Abstract
Effects of silencing ectopically expressed hSNCA in rat substantia nigra (SN) were examined as a novel therapeutic approach to Parkinson’s disease (PD). AAV-hSNCA with or without an AAV harboring a short-hairpin (sh)RNA targeting hSNCA or luciferase was injected into one SN. At 9wks, hSNCA-expressing rats had reduced SN dopamine (DA) neurons and exhibited a forelimb deficit. AAV-shRNA-SNCA silenced hSNCA and protected against the forelimb deficit. However, AAV-shRNA-SNCA also led to DA neuron loss suggesting undesirable effects of chronic shRNA expression. Effects on nigrostriatal-projecting neurons were examined using a retrograde tract tracer. Loss of striatal-projecting DA neurons was evident in the vector injection site, whereas DA neurons outside this site were lost in hSNCA-expressing rats, but not in hSNCA-silenced rats. These observations suggest that high levels of shRNA-SNCA were toxic to DA neurons, while neighboring neurons exposed to lower levels were protected by hSNCA gene silencing. Also, data collected on DA levels suggest that neurons other than or in addition to nigrostriatal DA neurons contributed to protection of forelimb use. Our observations suggest that while hSNCA gene silencing in DA neurons holds promise as a novel PD therapy, further development of silencing technology is required.
Keywords: neurodegeneration, RNAi, substantia nigra, gene therapy, tyrosine hydroxylase
1. Introduction:
Alpha-synuclein (SNCA) is a 140 amino acid presynaptic phosphoprotein that is abundant in neurons (Lee and Trojanowski, 2006). Findings from familial and sporadic Parkinson’s disease (PD) cases implicate SNCA in PD pathogenesis. Three point mutations in the human (h)SNCA gene, as well as multiplication of the SNCA gene, are included among familial forms of PD. Proteinacious inclusions termed Lewy bodies (Lee and Trojanowski, 2006), of which SNCA is a major component (Spillantini et al., 1997), are present in 90% of PD cases, suggesting that SNCA is also involved in sporadic PD. Further, specific SNCA promoter polymorphisms have been linked to PD (Maraganore et al., 2006).
Experimental SNCA over-expression models that result in loss of dopamine (DA) neurons are valuable for testing the hypothesis that SNCA is a therapeutic target for PD (Chesselet, 2008). Neurotoxin-induced PD models, including exposure to rotenone, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), 6-OHDA and paraquat, exhibit increased SNCA expression and aggregation as well as DA neuron loss (Cannon and Greenamyre, 2010). In addition, experimental expression of wild-type and mutant forms of SNCA have been shown to result in DA neuron degeneration and formation of aggregated SNCA cellular inclusions in drosophila (Feany and Bender, 2000), mouse (Chesselet, 2008), rat (Kirik et al., 2002) and monkey (Kirik et al., 2003). In SNCA transgenic (tg) mice, SNCA inclusion formation and DA-related deficits vary depending on the promoter and the form of SNCA used (Fernagut and Chesselet, 2004). Non-tg models of SNCA-induced PD-like symptoms also have been developed using lentiviral (LV, (Lo Bianco et al., 2002)) and adeno-associated viral (AAV, (Kirik et al., 2002; Kirik et al., 2003)) vector-mediated delivery of exogenous SNCA, which induces nigrostriatal DA neurodegeneration. In this study, we use AAV to express hSNCA in rat substantia nigra (SN), a model reported previously (Kirik et al., 2002).
Approaches that directly target aberrant SNCA expression may be promising for therapeutic development. Glial cell line-derived neurotrophic factor (GDNF) does not prevent DA neurodegeneration in a LV-based SNCA expression model (Lo Bianco et al., 2004), although GDNF has been shown to ameliorate neurodegeneration in neurotoxin models of PD (Choi-Lundberg et al., 1997; Kearns and Gash, 1995; Mandel et al., 1997; Sauer et al., 1995; Tomac et al., 1995). However, SNCA targeting using a ribozyme in a rat model of PD where 1-methyl-4-phenylpryridinium induced an increase in SNCA expression has been shown to protect against DA neuron loss in rat SN (Hayashita-Kinoh et al., 2006).
RNA interference (RNAi) is a conserved process whereby double-stranded RNA targets mRNA in a sequence-specific manner resulting in degradation or translational inhibition of target mRNA (Fire et al., 1998; Scherr and Eder, 2007). Development of RNAi as a therapeutic approach is expected to provide novel opportunities for treating a wide range of disorders. Exogenous interfering RNAs can be administered as synthetic small interfering (si)RNAs or as short hairpin (sh)RNAs, usually delivered using viral vectors. siRNAs incorporate into the cellular RNAi machinery at the RNA inhibitory silencing complex and have only transient effects. In contrast, shRNAs can be chronically expressed in cells where they undergo processing similar to that of endogenous pre-micro (mi)RNAs (Scherr and Eder, 2007). Several approaches have been used to silence SNCA expression, including use of ribozymes (Hayashita-Kinoh et al., 2006), intracelluarly expressed single chain antibodies (Yuan and Sierks, 2009), siRNAs (Lewis et al., 2008; McCormack et al.), miRNAs (Doxakis, 2010; Junn et al., 2009) and viral vector-mediated delivery of an shRNA (Sapru et al., 2006). AAV and LV are ideal viral vectors for delivery of gene silencing molecules to the CNS because they are capable of transducing neurons to elicit long-term transgene expression (Bjorklund et al., 2000).
We previously reported that an shRNA sequence targeting hSNCA effectively silences endogenous hSNCA in SH-SY5Y cells in vitro and ectopically expressed hSNCA in vivo in rat striatum (ST, (Sapru et al., 2006)). To further explore the potential of SNCA gene silencing for PD, we generated an AAV2 harboring this shRNA. This was used to determine whether SNCA gene silencing can protect SN DA neurons in a rat model of PD where hSNCA is ectopically expressed in SN, leading to DA neuronal degeneration (Kirik et al., 2002).
2. Results:
2.1. Silencing of ectopically expressed hSNCA in rat SN and ST:
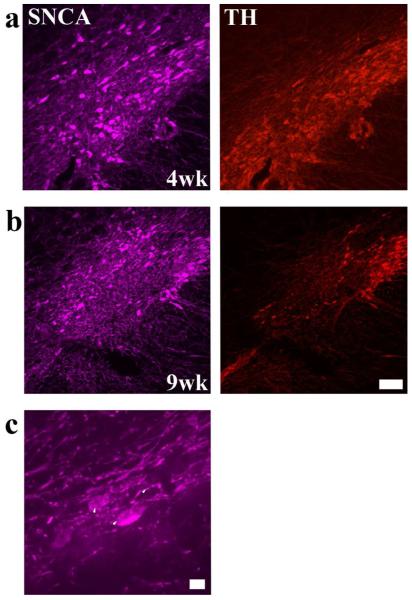
To confirm that ectopic hSNCA expression in rat SN using our AAV vector has a time-dependent toxic effect on nigral DA neurons as reported previously (Kirik et al., 2002), AAV-hSNCA was injected into one SN alone or with a control vector, AAV-humanized green fluorescent protein (hrGFP) or AAV-shRNA-luciferse (Luc)-hrGFP. Expression of hSNCA at 4 and 9wks after injection was studied by immunofluorescence using a hSNCA-specific antibody. hSNCA expression was abundant in SN of all SNCA-expressing groups at 4wks (Fig. 1a) and 9wks (Fig. 1b) after injection and cytoplasmic deposits that appeared to be aggregated SNCA were observed (Fig. 1c). Ectopic hSNCA expression resulted in a loss of neurons IR for tyrosine hydroxylase (TH), the rate-limiting enzyme in DA synthesis, 9wks after injection (Fig. 1b), but not 4wks after injection (Fig. 1a). hSNCA expression also was observed in the ventral tegmental area (VTA) where no apparent reduction in TH-IR neurons was observed (data not shown).
