Abstract
Post translational modifications, in particular glycosylation, represent critical structural attributes that govern both the pharmacodynamic and pharmacokinetic properties of therapeutic glycoproteins. To guarantee safety and efficacy of recombinant therapeutics, characterization of glycosylation present is a regulatory requirement. In the current paper, we applied a multidimensional strategy comprising a shallow anion exchange gradient in the first dimension, followed by analysis using the recently introduced 1.7 μm HILIC phase in the second dimension for the comprehensive separation of complex N-glycans present on the European Biological Reference Preparation (BRP) 3 erythropoietin standard. Tetra-antennary glycans with multiple sialic acids and poly-N-acetyl lactosamine extensions were the most abundant oligosaccharides present on the molecule. Site-specific glycan analysis was performed to examine microheterogeneity. Tetra-antennary glycans with up to four sialic acids and up to five poly-N-acetyl lactosamine extensions were observed at asparagine 24 and 83, while bi-antennary glycans were the major structures at asparagine 38. The combined AEC × UPLC HILIC allows for the rapid and comprehensive analysis of complex N-glycosylation present on therapeutic glycoproteins such as BRP3 erythropoietin.
Keywords: Erythropoietin, biopharmaceutical, oligosaccharides, glycan analysis, glycomics, proteomics, site specific glycosylation, ultra performance liquid chromatography, hydrophilic interaction liquid chromatography
Introduction
Glycosylation is a critical structural attribute that can affect both the pharmacodynamic and pharmacokinetic behaviour of therapeutic glycoproteins.1,2 Detailed characterization of the glycans covalently attached to the therapeutic protein is therefore a regulatory requirement.3 Moreover, recently introduced guidelines regarding acceptance criteria for biosimilar products require the manufacturer to demonstrate both comparability and similarity of quality, safety and efficacy to the innovator product being replicated.4 These requirements have created an increased demand for high performance analytical technologies capable of comprehensively characterizing protein glycosylation and thereby generating data of appropriate quality to allow the assessment of similarity.
Due to the inherent complexity of their branched structure, the full characterization of oligosaccharides poses a considerable analytical challenge. Separation techniques such as high performance liquid chromatography or capillary electrophoresis are commonly employed for the profiling and quantitation of glycans following their release from the parent protein and derivatization using a suitable fluorophore.5 Hydrophilic interaction (HILIC) liquid chromatography offers the advantage of predictable selectivity with retention increasing with increasing glycan size and associated hydrophilicity. Furthermore, each monosaccharide present in a given linkage contributes incrementally to the overall retention of the oligosaccharide on the HILIC phase. Currently available HILIC phases generally require excessive chromatographic run times and offer only moderate chromatographic efficiency.6 Porous graphitized carbon has also been investigated as an alternative stationary phase for liquid phase glycan separation due to its unique and attractive selectivity7, but issues of recovery especially for highly sialylated glycans can arise.8,9 Capillary electrophoretic analysis of intact glycoproteins and released glycans offers an alternative and complementary approach to chromatographic based techniques.10,11,12 Mass spectrometry using either matrix assisted laser desorption ionization or electrospray ionization has also been routinely employed for the characterization of oligosaccharides.5 A limitation of mass spectrometry based approaches is its inability to distinguish between isobaric monosaccharides and oligosaccharide isomers. Another concern when using mass spectrometry for oligosaccharide analysis is problems with quantitation due to ion suppression by contaminants present in the sample or competitive ionisation of different oligosaccharides within the ion source.13 Analyte derivatization, for example using permethylation, accompanied by multistage fragmentation may be necessary to characterize the oligosaccharides under study.14
The introduction of a 1.7 μm HILIC phase for ultra performance liquid chromatography (UPLC) represents a significant advance for glycomic characterization of therapeutic proteins, allowing for higher efficiency oligosaccharide separations and reduced analysis times.15,16 In the current study, the new 1.7 μm HILIC phase for UPLC is applied in the second dimension following fractionation using weak anion exchange chromatography with a shallow salt gradient to maximise peak capacity in the first dimension for the comprehensive characterization of the N-glycans present on the European Pharmacopeia Biological Reference Preparation (BRP) 3 erythropoietin standard, a mix of recombinant epoetin alpha and epoetin beta expressed in CHO cells.17 Erythropoietin contains three N-glycosylation sites at asparagine 24, 38 and 83 and a single O-glycosylation site at serine 126; the glycans account for up to 40% of the total molecular mass.18 In this study, N-glycosylation analysis of the BRP 3 reference standard revealed that tetra-antennary glycans, with multiple sialic acids and various numbers of poly-N-acetyl lactosamine extensions, represent the largest proportion of oligosaccharides present. O-glycosylation was not examined due to previous reports describing the minor biological significance of the O-glycan attached to EPO.19 Site-specific glycosylation analysis of the N-glycosylation sites was also performed using a combination of digestion with Lys-C and Glu-C, peptide mapping, glycan structural identification and site identification using LC/MS. The high performance HILIC column, in combination with a shallow gradient anion exchange chromatographic first dimension step, allowed the identification of the largest number of poly-N-acetyl lactosamine repeats present at asparagine 24 and 83 reported to date. An awareness of these large poly-N-acetyl lactosamine extensions and their relative amounts is important in assessing potential biological significance. These elongated structures may confer orthogonal properties to the drug allowing it to participate in secondary interactions such as high affinity interaction with galectins or may result in more rapid clearance.20,21
Experimental
Chemicals and reagents
Solvents, LC-MS Optima grade, were obtained from Thermo Fisher Scientific (Dublin, Ireland). All chemicals were from Sigma Aldrich (Dublin, Ireland) and were of the highest available quality. The European Pharmacopeia BRP 3 erythropoietin reference standard was purchased from LGC Standards, (Teddington, United Kingdom).
