Abstract
The human vitamin D receptor (hVDR) is a member of the nuclear receptor superfamily, involved in calcium and phosphate homeostasis; hence implicated in a number of diseases, such as Rickets and Osteoporosis. This receptor binds 1α,25-dihydroxyvitamin D3 (also referred to as 1,25(OH)2D3) and other known ligands, such as lithocholic acid. Specific interactions between the receptor and ligand are crucial for the function and activation of this receptor, as implied by the single point mutation, H305Q, causing symptoms of Type II Rickets. In this work, further understanding of the significant and essential interactions between the ligand and the receptor were deciphered, through a combination of rational and random mutagenesis. A hVDR mutant, H305F, was engineered with increased sensitivity towards lithocholic acid, with an EC50 value of 10 µM and 40 ± 14 fold activation in mammalian cell assays, while maintaining wild-type activity with 1,25(OH)2D3. Furthermore, via random mutagenesis, a hVDR mutant, H305F/H397Y, was discovered to bind a novel small molecule, cholecalciferol, a precursor in the 1α,25-dihydroxyvitamin D3 biosynthetic pathway, which does not activate wild-type hVDR. This variant, H305F/H397Y, binds and activates in response to cholecalciferol concentrations as low as 100 nM, with an EC50 value of 300 nM and 70 ± 11 fold activation in mammalian cell assays. In silico docking analysis of the variant displays a dramatic conformational shift of cholecalciferol in the ligand binding pocket in comparison to the docked analysis of cholecalciferol with wild-type hVDR. This shift is hypothesized to be due to the introduction of two bulkier residues, suggesting that the addition of these bulkier residues introduces molecular interactions between the ligand and receptor, leading to activation with cholecalciferol.
Keywords: human vitamin D receptor, cholecalciferol, lithocholic acid, protein engineering, random mutagenesis
1. Introduction
Nuclear receptors are ligand-activated transcription factors. Upon ligand binding, these receptors activate the transcription of essential genes required for a number of cellular and physiological processes including proliferation, differentiation, and development [1–3]. To date there are 49 known human nuclear receptors [4]. These receptors bind a wide variety of ligands, including bile acids, steroids, and non-steroidal ligands, such as 9-cis retinoic acid. Structurally, nuclear receptors consist of a DNA-binding domain (DBD) and a ligand-binding domain (LBD) connected by a hinge region [1, 2]. The DBD binds to short sequences of DNA known as response elements [3, 5–7]. The LBD is α-helical nature along with a few β-strands and includes a ligand binding pocket (LBP) responsible for binding the small molecule ligand. While nuclear receptor DBDs share ∼95% similarity, the differences in their LBDs’ account for the diverse ligand binding profiles of these nuclear receptors, leading to their unique structural identities and biological functions [5, 8].
As transcription factors, nuclear receptors function in a precise manner involving a series of molecular events that lead to regulation of essential genes [1, 8, 9]. In the absence of ligand, corepressors are bound to the nuclear receptors, leading to recruitment of histone deacetyltransferases (HDACs) involved in chromatin remodeling; thus transcription is repressed [8]. Upon ligand binding, a conformational change occurs in the LBD of these receptors, recruiting coactivators and histone acetyltransferases (HATs), inducing transcriptional activation of target genes [1, 8, 9].