Figure 1. Chronic expression of hSNCA in DA neurons in SN leads to SNCA accumulation and a time-dependent loss of DA neurons.
hSNCA expression (pseudo-colored purple) in rat SN at 4 weeks (a) and 9 weeks (b) after injection of AAV-CBA-hSNCA is shown. TH-IR is shown in red. Note reduced TH-IR at 9 weeks, but not at 4 weeks after injection. Size bar, 100 μm. (c) Higher magnification of DA neurons showing accumulation of hSNCA-IR in DA perikarya and fibers (arrowheads). Size bar, 10 μm
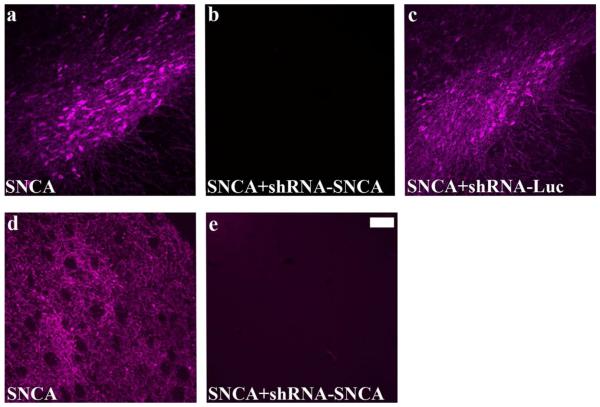
To determine whether it is possible to silence ectopically expressed hSNCA in rat SN using an AAV vector harboring an shRNA specifically targeting human and not rat SNCA (Han et al., 2011), AAV-hSNCA and AAV-shRNA-SNCA-hrGFP were co-injected into one SN. For controls, AAV-hSNCA was co-injected with AAV-hrGFP or AAV-shRNA-Luc-hrGFP. hSNCA expression was abundant in both control groups in SN (Fig. 2a,c). Both anterograde and retrograde transport of transgenes were observed as indicated by hSNCA expression in ST fibers (Fig. 2d) and perikarya, respectively. Effective silencing of hSNCA occurred because hSNCA-IR was observed only in rare cells and fibers in SN (Fig. 2b) and ST (Fig. 2e) of hSNCA-silenced rats. Co-injection of hSNCA and the control shRNA-Luc did not decrease hSNCA expression (Fig. 2c). Silencing of ectopically expressed hSNCA was also observed 9wks after injection and expression of hSNCA or hrGFP was not observed in the uninjected SN (data not shown).
Figure 2. Silencing of ectopically-expressed hSNCA in SN and ST using AAV2 vectors.
hSNCA-IR is shown in SN (a,b,c) and ST (d,e) of rats injected with AAV-CBA-SNCA (a,d), AAV-CBA-SNCA and AAV-shRNA-SNCA (b,e) or AAV-CBA-SNCA and AAV-shRNA-Luc (c) for 4 weeks. Note that expression and silencing of hSNCA was observed in both SN and ST as early as 4 weeks after injection and that the shRNA-Luc did not silence hSNCA expression. Size bar, 100 μm.
2.2. SNCA gene silencing protects against the forelimb deficit induced by hSNCA:
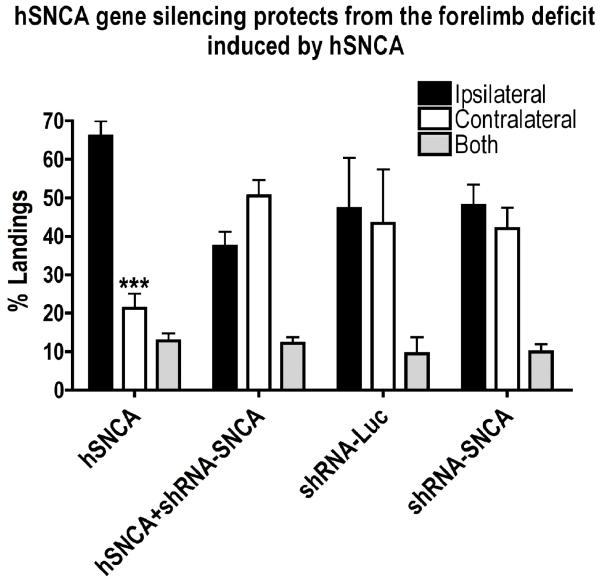
To determine whether silencing ectopic hSNCA expression in DA neurons has an effect on motor behavior, non-drug induced forelimb testing was performed at 4 and 8wks after injection. At 4wks post-injection, no preference for forelimb use was observed in any treatment group (data not shown). However, by 8wks (Fig. 3), hSNCA-expressing rats showed a strong preference for ipsilateral limb use and reduced contralateral limb use. Because forelimb use in the 3 treatment groups that received injection of AAV-hSNCA or AAV-hSNCA plus a control vector, either AAV-hrGFP or AAV-shRNA-Luc, were not statistically different using a Kruskal-Wallis one-way ANOVA followed by a Dunn’s post-hoc test, these data were combined into a single hSNCA-expressing group. Expression of hSNCA in rat SN led to a reduction in contralateral limb use (21.28±3.83%, n=27, p≤0.001) compared to ipsilateral limb use (65.99±3.89%, n=27), demonstrating a unilateral deficit in spontaneous motor behavior due to hSNCA expression. This ipsilateral forelimb use preference was not observed in rats where hSNCA was silenced by co-injection of AAV-shRNA-SNCA, suggesting a total protection of this deficit by hSNCA gene silencing. In addition, no effects on forelimb use were observed when the shRNAs were injected alone, in the absence of hSNCA expression. These observations suggest that preventing expression of ectopically expressed hSNCA using a hSNCA-specific shRNA protects against development of a hSNCA-induced deficit in contralateral forelimb use.
Figure 3. The hSNCA-specific shRNA protects against the deficit in forelimb use elicited by ectopic hSNCA expression in nigral DA neurons.
Rats underwent forelimb placement testing at 8 weeks after injection. Landings are expressed as percentage (number of landings for ipsilateral or contralateral forelimb, or both forelimbs, as a percent of total landings). Forelimb use in the three groups of rats that received injection of AAV-hSNCA or AAV-hSNCA plus a control vector, either AAV-hrGFP or AAV-shRNA-Luc, were not statistically different from each other using a Kruskal-Wallis one-way ANOVA followed by a Dunn’s post-hoc test, so these data were combined into a single hSNCA-expressing group. Note the preference for ipsilateral forelimb use in rats expressing hSNCA and protection of the contralateral forelimb deficit in rats in which hSNCA expression is silenced. No significant effect of either silencing vector alone was observed. Mean±SEM; data were analyzed using Kruskal-Wallis one-way ANOVA, followed by Dunn’s post-hoc tests. Asterisks denote significant differences between contralateral and ipsilateral forelimb use: ***=p≤0.001.
2.3. SNCA gene silencing does not protect against loss of DA neurons induced by hSNCA:
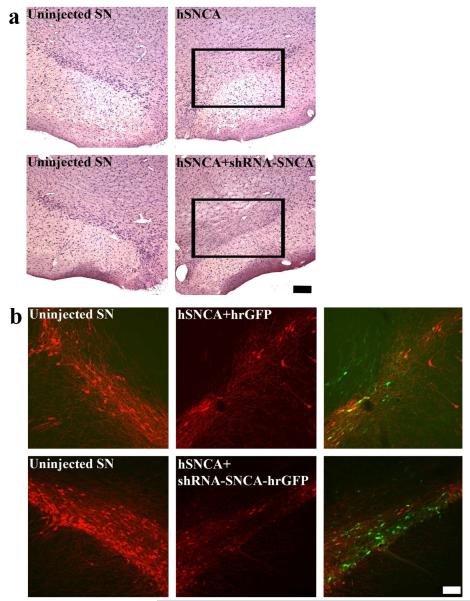
To further investigate the effect of hSNCA gene silencing in DA neurons, SN sections were examined at the 9wk time point. Sections were stained with hematoxylin and eosin (H&E) to examine neuron survival (Fig. 4a) and for TH-IR, using both a pan-TH antibody (Fig. 4b) and an antibody specific for Ser40 phosphorylated TH, an activated form of TH, since SNCA has been reported to affect the phosphorylation state of TH (Alerte et al., 2008).
Figure 4. hSNCA gene silencing does not protect against the loss of DA neurons induced by ectopic hSNCA expression.
Brains from rats that received co-injection of AAV-CBA-hSNCA with hrGFP (upper panels) or shRNA-SNCA-hrGFP (lower panels) into one SN for 9 weeks were examined for cell viability using H&E (a) and for TH-IR (b). In Figure 4A, panels show uninjected SN in the left column and injected SN in the right column. Note that both hSNCA expression and silencing resulted in reduced numbers of large neurons in SN. Size bar: 200 μm. Figure 4b shows uninjected SN in the left column and injected SN in the middle column. Merged images of TH-IR (red) and GFP fluorescence in the injected SN are shown in the right column. Note that both hSNCA expression and silencing resulted in loss of TH-IR neurons. Size bar, 100 μm.
