to humans, appropriate “humanized” animal models in which the mouse Rag1 locus is replaced by the human gene (in its wild-type or mutated sequence) must be developed before clinical trials can be proposed.
Zinc-finger nucleases
Zinc-finger nucleases (ZFNs) are a group of artificial fusion proteins that are generated by linking a zinc-finger DNA-binding domain that recognizes a specific DNA target to the nuclease domain of the endonuclease Fok1. ZFNs can be engineered to mediate specific targeting of a mutated gene. The introduction into the cell of a DNA wild-type template may induce gene repair by HR or through non homologous end-joining (NHEJ) (Fig 1), another cellular mechanism of DSB repair that however is error-prone. Using this approach, correction of a mutation in the IL2RG gene (mutated in X-linked SCID) was demonstrated in vitro in 5-17% of patient-derived cells14. Furthermore, ZFNs have been successfully used to mediate targeting of other disease loci in both human ES cells and iPSCs, thus opening the way for future studies and possible applications in human gene therapy15. However, for both HEs and ZFNs further studies are needed to investigate in greater detail the risks of undesired toxicity due to off-target cleavage. These risks include cell death and the formation of break-induced DNA sequence alterations due to NHEJ as well as chromosomal translocations possibly leading to oncogenic transformation. Finally, cellular delivery of HEs and ZFNs must be also considered. Use of RV or integrating LV vectors may re-introduce the risk of insertional mutagenesis; furthermore, prolonged expression of the HE or ZFN genes may cause deleterious effects as mentioned above. For this reason, NILV vectors that allow expression of the enzyme only for limited time appear attractive, although this type of vector allows less efficient expression of the gene than integrating vectors.
Transposons and transposases
Transposons are DNA elements that are able to spontaneously translocate from a specific chromosomal location to another16. They can either move directly as DNA or undergo transposition via an RNA intermediate (retrotransposons). This unique activity is facilitated by the transposase protein that targets any DNA cargo sequence flanked by the inverted terminal repeat (ITR) sequences (the transposon). As a result of this process, a DNA fragment is excised from the donor locus and is re-integrated in into another locus (Fig 1), making this system a good candidate to use as a possible gene delivery system. In order to transform this naturally occurring system into a novel gene delivery tool, a matrix that allows for the expression of the transposase as well as a donor vector containing the gene of interest (flanked by ITR sequences) need to be introduced (separately or jointly) into the host cell in order to achieve gene insertion.
Among transposon systems that are active in human cells and hence may have therapeutic relevance, the Sleeping Beauty (SB) transposon seems particularly promising, since it exhibits a random pattern of integration that is not biased toward integration into gene loci; furthermore, it is not associated with recombination or deletion events at the integration sites17.
The Tol2 system is considered less efficient then the SB; however, it has several advantages: a) it is able to transfer genes of up to 11 kb with minimal loss of transposition activity; b) it creates single-copy insertions; and, c) it is known not to cause gross rearrangements around the integration sites (reviewed in 18).
The PiggyBac is another transposon system that was recently shown to catalyze transposition in human and mouse somatic cells with higher transposition efficiency then SB or Tol2 in different cell lines. However, its genomic integration profile is similar to what observed for integrating viral vectors (reviewed in 18).
While the transposon technology has already entered clinical trials to induce expression of CD19 chimeric antigen receptors in cytotoxic T cells from patients with refractory B lymphoid malignancies, significant limitations remain for a possible use of this technology in gene therapy of PIDs. In particular, delivery of transposon and transposase constructs into somatic cells still depends on plasmids and/or viral vectors. Moreover, further studies are needed to gain better control of integration sites and to avoid intragenic insertion.
Limitations and Disadvantages
As for any novel technology, several limitations still exist that need to be addressed prior to clinical implementation of the novel approaches to gene therapy discussed here (table 1). Many of these imperfections have to do with specificity and efficiency. In particular, ZFN- and HE-specific recognition sites are not continuously distributed along the genome, thereby limiting the number of mutations that can be targeted and corrected. Furthermore, the introduction of DSBs mediated by ZFNs and HEs may result in a significant risk for chromosomal translocations; this risk is even higher if the enzyme shows poor specificity leading to off-site targeting. Of note, the frequency and nature of translocations (“translocatome”)19 caused by ZFNs and HEs will have to be assessed individually for each of these enzymes used to introduce DSBs. Finally, significant improvement is needed to increase the efficiency of intranuclear delivery of the desired machinery (ZFNs, HEs, etc) and the repair matrix, HE, ZFN etc), while limiting the risk of cellular transformation or dysregulation of gene expression. This goal might be achieved using NILVs to deliver HEs and ZFNs, however this approach currently suffers from low levels of expression mediated by NILVs.