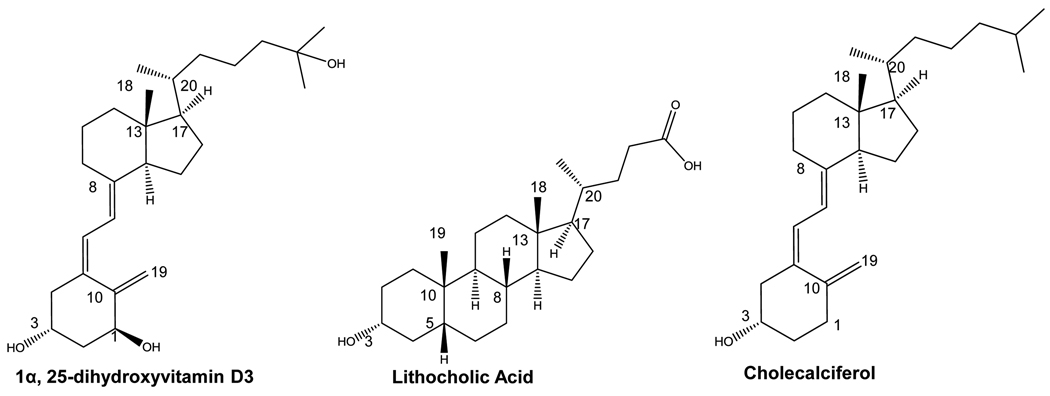
The human vitamin D receptor (hVDR) is a nuclear receptor primarily involved in biological processes such as apoptosis, immune responses, and calcium and phosphate homeostasis [10]. In addition to its natural ligand, 1α,25-dihydroxyvitamin D3 (1,25(OH)2D3), this receptor is known to bind a variety of 1,25(OH)2D3 analogs and bile acids, such as lithocholic acid (LCA) (Figure 1) [11–14]. The ability to activate VDR depends on a number of biological factors, such as the uptake of ligand across the membrane and ligand metabolism [15]. hVDR’s ligand binding domain is comprised of 303 amino acids with an elongated binding pocket, consisting of both polar and non-polar residues. These residues contribute essential molecular interactions between the ligand and receptor, required for receptor activation and function. Disruption of these key interactions reduces or inhibits the activity of a functional receptor, leading to adverse effects. An example of this is shown with a single point mutation in the LBD of hVDR, H305Q, which causes a 10-fold decrease in sensitivity of the receptor to 1,25(OH)2D3, causing Type II Rickets [16].
Figure 1. Structures of hVDR ligands and precursors.
1α,25-dihydroxyvitamin D3 (1,25(OH)2D3) and lithocholic acid (LCA) are known hVDR ligands. Cholecalciferol is a precursor in the 1,25(OH)2D3 biosynthetic pathway.
The effect of the H305Q mutation emphasizes the importance of understanding the structure and function relationship between the vitamin D receptor and various ligand. Previously, structural and mutational studies with hVDR were performed to assess the role of residues within the ligand binding pocket, deciphering important residues for ligand binding and activation [11, 17–25]. These findings, along with the crystal structures of hVDR with 1,25(OH)2D3 and other ligands, have provided useful preliminary information for interactions required for ligand binding. Via alanine scanning mutagenesis, hydrogen bonding residues were determined. Initially based on the alanine scanning results, Y143, S237, R274, S278, H305, and H397 were found to form crucial hydrogen bonds with 1,25(OH)2D3 [11, 19–22, 25, 26]. However, more recently mutational analysis has shown that the hydrogen bonds formed between H305, H397, and 1,25(OH)2D3 are not crucial for activation [27]. Mutating C288 to a glycine or alanine or mutating W286 to phenylalanine or alanine was also shown to cause loss of transcriptional activation [18, 23, 24, 26].
Using a combination of structural data and previous mutational research, this work focuses on further investigating and identifying key parameters in ligand binding and activation of hVDR with various ligands. Factors such as hydrogen bonding, residue size, and pocket volume were analyzed. Using both rational design and random mutagenesis, preliminary results of the mutational tolerance of the receptor was determined, and hVDR was engineered to bind and activate transcription in response to cholecalciferol, a 1,25(OH)2D3 precursor. Cholecalciferol, structurally lacks two of the hydroxyl groups present on 1,25(OH)2D3 and does not activate wild-type hVDR.
2. Methods and Materials
2.1 Ligands
Lithocholic acid, cholecalciferol, and 1α,25-dihydroxyvitamin D3 were ordered from MP Biomedicals, LLC (Solon, OH), Sigma (St. Louis, MO), and BIOMOL (Plymouth Meeting, PA), respectively. 10 mM stocks of lithocholic acid and cholecalciferol and a 13.3 µM stock of 1α,25-dihydroxyvitamin D3 were made with 80% ethanol: 20% DMSO and stored at 4 °C.