Enzymatic N-glycan release and labelling
N-glycans were enzymatically released from reduced and alkylated BRP 3 erythropoietin via in solution PNGase F digestion (Prozyme, Hayward, CA, USA). Glycans were fluorescently labelled with 2-aminobenzamide via reductive amination with sodium cyanoborohydride in 30% v/v acetic acid in DMSO at 65°C for two hours. The labelling reaction was quenched with the addition of 90 μL of water plus 900 μL of acetonitrile to the reaction vial with subsequent excess fluorophore removal by PhyNexus normal phase PhyTips (San Jose, CA, USA), using an adaptation of the method of Olajos et al.22 Exoglycosidase digestions were performed according to Royle et al.23N-glycan nomenclature and symbolic representations used throughout have been previously described by Harvey et al.24
UPLC-fluorescence N-glycan profiling
Fluorescently labelled N-glycans were separated on an Acquity™ UPLC instrument consisting of a binary solvent manager, sample manager and fluorescence detector under the control of Empower 2 chromatography workstation software, (Waters, Milford, MA, USA. Separations were performed using Waters BEH Glycan chromatography column, 100 × 2.1 mm i.d., 1.7 μm BEH particles, with a linear gradient of 70-53% acetonitrile at 0.56 ml/min in 16.5 minutes. An injection volume of 20 μL sample prepared in 80% v/v acetonitrile was employed throughout. Samples were maintained at 5°C prior to injection, and the separation temperature was 40°C. The fluorescence detection wavelengths were λex = 330 nm and λem = 420 nm with a data collection rate of 20 Hz. Integration parameters were as follows: signal to noise ratio >10 as a quantitation limit with a minimum area count of 10,000.
Anion exchange chromatography (AEC)
Anion exchange fractionation of the N-glycan pool was performed on a GlycoSep C 75 × 7.5 mm, 10 μm DEAE column (Prozyme, Hayward, CA, USA) using a Waters Alliance 2695 Separations Module with a Waters 474 Fluorescence Detector under the control of Empower Chromatography Workstation, (Waters Corporation, Milford, MA, USA). A linear gradient of 100 mM acetate, pH 7.0 in 20% v/v acetonitrile was used for the elution of charged oligosaccharides.
Protease digestion for N-glycosite identification and analysis
100 μg of BRP erythropoietin was reduced, alkylated and digested with Lys-C (Roche Diagnostics, Mannheim, Germany) in 25 mM Tris HCl, pH 8.5, 1 mM EDTA overnight at 37°C. The digest was reduced to dryness via vacuum centrifugation, reconstituted in 50 μL of 0.1% v/v trifluoroacetic acid (TFA) and the digest peptides separated, as described in the following section. For the generation of peptides containing a single glycosylation site, a further 100 μg of BRP erythropoietin was digested with Lys-C, peptide mapped, and the resulting glycopeptide peaks individually collected and subjected to further digestion with Glu-C (Roche Diagnostics, Mannheim, Germany) in 25 mM phosphate buffer, pH 7.8 overnight at 25°C. The digests were reduced to dryness via vacuum centrifugation, reconstituted in 50 μL of 0.1% v/v TFA, and the resulting peptides separated, collected and processed to characterize the N-glycosylation structures at each individual site.
LC-UV peptide mapping
Peptide mapping was performed using a Waters Alliance 2695 Separations Module with a Waters 2489 UV/Visible detector under the control of the Empower chromatography workstation software (Waters, Milford, MA, USA). Separations were performed using a Waters XBridge 3.5 μm, 135 Å C18 150 × 2.1 mm i.d. column. Peptides were eluted using a linear gradient of 5-65% acetonitrile containing 0.1% v/v TFA in 55 minutes at a flow rate of 200 μL/min with a column temperature of 40°C and detection at 214 nm. All resulting chromatographic peaks were individually collected, deglycosylated and subjected to glycosylation profiling by UPLC and peptide identification using LC-MS/MS.
Peptide identification using LC-MS/MS
LC-MS/MS analysis of enriched deglycosylated peptides was performed using an Agilent Technologies 1200 series instrument consisting of a ChipCube interface to a 6340 series ion trap mass spectrometer operated in the positive ion mode with a spray voltage of −1.8 kV (Agilent Technologies, Waldbronn, Germany). Separations were performed using a ProtID-Chip-43, consisting of a 40 nL enrichment column and a 43 mm analytical column packed with Zorbax StableBond 300 Å, 5 μm C18 particles. A linear gradient of 5-60% acetonitrile containing 0.1% v/v formic acid in six minutes, following a one-minute isocratic hold at 5 % acetonitrile, was used for peptide elution. The mass spectrometer was operated in the automatic data dependent mode with active exclusion. The three most abundant MS ions were selected for MS/MS analysis. MS/MS data were analyzed using Spectrum Mill Proteomics Workbench (Agilent Technologies, Santa Clara, CA, USA) and searched against the National Centre for Biotechnology Information database using the ‘human rodent’ taxonomic filter. Search parameters included a precursor ion mass tolerance of 2.5 Da, product ion mass tolerance of 0.7 Da, cysteine carbamidomethylation specified as a fixed modification, asparagine deamidation and methionine oxidation as variable modifications, and a maximum of two missed cleavage sites. Protein identifications were automatically validated using the autovalidation feature of Spectrum Mill, peptide scores > 6, protein scores > 11, and the percentage scored peak intensity > 60%.