As expected (Kirik et al., 2002), loss of large SN neurons (Fig. 4a) and reduced TH expression (Fig. 4b) were observed in hSNCA-injected SN. Co-injection of shRNA-SNCA and hSNCA did not protect against these reductions (Fig. 4). Neuron loss was also observed when sections were stained for phosphorylated TH-IR (data not shown). These observations suggest that although the shRNA-SNCA protected against the forelimb deficit induced by hSNCA, this was not due to protection against DA neuron loss induced by hSNCA.
2.4. shRNA-induced reduction in number of DA neurons in SN:
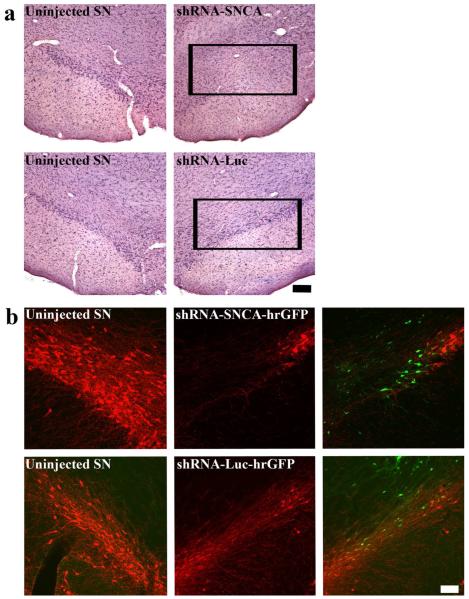
To determine whether silencing vectors alone show toxicity in SN DA neurons, brains from rats injected with AAV-shRNA-SNCA or AAV-shRNA-Luc alone were examined for cell survival by H&E (Fig. 5a) and TH phenotype (Fig. 5b) in the SN at 9wks. Injection of shRNA-SNCA alone resulted in a clear loss of neurons, whereas injection of shRNA-Luc (control) resulted in little to no loss of DA neurons (Fig. 5a). However, both silencing vectors resulted in a decreased number of TH-IR neurons in the injected SN, although this decrease was more pronounced with the shRNA-SNCA (Fig. 5b). Similar effects were observed for phosphorylated TH-IR (data not shown). These results suggest that both AAV vectors harboring shRNAs elicited undesirable effects on SN DA neurons in the vector-injected region.
Figure 5. Expression of shRNA-SNCA alone results in a greater loss of DA neurons than expression of shRNA-Luc alone.
Brains from rats that received stereotaxic injection of either AAV-CBA-shRNA-SNCA (upper panels) or AAV-CBA-shRNA-Luc (lower panels) into one SN for 9 weeks were examined for cell viability using H&E (a) and for TH-IR (b). Figure 5a shows brains that were examined for cell viability with H&E. Panels show uninjected SN in the left column and injected SN in the right column. Note the reduced number of large neurons in SN after injection of AAV-H1-shRNA-SNCA alone, but not after injection of AAV-H1-shRNA-Luc alone. Size bar: 200 μm. Figure 5b shows uninjected SN in the left column and injected SN in the middle column. Merged images of TH-IR (red) and GFP fluorescence in the injected SN are shown in the right column. Green fluorescence represents cells transduced with AAV-CMV-shRNA-SNCA (upper panel) or AAV-H1-shRNA-Luc (lower panel). Both silencing vectors led to loss of TH-IR neurons, but loss of TH-IR was more severe with the SNCA-specific silencing vector. Size bar, 100 μm.
2.5. hSNCA gene silencing protects ST-projecting DA neurons outside the main vector injection site:
To further examine the apparent conundrum of behavioral protection after SNCA gene silencing in the absence of DA neuron protection, effects on nigrostriatal-projecting DA neurons were determined using fluorogold (FG), a retrograde tracer. This approach permitted us to determine the number of DA neurons in SN and VTA that maintained axonal projections to a stereotaxically defined site in the ST at the end of the experiment. Further, these FG-labeled DA neurons could be counted without relying on TH-IR, which we had observed could be compromised by treatment. In order to examine whether nigrostriatal-projecting neurons were protected by hSNCA gene silencing, DA neurons with surviving ST projections were labeled with FG bilaterally at the end of treatment (~8wks after vector injection) and rats were allowed to survive for 5d to permit retrograde transport of FG to the SN. The number of FG-labeled DA neurons in both contralateral uninjected SN and ipsilateral SN injected with AAV-hSNCA, AAV-hSNCA & AAV-shRNA-SNCA-hrGFP or AAV-shRNA-SNCA-hrGFP was determined. At 9wks, all vectors induced a clear loss of FG positive neurons around the vector injection site (Fig. 6a). Together with the cell survival staining (Fig. 4a and 5a), these observations confirm that during the course of the experiment, many nigrostriatal-projecting DA neurons died. However, there were also DA neurons that survived and maintained projections to the ST.
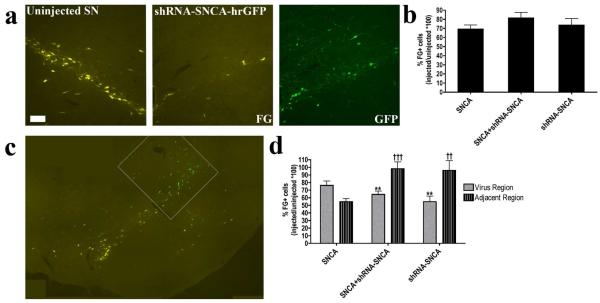
Figure 6. ST-projecting DA fibers in non-transduced SN regions are protected by hSNCA gene silencing.
Adult, male rats were stereotaxically injected into one SN with AAV vectors containing hSNCA and/or shRNA-SNCA plus hrGFP and DA neurons that maintained projections to the ST were bilaterally labeled with the retrograde tracer FG 5 days before euthanasia. Rats were euthanized 9 weeks after AAV injections and FG positive neurons in SN and VTA were counted. (a) AAV-CBA-hSNCA and/or AAV-H1-shRNA-SNCA reduced the number of neurons retrogradely labeled with FG in virus-transduced regions of the SN. Uninjected SN is shown in the left panel and AAV-injected SN is shown in the right two panels. FG is shown in yellow and GFP, representing shRNA-SNCA, is shown in green. Panels are representative of all treatment groups. Size bar, 100 μm. (b) FG positive DA neurons in the SN/VTA were counted. Total cell counts in SN/VTA, expressed as a ratio of injected SN/VTA / uninjected SN/VTA are shown in b. Data were analyzed using a Kruskal-Wallis one-way ANOVA, followed by Dunn’s post-hoc tests. In order to examine the effect of AAV injection on ST-projecting DA neurons more thoroughly, FG cell counts were divided into virus-transduced SN/VTA and adjacent un-transduced SN/VTA regions. c exemplifies the process by which cell counts were separated. The white outline represents the region of virus transduction in the injected SN. FG is shown in yellow and hrGFP, representing shRNA-SNCA is shown in green. Ratios of FG positive DA neurons in virus-transduced (gray bars) and adjacent un-transduced regions (striped bars) are shown in d. Mean±SEM; data were analyzed using two-way ANOVA followed by Bonferonni post-hoc tests. F2,44=11.48, p<0.0001. Asterisks show significant differences between virus-transduced and adjacent regions: **=p≤0.01. Significant differences compared to the adjacent region in hSNCA-injected rats: ††= p≤0.01, †††= p≤0.001.