2.2 Gene isolation and Yeast Expression Vector Construction
The human vitamin D receptor gene was cloned from human skin cDNA with PCR using the following primers: 5’-gcc gga att cat gga ggc aat ggc ggc-3’ and 5’-gga cta gtt cag gag atc tca ttg cca aac ac-3’. The underlined sequences denote EcoRI and SpeI restriction sites, respectively. The VDR gene and yeast expression vector (pGBT9), which contains a Gal4 DNA binding domain, were digested, ligated, and transformed into Z-competent XL-1 Blue E.coli cells (Zymo, USA). Fusing the hVDRLBD to the Gal4 DNA binding domain created the pGBDhVDR plasmid, which contains a tryptophan marker. The plasmid was sequenced for confirmation (Operon, USA).
2.3 Creating designed hVDR library
To create the hVDR insert cassette variants, eight oligonucleotides with overlapping ends, containing degenerate base codes at the seven positions, were pieced together using overlap extension PCR [28]. 100 ng of each oligo and 125 ng of each primer were combined in a PCR tube and subjected to a PCR program with the following conditions: 95 °C for 1 min, 59 °C for 1 min, 72 °C for 2 min, 15 cycles, 95 °C for 1 min, 56.6 °C for 1 min, 72 °C for 2 min, 15 cycles. The resulting insert cassette was 1014 bp.
To create a background vector cassette, site-directed mutagenesis was used to insert SacII and KpnI sites into pGBDhVDR 810 bp apart, resulting in the pGBDhVDRSacIIKpnI plasmid. This plasmid was digested with the two restriction enzymes, removing 810 bp of the wild-type VDR gene from the plasmid. A 145 bp segment of random DNA from another plasmid, pMSCVeGFP, was digested and ligated between the SacII and KpnI sites (pGBDhVDRSacIIKpnI), generating 3 stop codons in the open reading frame. The resulting plasmid was named pGBDhVDRbackground and confirmed by sequencing (Operon, USA).
2.4 Error-Prone PCR
pGBDhVDRH305F, the plasmid containing the hVDR mutant, was used as the template DNA, for error-prone PCR random mutagenesis with primers: 5’-gatcctgaagcggaaggagg-3’ and 5’-gtccaggcagggtggccagaacgggtgggcacaaaggatggactagttcaggagatctcattgccaaacacttcgag-3’. 0.5 µM of each primer, 500 µM dNTPs, 7 mM MgCl2, 20 µM MnCl2, 250 ng template DNA (pGBDhVDRH305F), 1x Taq buffer, and 5 units of Taq polymerase (Fermentas, USA) were used for each reaction. The PCR program used was: 95 °C for 5 min, 95 °C for 30 sec, 55 °C for 30 sec, 72 °C for 60 sec, 20 cycles yielding ∼ 1 µg of variant VDR insert cassette. The insert cassettes were then transformed into yeast using the 1x TRAFCO protocol and tested in liquid quantitation assays using chemical complementation as mentioned in Schwimmer et al. [5].
2.5 Yeast Transformation
Using the 1x TRAFCO yeast transformation protocol [29], 100 ng of the digested background plasmid (pGBDhVDRbackground) and 900 ng of insert cassette were transformed into the yeast strain PJ69-4A. A plasmid containing a fusion of the human co-activator ACTR and the Gal4 activation domain with a leucine marker was also co-transformed [7]. Transformants were plated onto synthetic complete (SC) agar plates lacking adenine, leucine, and tryptophan (SC-ALW) with various ligands. Transformants were also plated onto synthetic complete agar plates lacking leucine and tryptophan (SC-LW) containing no ligand, to determine transformation efficiency. Transformation efficiency was calculated based on the number of colonies on the plate divided by the µg of vector cassette that was plated onto each plate.
2.6 Liquid Quantitation Assay in Yeast
Variants were tested in liquid quantitation assays in 96-well plates with media lacking adenine, leucine, and tryptophan (SC-ALW), with or without LCA, cholecalciferol, or 1,25(OH)2D3 at varying concentrations (ranging from 10 nM - 10 µM for LCA and cholecalciferol, 1 nM - 1 µM for 1,25(OH)2D3). A 4:1 ratio of media (SC-ALW): cells (yeast resuspended in water) were aliquoted into 96-well plates. Plates were incubated at 30°C, with shaking at 170 rpm. Optical density (OD) readings at 630 nm were recorded at 0, 24, and 48 hours as a measure of growth density.