Table1.
Advantages and limitations of novel approaches to gene therapy
| Technology | Basic concept | Major advantages | Major limitations | Technical aspects that need to be improved prior to clinical implementation |
|---|---|---|---|---|
| Homing Endonucleases (HEs) |
Use of highly specific engineered endonuclease to induce a DSB in a desired locus and promote homologous recombination with a repair matrix. |
Gene correction within the endogenous locus, thus maintaining proper regulation of gene expression. |
The low number of HEs available limit applicability of this strategy for gene correction. Lack of animal models for pre- clinical trials. |
Low efficiency Off-target cleavage Inefficient intranuclear delivery of HE and repair matrix. |
| Zinc Finger Nucleases (ZFNs) |
Use of an artificial fusion protein that combines a nuclease with a Zinc-finger protein to induce site- specific DSBs and promote homologous recombination with a repair matrix. |
Gene correction within the natural locus allows for maintenance of endogenous regulation |
Need for a specific system for each gene and sometimes for different mutations with in the same gene. Lack of animal models for pre- clinical trials. |
Low efficiency Off target cleavage Inefficient intranuclear delivery of HE and repair matrix. |
| Safe Harbor targeting |
Targeting of specific and safe (ie, far from oncogenes) loci in the genome to introduce a normal copy of the desired gene. |
Safe and efficient insertion of a normal copy of the desired gene under any desired promoter. One system will work for various defects and diseases. |
Insertion of an extra copy of the desired gene. Does not utilize the natural regulatory elements in the endogenous locus. Significant limitation for use in dominant negative defects and leaky phenotypes |
Inefficient intranuclear delivery of HE and repair matrix. |
| Transposons and transposases |
Use of transposase protein that targets any DNA cargo sequence flanked by ITR sequences for random integration of a normal copy of the desired gene |
Efficient delivery and insertion of large sequences (up to 11 Kb). One system will work for various defects and diseases. |
Random integration may lead to insertional mutagenesis and/or intragenic disruption. Does not utilize the natural regulatory elements in the endogenous locus. Significant limitation for use in dominant negative defects and leaky phenotypes |
Inefficient intranuclear delivery of HE and repair matrix. |
DSB: double-strand DNA break; ITR: inverted terminal repeats
Perhaps the major limitation in the translation of these approaches from the bench to the bedside consists in the lack of appropriate animal models. Therapeutic ZFNs and HEs are designed to recognize and cleave human DNA sequences. Hence, both the efficiency and the safety profile of these approaches can be hardly studied in animal models. A possible strategy to overcome this impediment is the use of “humanized” animal models in which the mutated human gene of interest is introduced to replace the mouse orthologue gene. Despite these limitations, there is increasing interest in the development of novel strategies to gene therapy based on gene correction or targeting of safe harbors.
Conclusions
Twenty years after initial application, gene therapy has fulfilled its promise to correct human genetic diseases, and PIDs in particular. However, it has also shown that significant risks related to use of viral vectors remain to be addressed, in particular insertional mutagenesis. Development of novel and safer integrating vectors is expected to reduce, but probably not completely eliminate, this risk. There is growing interest for the development of novel strategies that aim to induce gene repair or to target the therapeutic gene into a safe harbor. While each of these new approaches currently has intrinsic limitations (in particular, efficiency), significant progress has been made to foresee use of these techniques in the future (table 1). In the meantime, development of suitable, patient-derived cellular models, such as iPSCs20 may permit in vitro testing and comparison of these various approaches and guide their development in the treatment of severe PIDs.