Cell counts of FG-labeled DA neurons that maintained projections to the ST at the end of the experiment revealed a loss of neurons in the injected SN compared to the uninjected contralateral SN and VTA in all 3 treatment groups, which ranged from 20-30% (Fig. 6b). All groups had reduced FG positive DA neuron numbers in the injected side compared to the uninjected side (p≤0.05), suggesting overall loss of ST-projecting neurons due to expression of hSNCA and/or shRNA-SNCA. These counts were then divided into FG-labeled neurons located in the virus-transduced region as defined by transgene expression (hrGFP or hSNCA-IR) and those that lay outside this region (adjacent region, Fig. 6c). In the vector-injected region, the number of FG-labeled neurons was reduced compared to those in a comparable area of the contralateral SN in all treatment groups (Fig. 6d). hSNCA expression also caused a large loss of ST-projecting neurons outside this region suggesting either that low levels of hSNCA transduction were toxic to these neurons or that loss of hSNCA-transduced neurons also led to loss of non-transduced neighboring neurons (Fig. 6d). In contrast, no significant loss of FG-labeled ST projection neurons was observed outside the vector injection region in rats injected with the shRNA-SNCA, either alone or with hSNCA (Fig. 6d; ratio of injected/contralateral SN FG cell counts: hSNCA alone, 54.6%±4.2%, n=9; hSNCA and shRNA-SNCA, 98.3%±8.7%, p≤0.01, n=10; shRNA-SNCA alone, 95.9%±1.3%, p≤0.05, n=6). These data suggest that hSNCA gene silencing protected nigrostriatal DA neurons lateral to the main area of viral transduction. There also was no obvious toxic effect of the shRNA-SNCA in this region.
2.6. Amphetamine-induced DA release and basal ST DA levels are unaffected after injection of AAV-CBA-hSNCA and/or silencing vector:
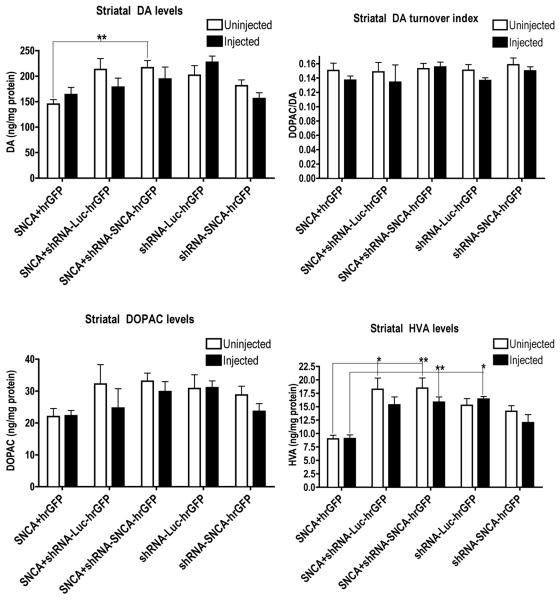
Two approaches were used to study possible effects of ectopic hSNCA expression and silencing on DA tone. As a histological measure of the amount of DA available for release in ST, rats were injected with D-amphetamine 2hrs before euthanasia and ST sections stained for c-fos-IR (Connor et al., 1999; Graybiel et al., 1990). The number of ST neurons with nuclear translocation of c-fos were counted on both sides of the brain. The percent of c-fos-IR nuclei was not significantly different (using Kruskal-Wallis one-way ANOVA followed by a Dunn’s post-hoc test) in rats co-injected with AAV-hSNCA and a control vector, either AAV-hrGFP or AAV-shRNA-Luc, so these data were combined into a single hSNCA-expressing group. No significant change in c-fos-IR nuclei between injected and uninjected ST was found in any treatment group suggesting that total amphetamine-releasable DA in ST was not changed by hSNCA expression or silencing (data not shown), despite the loss of nigrostriatal-projecting DA neurons. There was also no significant effect of unilateral SN injection of viral vectors on levels of DA and DA metabolites in injected ST compared with those in uninjected ST within each treatment group (Fig. 7).
Figure 7. Basal DA and DA metabolite levels in ST are not affected by expression of hSNCA and/or silencing vector 9 weeks after injection.
Rats were euthanized 9 weeks after injection and striatae were examined for DA and DA metabolite levels as a measure of SN DA neuron function. No effects on DA or DA metabolite levels in injected ST (closed bars) compared to uninjected ST (open bars) were observed, suggesting that hSNCA expression and silencing does not effect total basal levels of DA and DA turnover in the ST. Mean±SEM; comparison of injected and uninjected ST was analyzed using a Mann-Whitney t-test and comparison between treatments was analyzed using a Kruskal-Wallis one-way ANOVA, followed by Dunn’s post-hoc tests; asterisks denote significance as follows; *=p≤0.05, **=p≤0.01.
3. Discussion:
Development of techniques to specifically decrease SNCA toxicity holds promise for therapeutic applications in synucleinopathies, including PD. However, it remains a challenge to target aberrant SNCA expression in specific neurons in the brain. Here, we report the first study designed to silence ectopically expressed hSNCA in rat DA neurons. We investigated whether SNCA gene silencing using AAV-mediated delivery of a hSNCA-specific shRNA would protect DA neurons in rat SN from toxic effects of ectopic hSNCA expression. Confirming a previous report (Kirik et al., 2002), we observed progressive DA neuron loss due to hSNCA expression in rat SN. We found that hSNCA could be silenced to virtually undetectable levels in SN and ST by AAV-shRNA-SNCA, a vector that silences human, but not rat SNCA (Han et al., 2011; Sapru et al., 2006). Silencing also prevented formation of SNCA-IR intracellular inclusions that were observed in non-silenced DA neurons. Importantly, hSNCA silencing protected against the development of a hSNCA-induced forelimb deficit. However, hSNCA silencing did not fully protect against DA neuron loss. These paradoxical findings may partially be explained by effects on DA neurons outside the main virus-transduced region. However, effects of hSNCA expression and silencing appeared to also involve neurons other than DA neurons because levels of DA, DA metabolites, and releasable DA were not significantly different between injected and uninjected ST in any treatment group. One possibility is that retrograde and anterograde transport of the AAV vectors to other neurons in brainstem and/or ST contributed to this behavioral protection.
The observation that SNCA silencing protected against the hSNCA-induced forelimb deficit in rats suggests that SNCA gene silencing might also ameloriate movement deficits in PD patients. In this study, hSNCA and silencing vector were co-injected so that hSNCA accumulation in DA neurons was prevented rather than reversed. Demonstrating that hSNCA can be silenced in DA neurons by an AAV represents the first step towards developing a hSNCA gene silencing therapy. For further clinical development, the next step will be to determine whether hSNCA gene silencing can reverse SNCA-induced behavioral sequelae and cellular pathology after they have commenced. This goal remains for future experiments using a less toxic silencing vector (Han et al., 2011).
Our observations that chronic hSNCA expression in rat SN led to progressive nigrostriatal pathology and SNCA accumulation, accompanied by a motor deficit, are in agreement with those of Kirik et al who reported hSNCA-induced motor impairments in rat using three behavioral tests (Kirik et al., 2002). Rats that experienced over 50% of DA neuron loss exhibited motor impairments by apomorphine-induced rotational testing after 8wks and by the paw reaching test after 24wks of hSNCA expression. Kirik et al (2002) used the stepping test after administration of a TH inhibitor to assess ability of hSNCA-expressing rats to adjust forelimb movement. The cylinder test used in our study also assessed forelimb use by examining spontaneous vertical exploratory forelimb use. We observed a marked deficit in forelimb use after 8wks of hSNCA expression similar to the stepping test deficit reported by Kirik and co-workers (2002). Importantly, we also observed protection from this deficit in hSNCA-silenced rats. Behavioral protection has been reported after use of RNAi in models of other brain disorders including Alzheimer’s disease, spinocerebellar ataxia type 1, Huntington’s disease and prion disease (Boudreau and Davidson, 2010; Couto and High, 2010).
Although hSNCA silencing was found to offer behavioral protection, DA neurons in the main vector injection area still degenerated. Towne et al recently showed the reverse of this situation in an amyotrophic lateral sclerosis model where silencing mutant superoxide dismutase protected motor neurons and muscle, but did not affect behavior, survival, inflammation or ubiquitin deposition (Towne et al., 2011). Other reports have shown toxic shRNA effects (Boudreau et al., 2008; Boudreau et al., 2009; Castanotto et al., 2007; Grimm et al., 2006; McBride et al., 2008; Yi et al., 2005). McBride et al (2008) showed silencing huntingtin in vivo using two out of three shRNA target sequences also were toxic to ST neurons. Toxic shRNA effects are thought to result from saturation of endogenous RNAi machinery by high shRNA levels, leading to interference of miRNA processing (Boudreau et al., 2008; Boudreau et al., 2009; Castanotto et al., 2007; Diederichs et al., 2008; Grimm et al., 2006; McBride et al., 2008; Yi et al., 2005). Embedding the target sequence in a mir30 transcript reduces toxicity, suggesting that miRNA-embedded shRNAs are preferable for chronic in vivo applications (Boudreau et al., 2009; Castanotto et al., 2007; McBride et al., 2008). We have observed that the shRNA-SNCA used in the current study also induces cell death in the PC12 DA cell line, but that the same sequence embedded in a mir30 transcript does not (Han et al., 2011). When our data are considered with these previous reports, we speculate that use of a mir30-embedded hSNCA silencing vector may provide protection of both DA neurons and behavior in vivo. Another improvement might be use of a promoter that drives lower shRNA levels, such as the tRNA-val promoter (Rodriguez-Lebron et al., 2009).