2.7 Mammalian Cell Culture
HEK293T cells (ATCC, USA) were transfected with the following plasmids: pCMXwild-type hVDR, pCMXH305F, pCMXH305Y, and pCMXH305F/H397Y. These plasmids contain the Gal4DBD (GBD) fused to the corresponding VDR ligand binding domain (GBD:LBD fusion under the control of a cytomegalovirus (CMV) promoter). The reporter plasmids were p17*4TATAluc, containing the Renilla luciferase gene under the control of four Gal4 response elements located upstream from a minimal thymidine kinase promoter, and pCMXβgal, a plasmid containing the β-galactosidase gene under the control of the mammalian CMV promoter. Lipofectamine 2000 (Invitrogen, USA) served as the cationic lipid and transfection experimental details are described in Taylor et al. [30]. The ligands were added to the wells at various concentrations ((0.01 µM– 100 µM) LCA and (0.01 µM– 32 µM) cholecalciferol). Cells were harvested and analyzed for luciferase and β-galactosidase activity. All data points represent the average of triplicate experiments normalized against β-galactosidase activity. Error bars represent the standard deviation calculated using standard deviation: σ = Square root (Σ[(X-µ)2]). Fold activation was calculated by dividing the value at maximal activation by the value at the no ligand data point.
2.8 Docking hVDR mutants
The structures of the hVDR mutants were prepared in silico using the program TRITON 4.0.0 (National Centre for Biomolecular Research, Czech Republic) and its external program MODELLER (National Centre for Biomolecular Research, Czech Republic). These programs do not include any molecular dynamics. The computational site directed mutagenesis method, which is based on using the wild-type protein for homology modeling was employed [31, 32]. The wild-type hVDRLBD from the crystal structure 1DB1 was used as the template, which is missing residues 165–215for crystallization purposes [11]. These residues were not added computationally in the modeling process.
Variants were prepared for docking using the UCSF CHIMERA- interactive molecular graphics program by: (1) removing the ligand and water molecules, (2) adding polar hydrogens, and (3) assigning Gasteiger charges [33]. Ligands were created using ChemBioDraw Ultra 11.0 and ChemBio3D Ultra 11.0 (Cambridge Soft, USA) and modified with the AutoDockTools by adding Gasteiger charges, setting the partial charge property of each atom [34]. AutoDock Vina was used to perform docking simulations with default parameters [35]. In the Autodock simulations the hVDR ligand binding domain was held rigid while the ligand was allowed to rotate, based on quantum mechanical rotations that are incorporated into the algorithm. For the ligands the C3-OH bond, the carbon chain connecting the two ring systems, and the aliphatic chain extending off of C17 were allowed to rotate freely in the simulations. The solutions with the lowest free energy of binding were analyzed.