Peptides were also analysed using a Dionex Ultimate 3000 LC (Sunnyvale, CA, USA) connected to a LTQ Orbitrap XL mass spectrometer (ThermoFisher Scientific, Hemel Hempstead, UK). Separations were performed using an increasing linear gradient of acetonitrile on a Dionex C18 Pepmap, 0.075 × 150 mm column at 300 nL/min. The mass spectrometer was operated in the positive ion mode, the spray voltage was −1.8kV, and the transfer capillary temperature 200°C. Data was acquired in the automatic data dependent switching mode. A high resolution MS scan was performed using the Orbitrap to select the 5 most intense ions in the 300-2000 m/z range prior to MS/MS analysis in the linear ion trap. Resulting MS/MS data were searched using BioWorks 3.2 (ThermoFisher Scientifc) against the Uniprot-Swissprot database with the Homo sapiens taxonomic filter specified, database accessed August 5th 2010. The enzymatic deamidation of asparagine was manually annotated in a separate erythropoietin sequence FASTA file added to the database. Peptides were filtered according to the following parameters: XCorr ≥1.90 (+1), 2.0 (+2) and 2.5 (+3). Identified spectra were also verified manually.
Results and Discussion
N-Glycosylation on BRP 3 Erythropoietin: Identification of Highly Branched, Sialylated Structures with Poly-N-Acetyl Lactosamine Extensions
Erythropoietin is a cytokine expressed by fibroblasts of the renal cortex that stimulates the production of red blood cells upon binding to its receptor located on erythroid progenitor cells in the bone marrow. Glycosylation plays a crucial role in the stability and therapeutic potency of the glycoprotein. For example, intravenously administered erythropoietin (EPO), consisting of highly branched sialylated oligosaccharide structures, has been shown to result in a plasma half life of 5-6 hours as compared to desialylated EPO which is cleared within minutes.25 The rapid clearance of desialylated EPO is mainly due to the interaction of exposed galactose residues on tri- and tetra-antennary glycans with asialo glycoprotein receptors present on hepatocytes.26 Analysis of the glycosylation present on EPO is clearly required to ensure the safety and efficacy of the drug.
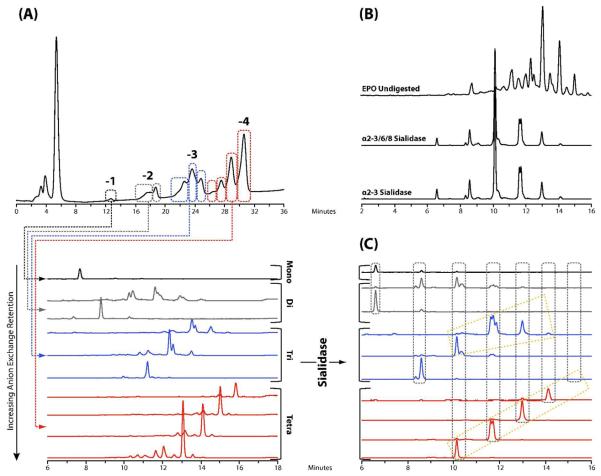
Due to the numerous members of the family of sialic acids that terminate the complex glycan antennae, a multidimensional separation strategy was developed to analyze the N-glycan pool released from EPO. In the first dimension, anion exchange chromatography with a shallow salt gradient was used to separate the N-glycans into 10 fractions. Each of the resulting fractions was then profiled in the second dimension by UPLC using the high-resolution 1.7 μm HILIC phase, as shown in Fig.1 (A). Separation by anion exchange was based on the number of sialic acids and, for the same net charge, glycan size, with larger glycans eluting before smaller oligosaccharide structures. Digestion of the N-glycan pool with Arthrobacter ureafaciens sialidase, an α2-3/6/8 sialidase and NANase I, a recombinant specific α2-3 sialidase revealed that all sialic acids were present in an α2-3 glycosidic linkage to the terminal galactose residues, in agreement with previous studies27, see Fig.1 (B). A repeating series of peaks was also observed in the UPLC separated fractions after sialidase digestion, as indicated in Fig.1 (C), suggesting antennae elongation of the tetra-antennary glycans present. Further exoglycosidase digestions with β1-4 galactosidase and β1-2/3/4/6 hexosaminidase (data not shown), confirmed these repeating units consisted of poly-N-acetyl lactosamine motifs (galactose β1-4 N-acetyl glucosamine). Exoglycosidase digestion also suggested that the oligosaccharides present in the EPO N-glycan pool contained an α1-6 linked core fucose on the reducing terminal N-acetyl glucosamine residue in agreement with previous reports which examined the glycosylation present on epoetins alpha, beta and delta.28,29
Fig.1.
(A): Two dimensional separation of the N-glycan pool of BRP 3 EPO using anion exchange chromatography followed by analysis of each of the fractions on 1.7 μm HILIC LC column. (B) Digestion of the N-glycan pool of BRP 3 EPO with Arthrobacter ureafaciens sialidase, an α2-3/6/8 sialidase, and NANase I, an α2-3 sialidase, revealing that all sialic acids are α2-3 linked. (C) Digestion of each of the anion exchange fractions with Arthrobacter ureafaciens sialidase for the identification of the glycans present. The vertical groupings indicate the presence of a common glycan structure with varying degrees of sialylation. Also indicated are groups of peaks displaying a periodic repeat that were identified by subsequent exoglycosidase digestion to correspond to the sequential addition of a lactosamine disaccharide unit to the parent glycan.