SNCA is physiologically important in synaptic function (Bellani et al., 2010), which is a concern for developing SNCA gene silencing as a therapy. Total silencing of endogenous SNCA may be deleterious to DA neurons as evidenced by a recent study where knockdown of endogenous SNCA in rat SN led to DA neuron loss and a motor deficit (Gorbatyuk et al., 2010). In our study, endogenous SNCA was probably not silenced because the shRNA-SNCA target sequence is specific for hSNCA with 3 mis-matches to that of rat, and in another study, we demonstrated that our shRNA silences only hSNCA and not rat SNCA in vitro (Han et al., 2011). Although we cannot completely exclude the possibility that rat SNCA was not reduced by chronic expression of the shRNA, the observed DA neuron loss likely led to decreased rat SNCA, thus making it impossible to differentiate neuron loss from an shRNA off target effect as the cause of decreased rat SNCA expression. However, the possible translation of this approach must consider the caveat that total SNCA gene silencing may compromise DA neuronal function. This concern could be addressed by development of a regulatable or somewhat leaky gene silencing approach.
Specific neuronal types may be more sensitive to shRNA-mediated gene silencing. Previously, LV-mediated SNCA silencing with the same shRNA used here showed no obvious neuronal death in ST (Sapru et al., 2006). Nigral DA neurons could be more sensitive to insult with shRNA than ST neurons. However, our previous study investigated in vivo silencing after a shorter duration (2wks) than that examined here (4 or 9wks). A longer survival could reveal toxicity on ST neurons, although expression from LV peaks by about 2wks (Blomer et al., 1997), which is faster than that from AAV. Further, DA neuron vulnerability is suggested by studies in cultured human DA neurons where DA increases susceptibility to SNCA-induced toxicity (Xu et al., 2002). Another report also demonstrates differential sensitivities of different neuron types in that AAV-mediated shRNA delivery leads to neurodegeneration in the red nucleus, but not in the dorsal root ganglion (Ehlert et al.).
Although SNCA gene silencing resulted in behavioral protection, ST levels of DA and its metabolites and releasable DA were unaffected. It is possible that focal DA metabolism effects in ST were masked by assaying the whole ST or that DA metabolism was upregulated in surviving neurons to compensate for lost neurons. Alternatively, neurons other than, or in addition to, nigral DA neurons may have been affected by SNCA expression and silencing and may have contributed to the behavioral deficit and protection. Other studies have shown that tg SNCA over-expression models often present with behavioral deficits in absence of nigrostriatal pathology or vice versa (Chesselet, 2008; Fernagut and Chesselet, 2004). Specifically, tg mice expressing hSNCA under the Thy1 promoter exhibit a forelimb deficit with no apparent nigrostriatal deficit, except for increased sensitivity to MPTP. SNCA expression is widespread in that model so effects on systems other than the nigrostriatal DA system may underlie the forelimb deficit (Fleming et al., 2004). In contrast, it is unlikely the forelimb deficit observed in the current study resulted from effects on neurons outside the ventral mesencephalon or ST due to the nature of our model where AAV was injected directly into the SN and transgene expression was confined to neurons in or projecting to the injection site. The majority of transduced neurons appeared to be DA neurons in SN and VTA, although retrograde transport to some ST neurons was observed and interneurons in the SN also may have been transduced. Although many transduced nigral DA neurons were lost, some survived to the end of the experiment and there was no apparent VTA neuron loss. In addition, abundant SNCA-IR fibers remained in ST of non-silenced rats. The cellular substrate involved in the behavioral protection observed here is unclear, but could involve interneurons, VTA neurons, non-transduced neighboring SN or VTA neurons that were synaptically affected by transduced neurons or large projection neurons in ST that innervate SN, which may have been less sensitive to toxic insult than the SN DA neurons.
Gene silencing approaches hold therapeutic potential for a wide range of diseases. Due to association of SNCA mutations and gene multiplications with PD, SNCA gene silencing is a promising genetically-based therapy for both sporadic and familial PD, diseases that currently have no treatment. In addition to decreasing SNCA expression, allele-specific gene silencing can be used as we have shown for A53T SNCA (Sapru et al., 2006). Nevertheless, the findings reported here emphasize the complexities inherent in an in vivo gene silencing approach in brain. Despite positive, protective effects of SNCA gene silencing, we observed loss of DA neurons. The positive SNCA gene silencing effects, however, are intriguing and suggest that refinement of the shRNA vector design may lead to an effective gene silencing approach for PD. Studies are currently in progress to test different SNCA gene silencing vector designs with the aim of protecting DA neurons at both behavioral and cellular levels.
4. Experimental Procedure:
4.1. Generation of AAV shuttle plasmids:
The dual expression pAAV gene silencing vectors used, pAAV-H1-shRNA-SNCA-CMV-hrGFP and pAAV-H1-shRNA-Luc-CMV-hrGFP, harbor the human H1 promoter driving a shRNA targeting either nucleotides 288-309 of hSNCA (GenBank Accession No. L08850) or nucleotides 153-173 of firefly GL2 Luc (GenBank Accession No X65323), respectively, joined by an 11-base pair duplex loop and the cellular reporter hrGFP under control of the cytomegalovirus (CMV) promoter. These were derived from plasmids pBCSK-H1 and pBCSK-H1-shRNA-SNCA as described previously (Sapru et al., 2006). To generate pBCSK-H1-shRNA-Luc-CMV-hrGFP, forward and reverse hairpin oligos (5′-GATCCCCCGTACGCGGAATACTTCGATTCAAGAGATCGAAGTATTCCGCGTACGTTTTTGGAAA-3′ and 5′- CTAGTTTCCAAAAACGTACGCGGAATACTTCGATCTCTTGAATCGAAGTATTCCGCGTACGGGG-3′) were annealed together and ligated into the BglII and XbaI sites of pBCSK-H1. The H1-shRNA-Luc or H1-shRNA-SNCA was PCR amplified from pBCSK-H1-shRNA-Luc/SNCA using the following primers: 5′-ATACGCGTAAGCTTGATATCGAATTCGAACGCTGAC-3′ (forward) and 5′-TTACTATTAATAACTAGCTCCTGGCGGCCGCTCTAGTTTCCAAAAAG-3′. This fragment was purified and digested with MluI and AseI. pAAV-MCS-hrGFP was cut into two fragments with MluI and ClaI. The smaller fragment (containing CMV-hrGFP) was further digested with AseI to allow for increased likelihood of H1-shRNA-Luc/SNCA fragment insertion. All three of these fragments were ligated to produce pAAV-H1-shRNA-Luc-CMV-hrGFP or pAAV-H1-shRNA-SNCA-CMV-hrGFP.
pAAV-CBA-SNCA was generated by first inserting the CBA (chicken β-actin) promoter into pAAV-MCS (Stratagene, San Diego, CA) digested with SnaBI and EcoRI. The SNCA fragment from pcDNA3-SNCA (a generous gift from Dr. Yong-Jian Liu) was then ligated into the XhoI and HindIII sites of pAAV-MCS-CBA.
Before virus preparation, plasmids were sequenced and in vitro transgene expression, including hSNCA gene silencing, was confirmed (Han et al., 2011).