3. Results
3.1 Chemical Complementation: A yeast selection system for protein engineering
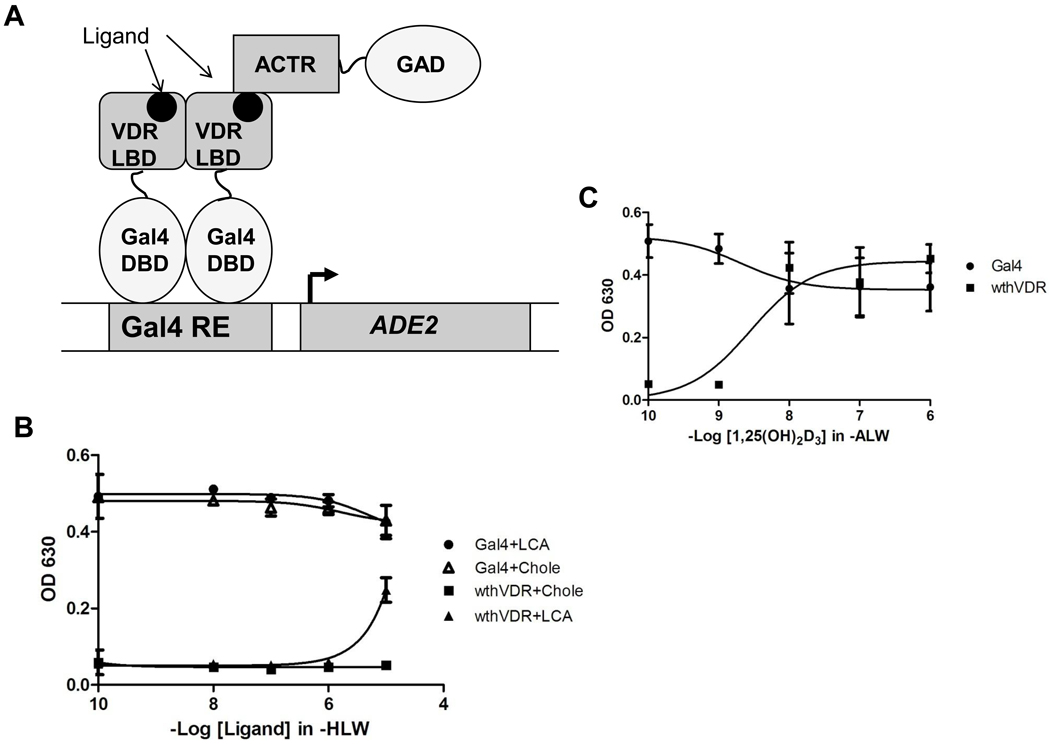
Chemical complementation is a multi-component system in which the survival of the Saccharomyces cerevisiae strain PJ69-4A is linked to the activation of a nuclear receptor by a ligand [5, 7, 30, 36]. The yeast strain used consists of Gal4 response elements (Gal4RE) controlling expression of genetic selection genes (i.e. HIS3, ADE2) [37]. In chemical complementation, a Gal4 DNA binding domain (GBD) is fused to a nuclear receptor ligand binding domain (LBD), and a nuclear receptor co-activator (CoAc) is fused to a Gal4 activation domain (GAD). The GBD: LBD fusion protein binds the Gal4RE, and upon ligand binding by the nuclear receptor LBD, a conformational change occurs, leading to the recruitment of the GAD:CoAc fusion protein. Transcription of the selective genes allows PJ69-4A to survive in media lacking these essential nutrients (Figure 2A) [5]. Thus, chemical complementation links the survival of yeast to the binding of a small molecule to the nuclear receptor of interest [7].
Figure 2. hVDR analysis.
(A) Chemical complementation is a genetic selection assay that links the survival of yeast to the presence of a small molecule ligand. The yeast strain that is used, PJ69-4A, contains Gal4 response elements controlling the expression of the ADE2, HIS3, and lacZ genes. A nuclear receptor ligand bind domain (LBD) is fused to a Gal4 DNA binding domain (GBD). This fusion protein is able to bind the Gal4 response element (Gal4RE), controlling expression of the ADE2 gene, required for adenine biosynthesis. Ligand binding by the nuclear receptor LBD leads to the recruitment of the corresponding co-activator (ACTR) fused to a Gal4 activation domain (GAD). Subsequent transcription of the regulated gene allows yeast to survive in media lacking adenine. (B) Results of chemical complementation of wild-type hVDR with LCA and cholecalciferol. Gal4 is a ligand independent transcription factor and is used as a positive control. Ligand-activated growth is observed with hVDR at 10 µM lithocholic acid. No ligand activated growth is observed for wild-type hVDR with cholecalciferol (Chole). Readings were taken at OD 630, measuring cell growth based on turbidity. (C) Results of chemical complementation of wild-type hVDR with 1,25(OH)2D3. Growth is observed at 10 nM 1,25(OH)2D3.