Abbreviations
- AAVS1
Adeno-associated virus integration site 1
- ADA
Adenosine deaminase
- CGD
Chronic granulomatous disease
- DSB
DNA double-stranded breaks
- ESC
Embryonic stem cell
- GvHD
Graft versus host disease
- HSCT
Hematopoietic stem cell transplantation
- HLH
Hemophagocytic lymphohistiocytosis
- HE
Homing Endonucleases
- HR
Homologous recombination
- iPSC
Induced pluripotent stem cell
- LV
Lentivirus
- LTR
Long terminal repeat
- NILV
Non-integrating lentivirus
- PID
Primary Immunodeficiencies
- RV
Retrovirus
- (SIN)
Self-inactivating
- SCID
Severe combined immunodeficiency
- SB
Sleeping Beauty
- WAS
Wiskott -Aldrich syndrome
- ZFN
Zinc Finger Nucleases
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Notarangelo LD, Fischer A, Geha RS, Casanova JL, Chapel H, Conley ME, et al. Primary immunodeficiencies: 2009 update. J Allergy Clin Immunol. 2009;124:1161–78. doi: 10.1016/j.jaci.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gennery AR, Slatter MA, Grandin L, Taupin P, Cant AJ, Veys P, et al. Transplantation of hematopoietic stem cells and long-term survival for primary immunodeficiencies in Europe: entering a new century, do we do better? J Allergy Clin Immunol. 2010;126:602–10 e1-11. doi: 10.1016/j.jaci.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 3.Buckley RH. Molecular defects in human severe combined immunodeficiency and approaches to immune reconstitution. Annu Rev Immunol. 2004;22:625–55. doi: 10.1146/annurev.immunol.22.012703.104614. [DOI] [PubMed] [Google Scholar]
- 4.Blaese RM, Culver KW, Miller AD, Carter CS, Fleisher T, Clerici M, et al. T lymphocyte-directed gene therapy for ADA- SCID: initial trial results after 4 years. Science. 1995;270:475–80. doi: 10.1126/science.270.5235.475. [DOI] [PubMed] [Google Scholar]
- 5.Ferrua F, Brigida I, Aiuti A. Update on gene therapy for adenosine deaminase-deficient severe combined immunodeficiency. Curr Opin Allergy Clin Immunol. 2010;10:551–6. doi: 10.1097/ACI.0b013e32833fea85. [DOI] [PubMed] [Google Scholar]
- 6.Fischer A, Hacein-Bey-Abina S, Cavazanna-Calvo M. Gene therapy for primary immunodeficiencies. Immunol Allergy Clin North Am. 2010;30:237–48. doi: 10.1016/j.iac.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- 8.Boztug K, Schmidt M, Schwarzer A, Banerjee PP, Diez IA, Dewey RA, et al. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N Engl J Med. 2010;363:1918–27. doi: 10.1056/NEJMoa1003548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qasim W, Gaspar HB, Thrasher AJ. Progress and prospects: gene therapy for inherited immunodeficiencies. Gene Ther. 2009;16:1285–91. doi: 10.1038/gt.2009.127. [DOI] [PubMed] [Google Scholar]
- 10.Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–22. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lagresle-Peyrou C, Yates F, Malassis-Seris M, Hue C, Morillon E, Garrigue A, et al. Long-term immune reconstitution in RAG-1-deficient mice treated by retroviral gene therapy: a balance between efficiency and toxicity. Blood. 2006;107:63–72. doi: 10.1182/blood-2005-05-2032. [DOI] [PubMed] [Google Scholar]
- 12.Silva G, Poirot L, Galetto R, Smith J, Montoya G, Duchateau P, et al. Meganucleases and other Tools for Targeted Genome Engineering: Perspectives and Challenges for Gene Therapy. Curr Gene Ther. 2010 Dec 24; doi: 10.2174/156652311794520111. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munoz IG, Prieto J, Subramanian S, Coloma J, Redondo P, Villate M, et al. Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus. Nucleic Acids Res. 2010 Sep 16; doi: 10.1093/nar/gkq801. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–51. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 15.Nakayama M. Homologous recombination in human iPS and ES cells for use in gene correction therapy. Drug Discov Today. 2010;15:198–202. doi: 10.1016/j.drudis.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Claeys Bouuaert C, Chalmers RM. Gene therapy vectors: the prospects and potentials of the cut-and-paste transposons. Genetica. 2010;138:473–84. doi: 10.1007/s10709-009-9391-x. [DOI] [PubMed] [Google Scholar]
- 17.Hackett PB, Largaespada DA, Cooper LJ. A transposon and transposase system for human application. Mol Ther. 2010;18:674–83. doi: 10.1038/mt.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.VandenDriessche T, Ivics Z, Izsvák Z, Chuah MK. Emerging potential of transposons for gene therapy and generation of induced pluripotent stem cells. Blood. 2009;114:1461–8. doi: 10.1182/blood-2009-04-210427. [DOI] [PubMed] [Google Scholar]
- 19.Gostissa M, Alt FW, Chiarle R. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annu Rev Immunol. 2010 Apr 5; doi: 10.1146/annurev-immunol-031210-101329. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 20.Pessach IM, Ordovas-Montanes J, Zhang SY, Casanova JL, Giliani S, Gennery AR, et al. Induced pluripotent stem cells: A novel frontier in the study of human primary immunodeficiencies. J Allergy Clin Immunol. 2010 Dec 23; doi: 10.1016/j.jaci.2010.11.008. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]