4.2. Virus packaging:
All vectors were packaged in the Children’s Memorial Viral Vector Core as AAV2 following the protocol of Zufferey et al (Zufferey et al., 1998; Zufferey and Trono, 2002) with modifications as described previously (Han et al., 2010). Briefly, 293T cells were transfected with shuttle plasmid and the appropriate packaging plasmids. At three days post-transfection, cells underwent three freeze-thaw cycles to induce lysis for collection of virus. Cellular debris was removed by centrifugation and the supernatant treated with octyl-β-D-glucopyranoside and benzonase. After centrifugation, the viral lysate was applied to a 15-60% iodixanol discontinuous gradient. The 40% layer was further purified either by FPLC affinity chromatography on a heparin sulfate column or by using a Mustang Q ion exchange membrane. Virus was then concentrated using a Centriplus 100,000 MS cut off membrane and stored in phosphate buffered saline (PBS), pH 7.4 containing 5% sorbitol and 0.001% PF-68. Viral titers were determined by real-time PCR (rtPCR) and the purity of preparations examined by electron microscopy. Viral titers were: AAV-CBA-hSNCA – 1.5×1012 vector genomes (vg)/ml, AAV-hrGFP – 2×1012 vg/ml, AAV-SNCA shRNA-hrGFP – 2.6×1012 vg/ml, AAV-Luc shRNA-hrGFP – 1.2×1012 vg/ml.
4.3. Animals:
Adult male Sprague Dawley rats weighing 175-200g were purchased from Harlan Laboratories (Indianapolis, IN) and housed in the Children’s Memorial Research Center animal facility on a 12 hr light/dark cycle with food and water available ad libitum. Animal care and use procedures were conducted in accordance with NIH, USDA and institutional guidelines.
4.4. Treatment groups and sample sizes:
Rats were generated for (1) morphological examination, (2) biochemical evaluation, and (3) examination of nigrostriatal projections. Table 1 shows the number of rats used for each experimental group. Overall, 9 rats died during or shortly after surgery and additional rats were excluded from data analyses if the injection site was not precisely in the SN. (1) For morphological examination, two survivals after viral vector injection were examined. Treatment groups included rats that were injected with AAV-CMV-hrGFP+AAV-CBA-hSNCA (4wk, n=10; 9wk, n=7), AAV-H1-shRNA-Luc-hrGFP-CMV-hrGFP+AAV-CBA-hSNCA (4wk, n=4; 9wk, n=5), AAV-H1-shRNA-SNCA-CMV-hrGFP+AAV-CBA-hSNCA (4wk, n=3; 9wk, n=8), AAV-H1-shRNA-Luc-CMV-hrGFP (9wk, n=4), or AAV-H1-shRNA-SNCA-CMV-hrGFP (9wk, n=9) and euthanized at 4 or 9 weeks post-injection. (2) Treatment groups used to examine ST DA and DA metabolite levels included rats that were injected with AAV-CMV-hrGFP+AAV-CBA-hSNCA (n=5), AAV-H1-shRNA-Luc-CMV-hrGFP+AAV-CBA-hSNCA (n=3), AAV-H1-shRNA-SNCA-CMV-hrGFP+AAV-CBA-hSNCA (n=6), AAV-H1-shRNA-Luc-CMV-hrGFP (n=4), or AAV-H1-shRNA-SNCA-CMV-hrGFP (n=6) and euthanized at 9 weeks after injection. (3) For examination of nigrostriatal-projecting DA neurons, rats were injected with AAV-CBA-hSNCA (2ul; n=9), AAV-CBA-hSNCA+ AAV-H1-shRNA-SNCA-CMV-hrGFP (n=10), or AAV-H1-shRNA-SNCA-CMV-hrGFP (n=6) and received bilateral injection of FG into the ST 5 days before perfusion at 9 weeks after injection.
Table 1.
Experimental sample sizes
| Parameters for analysis: | ||||
|---|---|---|---|---|
| AAV Treatment Group |
Morphology | Forelimb Use – 9 wk |
Nigrostriatal- projecting DA neurons – 9 wk |
DA/DA metabolites – 9 wk |
| hSNCA | N/A | 16 | 9 | N/A |
| hSNCA+hrGFP | 4wk-n=10 9wk-n=7 |
7 | n.d. | 5 |
|
hSNCA+shRNA-
Luc |
4wk-n=4 9wk-n=5 |
4 | n.d. | 3 |
|
hSNCA+shRNA-
SNCA |
4wk-n=3 9wk-n=8 |
28 | 10 | 6 |
| shRNA-Luc | 9wk-n=4 | 5 | n.d. | 4 |
| shRNA-SNCA | 9wk-n=9 | 21 | 6 | 6 |
n.d., not determined
4.5. Stereotaxic surgery and post-operative animal care:
The skin overlying the skull of isoflurane-anesthetized rats was shaven. After placing rats in a Stoelting stereotaxic apparatus equipped with a Stoelting quintessential stereotaxic injector holding a 10μl Hamilton syringe with a 26 gauge needle, a hole was drilled in the skull over the appropriate injection site. The needle was slowly inserted at a speed of ~1 mm/min and then allowed to sit for 2 min before injection. 1.5 μl of each virus was injected unilaterally into the SN at a rate of 0.5 μl/min. The stereotaxic coordinates for SN injection were 5.5 mm posterior, −1.9 mm lateral and 7.4 mm ventral from Bregma. A group of rats also received bilateral stereotaxic injection of the retrograde tracer FG 5d before euthanasia. 0.3 μl of 2% FG was injected into both ST at a rate of 0.05 μl/min at stereotaxic coordinates of 1.0 mm anterior, +/−3.0 mm lateral and 5.0 mm ventral from Bregma. After injection(s), injected fluid was allowed to settle for 5 minutes to prevent diffusion up the needle track. Then the syringe was withdrawn at ~1 mm/minute, the drill hole in the skull was plugged with gel foam to control bleeding and 0.1 ml of a 1:1 dilution of the analgesic Marcaine was spread around the surgical site. The skin was then sutured and treated with topical antibiotic ointment. Rats were monitored until they recovered from anesthesia and then were returned to the vivarium. 24 hours following surgery, rats were moved to a clean cage and the bedding autoclaved.
4.6. Non-drug-induced forelimb preference:
For behavioral testing, animals were kept in a quiet, dark room. Rats were placed inside a glass cylinder, partially surrounded by mirrors and their activity was recorded on videotape under red light for a 10-min period. A technician blinded to treatment group then recorded which paw was used for landing on each of 25 rearings for each rat, to determine forelimb use preference as previously published (Schallert et al., 2000).
4.7. Tissue preparation:
At either 4 or 9wks after vector injection, rats were injected with 2.5 mg/kg free base weight D-amphetamine hemisulfate salt ~2hrs before perfusion to examine amphetamine-induced ST nuclear c-fos translocation as a measure of DA neuron function in rats used for morphological assessment (Connor et al., 1999; Graybiel et al., 1990). Rats were weighed and anesthetized with sodium pentobarbital (75 mg/kg). For histological purposes, rats were perfused with 0.9% saline containing 0.002% Na nitrite, followed by 4% phosphate buffered paraformaldehyde (pH=7.4) and brains were removed and cut coronally into tissue blocks containing either SN or ST. These tissue blocks were post-fixed overnight in 5% sucrose-4% paraformaldehyde and then cryoprotected by an increasing gradient of sucrose concentrations (10%-30%) in 0.1 M PBS. Six sets of serial frozen coronal tissue sections were made at 40 μm using a sliding microtome (Leica SM2000 R) and placed in cryoprotectant solution until used. For measurements of DA and DA metabolites, rats were perfused with 0.9% saline only and brains were removed and cut coronally into tissue blocks containing either SN or ST. ST were placed in 0.4 N perchloric acid (1:20 ratio by weight) for 5-10 min on wet ice and then stored at −80°C until use.
4.8. Immunocytochemistry:
Free-floating tissue sections were rinsed of cryoprotectant using 0.1M tris-buffered saline (TBS). For immunoperoxidase staining, sections were also incubated in 0.3% H2O2 for 15min to inhibit endogenous peroxidase activity. Sections were incubated for 1 hour in normal goat serum (NGS) to block nonspecific antibody binding. Directly after blocking, sections were incubated overnight at RT in primary antibody (rabbit α pan TH, Millipore, 1:500, Billerica, MA; rabbit α P-Ser40 TH, 1:500, Millipore; mouse α hSNCA, 1:50, Invitrogen, Carlsbad, CA; rabbit α c-fos, 1:1000, Santa Cruz Biotechnology, Santa Cruz, CA). After rinsing with 0.1% Triton-TX in 0.1M TBS, sections were incubated in secondary antibody (Cy3- or Cy2-conjugated goat α mouse, 1:100, Jackson Immunoresearch, West Grove, PA; Cy3-conjugated goat α rabbit, 1:200, Jackson Immunoresearch; biotinylated-goat α rabbit, 1:500, Vector laboratories, Burlingame, CA) for 2.5 hrs at room temperature. For immunofluorescence, sections were mounted on slides, air dried overnight and coverslipped with Fluorosave (Calbiochem, La Jolla, CA). For immunoperoxidase staining, sections were incubated in an avidin-biotin complex solution (Vectastain kit, Vector laboratories) and immunoreactivity (IR) was visualized through development with a diaminobenzedine/nickel sulfate solution. Sections were then mounted, dehydrated and coverslipped using Permount. Immunostaining was evaluated using a Leica DMR upright microscope equipped for epifluorescence microscopy.