To determine the activity of the human vitamin D receptor (hVDR) in yeast, the hVDR gene was cloned from skin cDNA into a yeast expression vector, creating a fusion protein with the Gal4 DBD (pGBDhVDR). The resulting plasmid was tested in chemical complementation with the steroid receptor co-activator 1 (SRC-1) fused to the Gal4 activation domain. The two plasmids were tested with 1,25(OH)2D3, lithocholic acid (LCA), and cholecalciferol. As shown in Figure 2B, growth was observed at 10 µM LCA in histidine selective media (SC-HLW + 0.1 mM 3AT). Due to the leaky expression of the HIS3 gene, 3-amino-1,2,4-triazole (3AT) is added to reduce the background and therefore creates a less stringent selection marker in comparison to ADE2 [38]. hVDR showed growth at 10 nM 1,25(OH)2D3 in adenine selective media (SC-ALW) (Figure 2C). Ligand-activated growth was not observed with cholecalciferol at any ligand concentration (Figure 2B). This could be due to the lack of two hydroxyl groups that are present in 1,25(OH)2D3.
3.2 Creating and testing variants in yeast
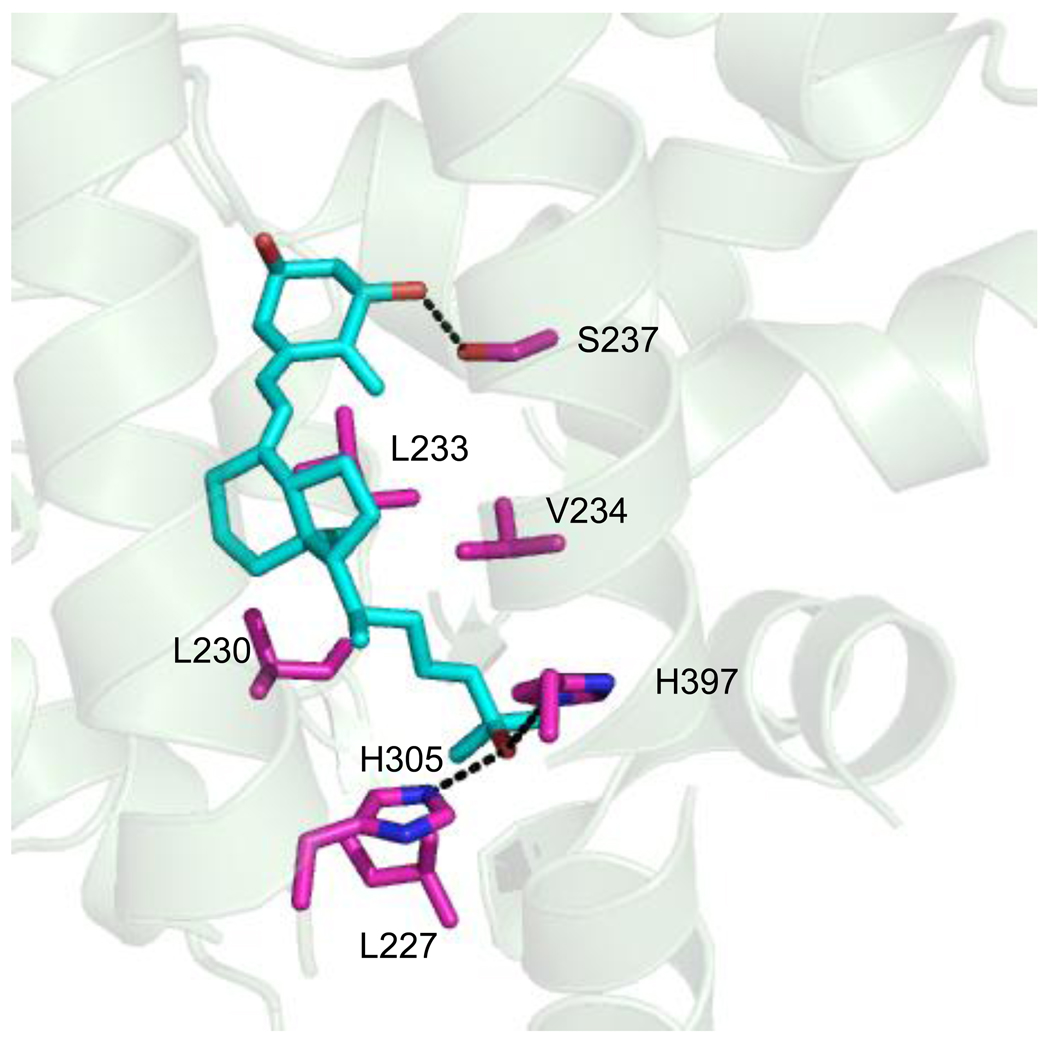
To determine the mutational tolerance of the residues within the ligand binding pocket of hVDR, a library was designed based on the crystal structure of hVDR bound to 1,25(OH)2D3, as well as previous mutational analysis and sequence alignments of the human VDR with other VDR orthologs. Using the crystal structure of wild-type hVDR with 1,25(OH)2D3 (PDB:1DB1), residues within four angstroms of the ligand were determined [11]. Four angstroms was chosen as the optimal range for contacts between the ligand and receptor. Twenty amino acid residues were found within this distance range, and seven of the twenty residues were chosen for mutagenesis (Figure 3).