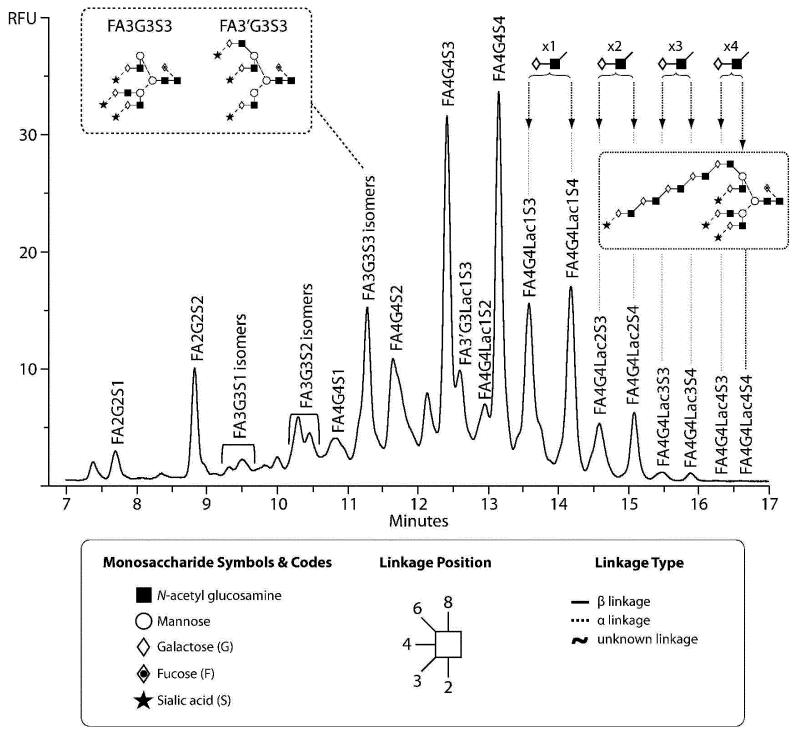
The combination of the data from the anion exchange separation, exoglycosidase digestion and profiling on the 1.7 μm HILIC phase allowed the annotation of the peaks present in the total N-glycan pool of EPO, as seen in Fig.2. The largest proportion of oligosaccharides are tetraantennary structures carrying up to four sialic acids, F(6)A4G(4)4S(3)4, representing 21% of the total glycan pool. Tetra-antennary glycans carrying one lactosamine extension represented 19% of the total glycan pool, those carrying two lactosamine repeats represented 6%, three repeats 1% and four repeats 0.1% of the total glycan pool, respectively. Previous studies have reported only one,29 two28 or a maximum of three poly-N-acetyl lactosamine repeats on recombinant EPO.30 The observation of the greater number of poly-N-acetyl lactosamine repeats in this instance are undoubtedly the result of the increased resolving power of the 2D separation including the 1.7 μm HILIC phase. These extended oligosaccharides appear to be a critical structural feature of recombinant EPO, previous characterisation of EPO from human serum reported the absence of these large tetra-antennary tetrasialylated glycans on the endogenous protein.31
Fig.2.
N-glycans present on the BRP 3 EPO reference standard as annotated using the combination of anion exchange fractionation, exoglycosidase digestion and analysis on the 1.7 μm HILIC phase, as described in the text. Nomenclature from ref (24). All fucose residues present are α1-6 linked, all galactose residues are β1-4 linked, and all sialic acid residues are α2-3 linked. The inset shows the two positional tri-antennary isomers present. The presence of the polylactosamine extension within the glycan is assumed based upon the biosynthetic pathway.
Poly-N-acetyl lactosamine repeats are synthesized by the sequential enzymatic action of a β1,4-galactosyltransferase and a β1,3-N-acetylglucosaminyltransferase (β3GnT) located within the cisternae of the trans Golgi.32 The action of the GnTV N-acetylglucosylaminyl transferase, which synthesizes the β1-6 linked antennary GlcNAc connected to the α1-6 linked core mannose residue, will generate the preferred substrate for the subsequent action of β3GnT, the transferase which adds the β1-3 linked N-acetyl glucosamine residues to galactose residues, leading to the extension of the glycan with poly-N-acetyl lactosamine repeats from this β1-6 branch.33 In addition to the residence time of glycoproteins within the Golgi,33 the flux of UDP-GlcNAc into the Golgi has also been demonstrated to be critically important for the processing of complex branched oligosaccharides and the addition of poly-N-acetyl lactosamine repeats.34 An in depth proteomic understanding of cellular processes would be useful to understand more completely the factors that can potentially give rise to such complex variation in product glycosylation as observed here.
Elongated poly-N-acetyl lactosamine motifs on EPO may result in the non-specific recognition by other receptors including secreted or membrane bound S-type lectins or galectins, a family of proteins that have biological functions including metabolic activation and cellular development.20 For example, galectin-1, involved in the modulation of the adaptive immune response including the induction of apoptosis of T-cells has been demonstrated to bind poly-N-acetyl lactosamine residues with higher efficiency that a single Gal β1-4 GlcNAc repeat.35,36 On the other hand, at the same time, the presence of glycans carrying large poly-N-acetyl lactosamine repeats may potentially be beneficial in minimizing the risk of induction of pure red cell aplasia (PRCA) by virtue of their size and their ability to mask potentially immunogenic conformational epitopes present on EPO through their molecular dynamics.37,38 However, enriched EPO bearing poly-N-acetyl lactosamine repeats using Lycopersicon Esculentum lectin affinity chromatography were found to undergo faster clearance when compared with EPO that did not bind to the lectin column following intravenous administration.20 The increased clearance of the EPO glycoforms bearing sialylated poly-N-acetyl lactosamine repeats was suggested to be due to recognition of these elongated glycan motifs by galactose binding proteins present on hepatocytes.20 Although these particular glycoforms are present are relatively low levels, differences in the length of the poly-N-acetyl lactosamine chains present on the N-glycans covalently attached to EPO may result in potential alterations in potency of the drug.
Site-Specific characterisation of BRP 3 EPO glycosylation
Having examined the total N-glycosylation present on the BRP 3 EPO reference standard, site-specific structural microheterogeneity was next explored. For therapeutic proteins such as EPO with more than one N-glycosylation site, the population and relative amount of oligosaccharides present at each site should be determined for a full comparison between the innovator and biosimilar product. Due to the close proximity of asparagine 24 and 38, a double round of protease digestion (Lys-C followed by Glu-C) with subsequent isolation of individual glycopeptides was performed. First, BRP 3 EPO was digested with Lys-C, and the resulting peptides were separated using microbore reversed-phase LC with each chromatographic peak collected. Subsequently, each fraction was digested with PNGaseF to release the N-glycans, which were then analysed on the 1.7 μm HILIC phase, as above.