4.9. H&E staining:
Tissue sections were rinsed of cryoprotectant, mounted on gel-subbed slides and dried for 48 hrs in a vacuum desiccator. Sections were dehydrated using 95% and 100% ethanol (2 minutes each) and lipids were removed using xylene (5 min). Sections were then rehydrated through a gradient of ethanols and tap water, followed by 5 min in Harris hematoxylin to stain the chromatin. After a tap water rinse, sections were dipped in 1% lithium carbonate for 30 seconds. Sections were rinsed in running tap water for 10 minutes, rinsed in 70% ethanol for 30 seconds and dipped in eosin twice. Sections were then dehydrated and coverslipped with Permount.
4.10. FG cell counts:
FG positive cells in the SN and VTA were quantified using Neurolucida software. Only large FG-containing neurons (greater than ~15 μm in diameter) were counted to ensure that only DA neurons, not dying neurons or microglia, were counted. Five sections centered around the injection site (~240 μm apart) were counted for each brain. Separate cell counts were made in virus-transduced and adjacent SN/VTA. The equivalent area on the uninjected side was determined by flipping the virus region contour and placing it in the same position on the uninjected SN/VTA.
4.11. c-fos cell counts:
Nuclei IR for c-fos in injected and uninjected ST were counted at one level of the ST (at about Bregma) using Neurolucida software. Five 400 μm × 400 μm boxes were placed on each ST and c-fos IR nuclei were counted within each. The boxes included a central, dorsal, ventral, lateral and medial box.
4.12. Imaging:
Images of SNCA-IR, TH-IR, GFP fluorescence, FG fluorescence, and H&E staining were taken using a Retiga 4000R digital camera on a Leica DMR upright microscope. Adobe Photoshop CS2 was used to generate composites.
4.13. Measurement of DA and DA metabolites:
ST samples incubated in 0.4M perchloric acid were sonicated and centrifuged for 15 min at 14,000 rpm at 4°C. Supernatant was analyzed for DA, dihydroxyphenylacetic acid (DOPAC) and homovanillic acid (HVA) using an HPLC system coupled to electrochemical detection (Coularray, ESA Inc., Chelmsford, MA). The Lowry method was used to determine protein content.
4.14. Statistical analysis:
Data are expressed as mean ± SEM and were analyzed using Prism™ software. Forelimb behavioral data were analyzed using a Kruskal-Wallis one-way analysis of variance (ANOVA) followed by Dunn’s post-hoc tests. Comparison between injected and uninjected side data within a single treatment group, specifically for examination of nigrostriatal-projecting DA neurons and basal DA and DA metabolite levels, was analyzed using a Mann-Whitney t-test. Ratios of FG counts, and comparison of FG cell numbers in injected and uninjected SN for each region (i.e. total, virus or adjacent) were analyzed using a two-way ANOVA followed by Bonferroni post-hoc tests. Statistically significant differences were set at p≤0.05.
Acknowledgements:
This study was supported by the Department of Defense Neurotoxicology Program (NO06079001) and NIH grants (NS31957 and NS054989 to MCB and T32 NS041234 to CEK), the Harry F and Elaine M Chaddick Foundation and the Medical Research Institute Council of Children’s Memorial Hospital. Support from the Chicago Biomedical Consortium and the Illinois Excellence in Academic Medicine to the CMRC Viral Vector Core is acknowledged. The technical assistance of Jianping Xie, Xue Song Wang, David George and Brian Corstange (Children’s Memorial Research Center) is appreciated. The authors thank James Surmeier (Northwestern University) for his insightful comments on the manuscript. MCB and MKS hold a European patent on this technology and have a pending U.S. patent application.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Alerte TN, Akinfolarin AA, Friedrich EE, Mader SA, Hong CS, Perez RG. Alpha-synuclein aggregation alters tyrosine hydroxylase phosphorylation and immunoreactivity: lessons from viral transduction of knockout mice. Neurosci Lett. 2008;435:24–9. doi: 10.1016/j.neulet.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellani S, Sousa VL, Ronzitti G, Valtorta F, Meldolesi J, Chieregatti E. The regulation of synaptic function by alpha-synuclein. Commun Integr Biol. 2010;3:106–9. doi: 10.4161/cib.3.2.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund A, Kirik D, Rosenblad C, Georgievska B, Lundberg C, Mandel RJ. Towards a neuroprotective gene therapy for Parkinson’s disease: use of adenovirus, AAV and lentivirus vectors for gene transfer of GDNF to the nigrostriatal system in the rat Parkinson model. Brain Res. 2000;886:82–98. doi: 10.1016/s0006-8993(00)02915-2. [DOI] [PubMed] [Google Scholar]
- Blomer U, Naldini L, Kafri T, Trono D, Verma IM, Gage FH. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J Virol. 1997;71:6641–9. doi: 10.1128/jvi.71.9.6641-6649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau RL, Monteys AM, Davidson BL. Minimizing variables among hairpin-based RNAi vectors reveals the potency of shRNAs. Rna. 2008;14:1834–44. doi: 10.1261/rna.1062908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau RL, Martins I, Davidson BL. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol Ther. 2009;17:169–75. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau RL, Davidson BL. RNAi therapeutics for CNS disorders. Brain Res. 2010;1338:112–21. doi: 10.1016/j.brainres.2010.03.038. [DOI] [PubMed] [Google Scholar]
- Cannon JR, Greenamyre JT. Neurotoxic in vivo models of Parkinson’s disease recent advances. Prog Brain Res. 2010;184:17–33. doi: 10.1016/S0079-6123(10)84002-6. [DOI] [PubMed] [Google Scholar]
- Castanotto D, Sakurai K, Lingeman R, Li H, Shively L, Aagaard L, Soifer H, Gatignol A, Riggs A, Rossi JJ. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–64. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesselet MF. In vivo alpha-synuclein overexpression in rodents: a useful model of Parkinson’s disease? Exp Neurol. 2008;209:22–7. doi: 10.1016/j.expneurol.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi-Lundberg DL, Lin Q, Chang YN, Chiang YL, Hay CM, Mohajeri H, Davidson BL, Bohn MC. Dopaminergic neurons protected from degeneration by GDNF gene therapy. Science. 1997;275:838–41. doi: 10.1126/science.275.5301.838. [DOI] [PubMed] [Google Scholar]
- Connor B, Kozlowski DA, Schallert T, Tillerson JL, Davidson BL, Bohn MC. Differential effects of glial cell line-derived neurotrophic factor (GDNF) in the striatum and substantia nigra of the aged Parkinsonian rat. Gene Ther. 1999;6:1936–51. doi: 10.1038/sj.gt.3301033. [DOI] [PubMed] [Google Scholar]
- Couto LB, High KA. Viral vector-mediated RNA interference. Curr Opin Pharmacol. 2010;10:534–42. doi: 10.1016/j.coph.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Diederichs S, Jung S, Rothenberg SM, Smolen GA, Mlody BG, Haber DA. Coexpression of Argonaute-2 enhances RNA interference toward perfect match binding sites. Proc Natl Acad Sci U S A. 2008;105:9284–9. doi: 10.1073/pnas.0800803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem. 2010;285:12726–34. doi: 10.1074/jbc.M109.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlert EM, Eggers R, Niclou SP, Verhaagen J. Cellular toxicity following application of adeno-associated viral vector-mediated RNA interference in the nervous system. BMC Neurosci. 11:20. doi: 10.1186/1471-2202-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404:394–8. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- Fernagut PO, Chesselet MF. Alpha-synuclein and transgenic mouse models. Neurobiol Dis. 2004;17:123–30. doi: 10.1016/j.nbd.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fleming SM, Salcedo J, Fernagut PO, Rockenstein E, Masliah E, Levine MS, Chesselet MF. Early and progressive sensorimotor anomalies in mice overexpressing wild-type human alpha-synuclein. J Neurosci. 