Figure 3. hVDR library design.
The crystal structure of wild-type hVDR with its natural ligand 1,25(OH)2D3 (PDB:1DB1). L227, L230, L233, V234, S237, H305, and H397 were the residues chosen for the rationally designed library. (---) shows residues with hydrogen bonding potential in the library design.
Mutations were chosen based on polarity changes, as well as the shape and volume (bulkiness) of the residues within the pocket. L227, L230, L233, and V234 were targeted for mutation based on their proximity to the ligand and were modified to other hydrophobic residues, such as leucine, isoleucine, valine, phenylalanine, and methionine altering the pocket volume. S237, forming a hydrogen bond with the 1α-hydroxyl group of 1,25(OH)2D3, was modified to the hydrophobic residues leucine, isoleucine, valine, phenylalanine, methionine, alanine, as well as hydrogen bonding residues serine and threonine to determine if modifying the polarity of this residue would continue to allow binding and activation with LCA and 1,25(OH)2D3 [11]. Additionally, two histidines were also target residues: H305 which is located in a loop between helices 6 and 7, and H397 which is at the C-terminal of helix 11. Previously, the role of H305 in the LBD of hVDR was determined to be significant for the binding of 1,25(OH)2D3, as it forms a hydrogen bond with the 25-hydroxyl group of the ligand. As a result, it was decided that this position would be saturated [11, 18]. In the case of H397, which also forms a hydrogen bond with the 25-hydroxyl group of 1,25(OH)2D3, selected residues (tyrosine, lysine, glutamine, and asparagine) were chosen based on amino acid sequence alignments of various VDR orthologs [11, 39]. Overall, the combination of these designed mutations yields a theoretical library size (based on amino acids) of 4.5 × 106 variants.
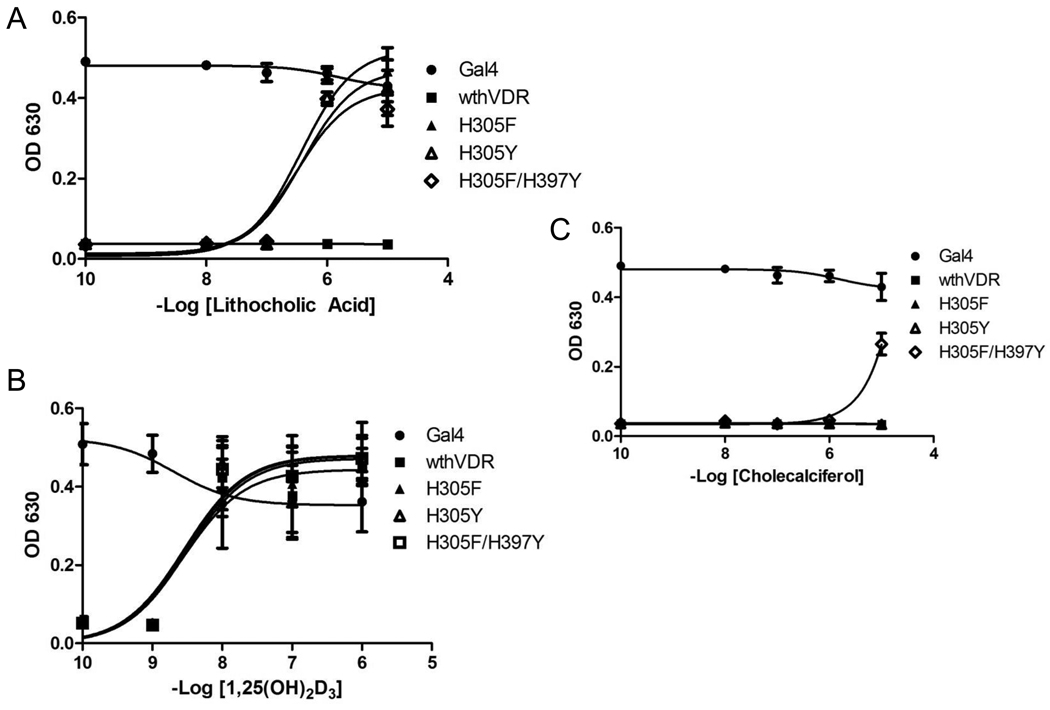
Individual oligonucleotides (oligos) containing degenerate base codes at the specific codon locations were designed and combined through hybridization and overlap extension PCR, to generate a library of hVDR mutant insert cassettes [28]. These cassettes were then transformed into yeast with a linearized vector cassette (see Materials and Methods section for details) and subjected to chemical complementation. A library size of 1.0 × 106 variants was achieved. Only two variants, H305F and H305Y, were found to have enhanced sensitivity to LCA when compared to wild-type hVDR. As shown in Figure 4A, these two variants grew in adenine selective media (-ALW) at 1 µM LCA, whereas wild-type hVDR showed