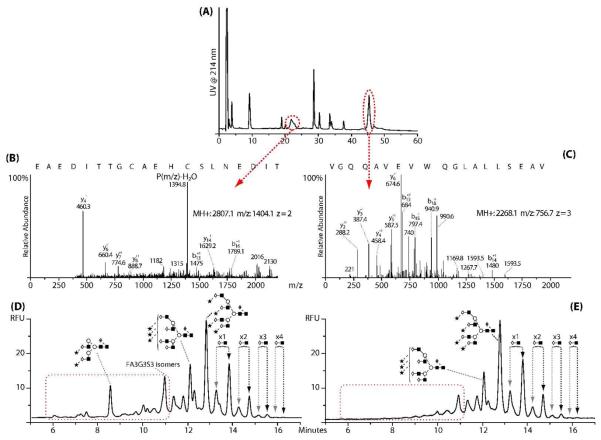
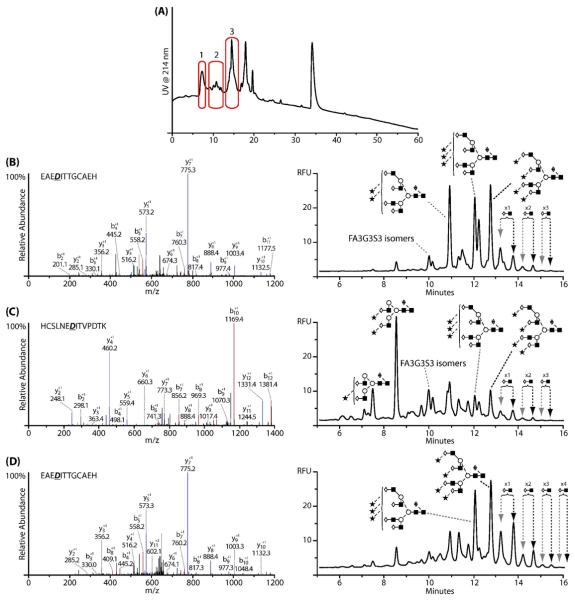
Two broad peaks were identified in the Lys-C digest of EPO, as shown in Fig.3 (A), the first corresponding to the glycopeptides with asparagine 24 and 38 and the second containing asparagine 83. The assignments were confirmed by LC-MS/MS, see Fig.3 (B and C). The resulting annotated fluorescence profiles for the N-glycans are shown in Fig.3 (D and E). Using the results from the total glycosylation analysis, it can be seen from Fig.3 (D) that the peptide with asparagine 24 and 38 contains bi-, tri- and tetra-antennary glycans with poly-N-acetyl lactosamine repeats. Primarily tetra-antennary type glycans with poly-N-acetyl lactosamine repeats were detected on the peptide containing asparagine 83 as seen in Fig.3 (E).
Fig.3.
(A) LC-UV analysis of Lys-C peptide digest of BRP 3EPO, (B) MS/MS spectra of the deglycosylated peptide containing the sequons at asparagine 24 and 38, (C) MS/MS spectra of the deglycosylated peptide containing the sequon at asparagine 83. For B and C, the black text corresponds to the portion of the peptide sequence as annotated from the MS/MS spectra. (D) N-glycans released from the peptide containing asparagine 24 and 38 indicating the presence of bi-, tri- and tetra-antennary N-glycans, (E) N-glycans released from the peptide containing asparagine 83 indicating the presence of mainly tetra-antennary N-glycans with poly-N-acetyl lactosamine extensions.
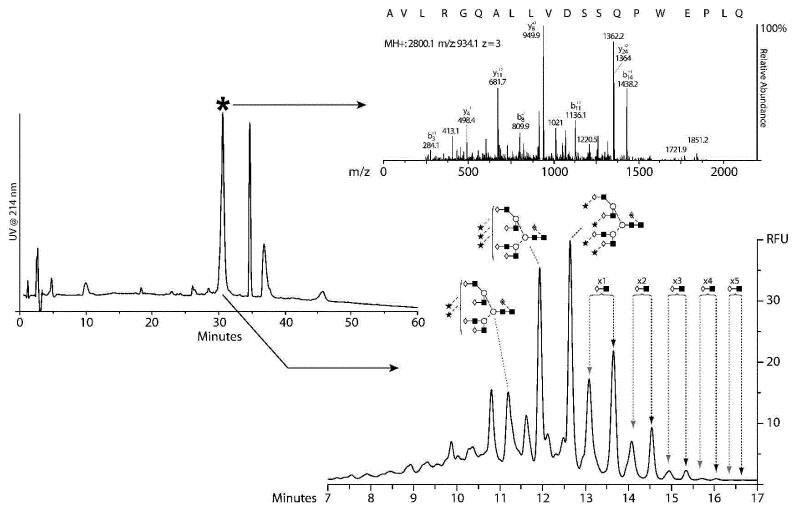
To separate asparagine 24 from asparagine 38 and also to digest the large glycopeptide containing asparagine 83 further, these glycopeptides were next digested with Glu-C. Fig.4 (A) displays the resulting Glu-C peptide separation of the glycopeptide containing asparagine 83. Using LC-MS/MS, the highlighted peak was found to contain the N-glycosylation sequon, Fig.4 (B). The asparagine in the sequon (N83SS) was confirmed to have undergone deamidation to yield aspartic acid during the deglycosylation process. Furthermore, since nonglycosylated peptides were not detected, it is assumed that the site was completely occupied. Fig.4 (C) displays the resulting annotated N-glycan profile of the released glycans on the 1.7 μm HILIC phase. The N-glycans at asparagine 83 are predominantly tetra-antennary with up to five poly-N-acetyl lactosamine repeats on both the tri- and tetrasialylated analogues. To our knowledge, this is the first report of five poly-N-acetyl lactosamine repeats on EPO N-glycans, the largest previously reported was three repeats.30 F(6)A4G(4)S(3)3 and F(6)A4G(4)S(3)4 represented 12% and 14%, respectively, of the total glycan pool at asparagine 83. For both F(6)A4G(4)S(3)3 and F(6)A4G(4)S(3)4 structural analogues containing one, two and three poly-N-acetyl lactosamine repeats represented ~9%, ~3% and ~1% of the total glycan pool, respectively. Those glycans carrying four and five poly-N-acetyl lactosamine repeats were each present at 0.1% of the total glycan pool. From the EPO polypeptide configuration, asparagine 83 is geometrically isolated from the other glycosylation sites on the molecule.18 Therefore, the presence of these large highly branched oligosaccharide structures is not surprising as there is easy access of the glycosyltransferase enzymes responsible for generating these structures.