2004;24:9434–40. doi: 10.1523/JNEUROSCI.3080-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk OS, Li S, Nash K, Gorbatyuk M, Lewin AS, Sullivan LF, Mandel RJ, Chen W, Meyers C, Manfredsson FP, Muzyczka N. In Vivo RNAi-Mediated alpha-Synuclein Silencing Induces Nigrostriatal Degeneration. Mol Ther. 2010 doi: 10.1038/mt.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybiel AM, Moratalla R, Robertson HA. Amphetamine and cocaine induce drug-specific activation of the c-fos gene in striosome-matrix compartments and limbic subdivisions of the striatum. Proc Natl Acad Sci U S A. 1990;87:6912–6. doi: 10.1073/pnas.87.17.6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–41. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Han Y, Chang QA, Virag T, West NC, George D, Castro MG, Bohn MC. Lack of humoral immune response to the tetracycline (Tet) activator in rats injected intracranially with Tet-off rAAV vectors. Gene Ther. 2010;17:616–25. doi: 10.1038/gt.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Khodr CE, Sapru M, Pedapati J, Bohn MC. A microRNA embedded AAV alpha-synuclein gene silencing vector for dopaminergic neurons. Brain Res. 2011;1386:15–24. doi: 10.1016/j.brainres.2011.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashita-Kinoh H, Yamada M, Yokota T, Mizuno Y, Mochizuki H. Down-regulation of alpha-synuclein expression can rescue dopaminergic cells from cell death in the substantia nigra of Parkinson’s disease rat model. Biochem Biophys Res Commun. 2006;341:1088–95. doi: 10.1016/j.bbrc.2006.01.057. [DOI] [PubMed] [Google Scholar]
- Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci U S A. 2009;106:13052–7. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns CM, Gash DM. GDNF protects nigral dopamine neurons against 6-hydroxydopamine in vivo. Brain Res. 1995;672:104–11. doi: 10.1016/0006-8993(94)01366-p. [DOI] [PubMed] [Google Scholar]
- Kirik D, Rosenblad C, Burger C, Lundberg C, Johansen TE, Muzyczka N, Mandel RJ, Bjorklund A. Parkinson-like neurodegeneration induced by targeted overexpression of alpha-synuclein in the nigrostriatal system. J Neurosci. 2002;22:2780–91. doi: 10.1523/JNEUROSCI.22-07-02780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik D, Annett LE, Burger C, Muzyczka N, Mandel RJ, Bjorklund A. Nigrostriatal alpha-synucleinopathy induced by viral vector-mediated overexpression of human alpha-synuclein: a new primate model of Parkinson’s disease. Proc Natl Acad Sci U S A. 2003;100:2884–9. doi: 10.1073/pnas.0536383100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VM, Trojanowski JQ. Mechanisms of Parkinson’s disease linked to pathological alpha-synuclein: new targets for drug discovery. Neuron. 2006;52:33–8. doi: 10.1016/j.neuron.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Lewis J, Melrose H, Bumcrot D, Hope A, Zehr C, Lincoln S, Braithwaite A, He Z, Ogholikhan S, Hinkle K, Kent C, Toudjarska I, Charisse K, Braich R, Pandey RK, Heckman M, Maraganore DM, Crook J, Farrer MJ. In vivo silencing of alpha-synuclein using naked siRNA. Mol Neurodegener. 2008;3:19. doi: 10.1186/1750-1326-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Bianco C, Ridet JL, Schneider BL, Deglon N, Aebischer P. alpha - Synucleinopathy and selective dopaminergic neuron loss in a rat lentiviral-based model of Parkinson’s disease. Proc Natl Acad Sci U S A. 2002;99:10813–8. doi: 10.1073/pnas.152339799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Bianco C, Deglon N, Pralong W, Aebischer P. Lentiviral nigral delivery of GDNF does not prevent neurodegeneration in a genetic rat model of Parkinson’s disease. Neurobiol Dis. 2004;17:283–9. doi: 10.1016/j.nbd.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Mandel RJ, Spratt SK, Snyder RO, Leff SE. Midbrain injection of recombinant adeno-associated virus encoding rat glial cell line-derived neurotrophic factor protects nigral neurons in a progressive 6-hydroxydopamine-induced degeneration model of Parkinson’s disease in rats. Proc Natl Acad Sci U S A. 1997;94:14083–8. doi: 10.1073/pnas.94.25.14083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraganore DM, de Andrade M, Elbaz A, Farrer MJ, Ioannidis JP, Kruger R, Rocca WA, Schneider NK, Lesnick TG, Lincoln SJ, Hulihan MM, Aasly JO, Ashizawa T, Chartier-Harlin MC, Checkoway H, Ferrarese C, Hadjigeorgiou G, Hattori N, Kawakami H, Lambert JC, Lynch T, Mellick GD, Papapetropoulos S, Parsian A, Quattrone A, Riess O, Tan EK, Van Broeckhoven C. Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson disease. Jama. 2006;296:661–70. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, Carter BJ, Davidson BL. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A. 2008;105:5868–73. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack AL, Mak SK, Henderson JM, Bumcrot D, Farrer MJ, Di Monte DA. Alpha-synuclein suppression by targeted small interfering RNA in the primate substantia nigra. PLoS One. 5:e12122. doi: 10.1371/journal.pone.0012122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Lebron E, Gouvion CM, Moore SA, Davidson BL, Paulson HL. Allele-specific RNAi mitigates phenotypic progression in a transgenic model of Alzheimer’s disease. Mol Ther. 2009;17:1563–1573. doi: 10.1038/mt.2009.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapru MK, Yates JW, Hogan S, Jiang L, Halter J, Bohn MC. Silencing of human alpha-synuclein in vitro and in rat brain using lentiviral-mediated RNAi. Exp Neurol. 2006;198:382–90. doi: 10.1016/j.expneurol.2005.12.024. [DOI] [PubMed] [Google Scholar]
- Sauer H, Rosenblad C, Bjorklund A. Glial cell line-derived neurotrophic factor but not transforming growth factor beta 3 prevents delayed degeneration of nigral dopaminergic neurons following striatal 6-hydroxydopamine lesion. Proc Natl Acad Sci U S A. 1995;92:8935–9. doi: 10.1073/pnas.92.19.8935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallert T, Fleming SM, Leasure JL, Tillerson JL, Bland ST. CNS plasticity and assessment of forelimb sensorimotor outcome in unilateral rat models of stroke, cortical ablation, parkinsonism and spinal cord injury. Neuropharmacology. 2000;39:777–87. doi: 10.1016/s0028-3908(00)00005-8. [DOI] [PubMed] [Google Scholar]
- Scherr M, Eder M. Gene silencing by small regulatory RNAs in mammalian cells. Cell Cycle. 2007;6:444–9. doi: 10.4161/cc.6.4.3807. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–40. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- Tomac A, Lindqvist E, Lin LF, Ogren SO, Young D, Hoffer BJ, Olson L. Protection and repair of the nigrostriatal dopaminergic system by GDNF in vivo. Nature. 1995;373:335–9. doi: 10.1038/373335a0. [DOI] [PubMed] [Google Scholar]
- Towne C, Setola V, Schneider BL, Aebischer P. Neuroprotection by Gene Therapy Targeting Mutant SOD1 in Individual Pools of Motor Neurons Does not Translate Into Therapeutic Benefit in fALS Mice. Mol Ther. 2011;19:274–83. doi: 10.1038/mt.2010.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Kao SY, Lee FJ, Song W, Jin LW, Yankner BA. Dopamine-dependent neurotoxicity of alpha-synuclein: a mechanism for selective neurodegeneration in Parkinson disease. Nat Med. 2002;8:600–6. doi: 10.1038/nm0602-600. [DOI] [PubMed] [Google Scholar]
- Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. Rna. 2005;11:220–6. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Sierks MR. Intracellular targeting and clearance of oligomeric alpha-synuclein alleviates toxicity in mammalian cells. Neurosci Lett. 2009;459:16–8. doi: 10.1016/j.neulet.2009.04.046. [DOI] [PubMed] [Google Scholar]
- Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–80. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Trono D. Current Protocols in Human Genetics. John Wiley & Sons, Inc; New York: 2002. Production of high-titer lentiviral vectors. [Google Scholar]