Fig.4.
(A) Peptide map of the Lys-C glycopeptide containing asparagine 83 after digestion with Glu-C, (B) MS/MS spectrum of the peptide identified to contain the N-glycan sequon; full asparagine to aspartic acid conversion was observed indicating complete site occupancy. (C) N-glycan profile of the oligosaccharides present at asparagine 83; tetra-antennary glycans with poly-N-acetyl lactosamine extensions were observed, including small levels of tetra-antennary glycans with three or four sialic acids and up to five poly-N-acetyl lactosamine extensions.
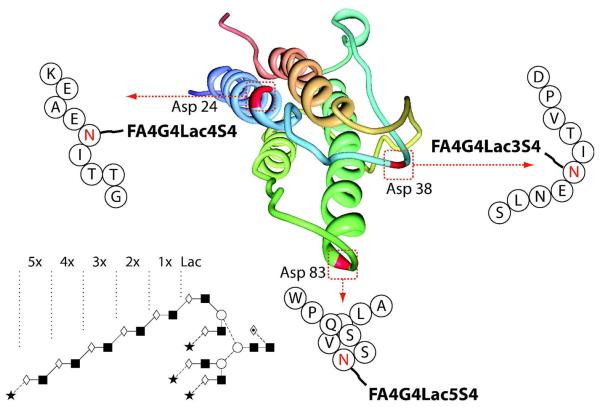
In an analogous manner, the glycopeptide containing asparagines 24 and 38 was further digested with Glu-C. Fig.5 (A) shows the resulting peptide map with the peaks found to contain N-glycans highlighted. The MS/MS spectrum used for sequence annotation along with the corresponding N-glycan profile are shown in Fig.5 (C and D, respectively). The identities of the peptides contained within these peaks were determined using high mass accuracy on the Orbitrap. As observed previously, all asparagine residues in each glycosylation sequon, (N24IT and N38IT) had undergone deamidation. Again, nonglycosylated peptides were not detected, suggesting complete occupancy for both sites. Two peptide fragments were found to contain asparagine 24, thought to be due to non-specific cleavage by Glu-C, Fig.5 (B and D). Despite this, both peptides were found to contain tri- and tetrasialylated tetra-antennary N-glycans with up to four poly-N-acetyl lactosamine repeats as the major structures, with low proportions of bi-antennary glycans. In contrast, the glycosylation profile of the peptide that contained asparagine 38 indicated that the major glycan present at this site was a desialylated bi-antennary glycan with tri- and tetraantennary glycans present at much lower levels, Fig.5 (C). Poly-N-acetyl lactosamine repeats detected on the tetra-antennary glycans at this site were shorter than those at asparagine 24 or 83, with a maximum of only three repeating lactosamine units. Considering further the structure of EPO,18 the N-glycosylation sites at asparagine 24 and 38 are in close proximity due to the formation of a disulfide bond between the cysteine residues at positions 29 and 33. It is therefore likely that the higher proportion of bi-antennary and shorter poly-N-acetyl lactosamine structures at asparagine 38 is a result of steric hindrance due to the presence of the neighbouring disulfide bond which appears to pull asparagine 38 into a pocket that reduces the accessibility of the glycosyltransferase enzymes required for processing and oligosaccharide elongation. Glycosyltransferase access to asparagine 38 may be further reduced by the more facilitated processing and elongation of the oligosaccharide present at asparagine 24. Fig.6 is a schematic of the three-dimensional EPO polypeptide annotated with the largest N-glycan as determined at each individual N-glycosylation site.39
Fig.5.
(A): Peptide map of the Lys-C glycopeptide containing asparagine 24 and 38 after digestion with Glu-C, (B) Orbitrap MS/MS spectra and corresponding annotated N-glycan profile of peak 1 in (A) determined to correspond to the sequon containing asparagine 24. (C) Orbitrap MS/MS spectra and corresponding annotated N-glycan profile of peak 2 in (A) determined to correspond to the sequon containing asparagine 38. (D) Orbitrap MS/MS spectra and corresponding annotated N-glycan profile of peak 3 in (A) determined to correspond to the sequon containing asparagine 24.
Fig.6.
Schematic of the EPO polypeptide annotated with the largest N-glycan as determined at each individual N-glycosylation site. The polypeptide structure is as described in ref (39) and is available online through the Protein Data Bank. The presence of the polylactosamine extension within the glycan is assumed based upon the biosynthetic pathway.
The identification of N-glycans containing long poly-N-acetyl lactosamine repeats at specific N-glycosylation sites significantly adds to data previously reported following CZE-ESI-Tof-MS characterisation of intact BRP 2 EPO glycoforms.40 A maximum deconvoluted mass of 32,448 Da was observed corresponding to three tetrasialylated tetra-antennary glycans with up to nine poly-N-acetyl lactosamine repeats distributed across the three N-glycosylation sites. From the intact analysis, it was not possible to determine site-specific glycosylation. In the work of the current study, a maximum of three tetrasialylated tetra-antennary glycans with up to twelve poly-N-acetyl lactosamine repeats distributed across the three N-glycosylation sites were found, but at potentially low levels that were beyond the sensitivity limit of the CZE-ESI-Tof-MS approach in reference (40).
Conclusions
Using a 2D LC approach with a shallow weak anion exchange gradient in the first dimension followed by profiling of each fraction using the 1.7 μm HILIC column in the second dimension, the glycosylation structures on the BRP 3 EPO standard were comprehensively analyzed. Tetraantennary glycans with up to four sialic acids and multiple poly-N-acetyl lactosamine repeats were determined to be the major structures present, with bi- and tri-antennary glycans at much lower levels. Site-specific glycosylation profiling showed tetra-antennary glycans to be the primary oligosaccharides present at asparagine 24 and 83 with up to five poly-N-acetyl lactosamine repeats observed on a tetrasialylated glycan present at asparagine 83, albeit at <0.1% of the total glycan pool. In contrast, bi-antennary glycans were the major structure present at asparagine 38. The determination of up to twelve poly-N-acetyl lactosamine repeats covalently attached to EPO is a novel finding. Although present at low levels, these large repeating glycan motifs may potentially infer additional functional interactions for EPO including recognition by galectins or galactose binding proteins on hepatocytes resulting in either secondary biological activity or more facilitated clearance. On the other hand, such large oligosaccharide structures could potentially be beneficial in masking immunogenic conformational epitopes on the polypeptide that can cause unwanted recognition and activation. Therefore, characterisation of the different lengths of the poly-N-acetyl lactosamine extensions present on EPO N-glycans is important and requires further research to fully understand their potential role.
The combined 2D AEC × UPLC HILIC method represents a considerable advance for the liquid phase glycomic analysis of therapeutic glycoproteins in terms of both resolving power and separation speed, allowing us, in this instance, to perform a rapid, in depth, comprehensive analysis of the complex N-glycosylation present of the BRP3 EPO reference standard. Such detailed glycosylation analysis is important as biosimilar products become of increasing interest.
Acknowledgement
This research was supported by the European Union via the EU FP6 GLYFDIS research program, grant reference 037661 (JB and PMR) and NIH GM15847 (BLK). JB and PMR gratefully acknowledge Waters Corporation for the donation of the Acquity™ UPLC instrument and Agilent Technologies for the donation of the ChipCube interface. Stefan Mittermayr is acknowledged for his assistance in the preparation of the manuscript. Contribution number 981 from the Barnett Institute.
References
- 1.Walsh G, Jefferis R. Nat. Biotechnol. 2006;24:1241–1252. doi: 10.1038/nbt1252. [DOI] [PubMed] [Google Scholar]
- 2.Li H, d'Anjou M. Curr. Opin. Biotechnol. 2009;20:678–684. doi: 10.1016/j.copbio.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 3.ICH Harmonised Tripartite Guideline Q6B [last viewed January 12th 2011];Specifications: Test procedures and acceptance criteria for biotechnological/biological products. available online at: http://www.ich.org/LOB/media/MEDIA432.pdf.
- 4.European Parliament and Council Off. J. Eur. Union. 2004;47:34–57. [Google Scholar]
- 5.Domann PJ, Pardos-Pardos AC, Fernandes DL, Spencer DI, Radcliffe CM, Royle L, Dwek RA, Rudd PM. Proteomics. 2007;7:70–76. doi: 10.1002/pmic.200700640. [DOI] [PubMed] [Google Scholar]
- 6.Alpert AJ, Shukla M, Shukla AK, Zieske LR, Yuen SW, Ferguson MA, Mehlert A, Pauly M, Orlando R. J. Chromatogr. A. 1994;676:191–202. doi: 10.1016/0021-9673(94)00467-6. [DOI] [PubMed] [Google Scholar]
- 7.Stadlmann J, Pabst M, Kolarich D, Kunert R, Altmann F. Proteomics. 2008;8:2858–2871. doi: 10.1002/pmic.200700968. [DOI] [PubMed] [Google Scholar]
- 8.Melmer M, Stangler T, Premstaller A, Lindner W. J. Chromatogr. A. 2010;1217:6092–6096. doi: 10.1016/j.chroma.2010.07.059. [DOI] [PubMed] [Google Scholar]
- 9.Melmer M, Stangler T, Premstaller A, Lindner W. J. Chromatogr. A. 2010;1217:6097–6101. doi: 10.1016/j.chroma.2010.06.069. [DOI] [PubMed] [Google Scholar]
- 10.Balaguer E, Neusüss C. Anal. Chem. 2006;78:5384–5393. doi: 10.1021/ac060376g. [DOI] [PubMed] [Google Scholar]
- 11.Balaguer E, Demelbauer U, Pelzing M, Sanz-Nebot V, Barbosa J, Neusüss C. Electrophoresis. 2006;27:2638–2650. doi: 10.1002/elps.200600075. [DOI] [PubMed] [Google Scholar]
- 12.Thakur D, Rejtar T, Karger BL, Washburn NJ, Bosques CJ, Gunay NS, Shriver Z, Venkataraman G. Anal. Chem. 2009;81:8900–8907. doi: 10.1021/ac901506p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alley WR, Jr, Madera M, Mechref Y, Novotny MV. Anal Chem. 2010;82:5095–5106. doi: 10.1021/ac100131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mechref Y, Kang P, Novotny MV. Methods Mol. Biol. 2009;534:53–64. doi: 10.1007/978-1-59745-022-5_4. [DOI] [PubMed] [Google Scholar]
- 15.Ahn J, Bones J, Yu YQ, Rudd PM, Gilar M. J. Chromatogr. B. 2010;878:403–408. doi: 10.1016/j.jchromb.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 16.Bones J, Mittermayr S, O'Donoghue N, Guttman A, Rudd PM. Anal Chem. 2010;82:10208–10215. doi: 10.1021/ac102860w. [DOI] [PubMed] [Google Scholar]
- 17.Behr-Gross ME, Daas A, Burns C, Bristow AF. Pharmeuropa Bio. 2007 Dec;(1):49–66. 2007. [PubMed] [Google Scholar]
- 18.Sasaki H, Ochi N, Dell A, Fukuda M. Biochemistry. 1988;27:8618–8626. doi: 10.1021/bi00423a017. [DOI] [PubMed] [Google Scholar]
- 19.Wasley LC, Timony G, Murtha P, Stoudemire J, Dorner AJ, Caro J, Krieger M, Kaufman RJ. Blood. 1991;77:2624–32. [PubMed] [Google Scholar]
- 20.Delacour D, Koch A, Jacob R. Traffic. 2009;10:1405–1413. doi: 10.1111/j.1600-0854.2009.00960.x. [DOI] [PubMed] [Google Scholar]
- 21.Fukuda MN, Sasaki H, Lopez L, Fukuda M. Blood. 1989;73:84–89. [PubMed] [Google Scholar]
- 22.Olajos M, Hajós P, Bonn GK, Guttman A. Anal. Chem. 2008;80:4241–4246. doi: 10.1021/ac8002598. [DOI] [PubMed] [Google Scholar]
- 23.Royle L, Radcliffe CM, Dwek RA, Rudd PM. Methods Mol. Biol. 2006;347:125–43. doi: 10.1385/1-59745-167-3:125. [DOI] [PubMed] [Google Scholar]
- 24.Harvey DJ, Merry AH, Royle L, Campbell MP, Dwek RA, Rudd PM. Proteomics. 2009;9:3796–3801. doi: 10.1002/pmic.200900096. [DOI] [PubMed] [Google Scholar]
- 25.Delorme E, Lorenzini T, Giffin J, Martin F, Jacobsen F, Boone T, Elliott S. Biochemistry. 1992;31:9871–9876. doi: 10.1021/bi00156a003. [DOI] [PubMed] [Google Scholar]
- 26.Agoram B, Aoki K, Doshi S, Gegg C, Jang G, Molineux G, Narhi L, Elliott S. J. Pharm Sci. 2009;98:2198–2211. doi: 10.1002/jps.21578. [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi M, Takasaki S, Miyazaki H, Kato T, Hoshi S, Kochibe N, Kobata A. J. Biol. Chem. 1988;263:3657–3663. [PubMed] [Google Scholar]
- 28.Llop E, Gutiérrez-Gallego R, Segura J, Mallorquí J, Pascual JA. Anal. Biochem. 2008;383:243–254. doi: 10.1016/j.ab.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 29.Shahrokh Z, Royle L, Saldova R, Bones J, Abrahams JL, Artemenko NV, Flatman S, Davies M, Baycroft A, Sehgal S, Heartlein M, Harvey DJ, Rudd PM. Mol. Pharm. 2010;8:286–296. doi: 10.1021/mp100353a. [DOI] [PubMed] [Google Scholar]
- 30.Kawasaki N, Ohta M, Hyuga S, Hyuga M, Hayakawa T. Anal. Biochem. 2000;285:82–91. doi: 10.1006/abio.2000.4739. [DOI] [PubMed] [Google Scholar]
- 31.Skibeli V, Nissen-Lie G, Torjesen P. Blood. 2001;98:3626–3634. doi: 10.1182/blood.v98.13.3626. [DOI] [PubMed] [Google Scholar]
- 32.Togayachi A, Kozono Y, Ishida H, Abe S, Suzuki N, Tsunoda Y, Hagiwara K, Kuno A, Ohkura T, Sato N, Sato T, Hirabayashi J, Ikehara Y, Tachibana K, Narimatsu H. Proc. Natl. Acad. Sci. USA. 2007;104:15829–15834. doi: 10.1073/pnas.0707426104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nabi IR, Dennis JW. Glycobiology. 1998;8:947–953. doi: 10.1093/glycob/8.9.947. [DOI] [PubMed] [Google Scholar]
- 34.Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 35.Stowell SR, Dias-Baruffi M, Penttilä L, Renkonen O, Nyame AK, Cummings RD. Glycobiology. 2004;14:157–167. doi: 10.1093/glycob/cwh018. [DOI] [PubMed] [Google Scholar]
- 36.He J, Baum LG. J. Biol. Chem. 2004;279:4705–4712. doi: 10.1074/jbc.M311183200. [DOI] [PubMed] [Google Scholar]
- 37.Rossert J, Casadevall N, Eckardt KU. J. Am. Soc. Nephrol. 2004;15:398–406. doi: 10.1097/01.asn.0000107561.59698.42. [DOI] [PubMed] [Google Scholar]
- 38.Casadevall N, Nataf J, Viron B, Kolta A, Kiladjian JJ, Martin-Dupont P, Michaud P, Papo T, Ugo V, Teyssandier I, Varet B, Mayeux P. N. Engl. J. Med. 2002;346:469–475. doi: 10.1056/NEJMoa011931. [DOI] [PubMed] [Google Scholar]
- 39.Cheetham JC, Smith DM, Aoki KH, Stevenson JL, Hoeffel TJ, Syed RS, Egrie J, Harvey TS. Nat. Struct. Biol. 1998;5:861–866. doi: 10.1038/2302. [DOI] [PubMed] [Google Scholar]
- 40.Neusüss C, Demelbauer U, Pelzing M. Electrophoresis. 2005;26:1442–1450. doi: 10.1002/elps.200410269. [DOI] [PubMed] [Google Scholar]