no growth in adenine selective media. Both variants display growth at 100 nM LCA in histidine selective media (data not shown). Wild-type hVDR and the variants H305F and H305Y were analyzed for activation with 1,25(OH)2D3, for which growth was observed at 10 nM in adenine selective media for each (Figure 4B). Originally, based on alanine scanning mutagenesis, H305 was thought to form a crucial bond to 1,25(OH)2D3. However, Mizwicki et al., observed that the replacement of H305 with a non-polar romatic residue, phenylalanine, leads to a similar activation profile with 1,25(OH)2D3 as observed with wild-type hVDR [27, 40]. In addition this mutation was shown to turn the antagonist, MK, into a superagonist [27, 40]. Thus, MK and LCA take up a similar amount of space in the ligand binding pocket and therefore the assumption would be that the H305F mutation should lead to an increase in sensitivity towards LCA. This is most likely due to the larger molar volume of the phenylalanine (189.9 Å3) and tyrosine (196.3 Å3) in place of the histidine (153.2 Å3), leading to increased bulk/decreased volume in the pocket and a shift of the ligand [40, 41].
Figure 4. Testing variants with Chemical Complementation.
Chemical complementation in adenine selective media (-ALW) with wild-type hVDR and hVDR mutants. The variants tested are H305F, H305Y, and H305FH397Y. Gal4 is a ligand-independent transcription factor used as a positive control. All variants were tested for growth in the presence of LCA (A), 1,25(OH)2D3 (B), and cholecalciferol (C) with ligand concentrations ranging from 10−9 M − 10−5 M. With lithocholic acid all three variants display growth at 1 µM, whereas with cholecalciferol growth is only observed with H305F/H397Y at 10 µM. All of the variants and wild-type hVDR show growth at 10 nM 1,25(OH)2D3. The 10−10 M data point actually contains no ligand.
In an attempt to obtain a hVDR mutant that could bind cholecalciferol, a precursor in the 1,25(OH)2D3 biosynthetic pathway, random mutagenesis was performed with H305F and H305Y as templates. As mentioned previously, cholecalciferol lacks the hydrogen bonding potential of the 1,25(OH)2D3 ligand. H305F and H305Y both showed no growth in response to cholecalciferol in adenine selective media (Figure 4C). Random mutagenesis, specifically error-prone PCR, was chosen over rational design to determine if mutations beyond the four angstrom bias, could lead to an ideal conformation causing activation with cholecalciferol [42]. A library size of 4.2 × 103 variants was achieved. One hVDR mutant, H305F/H397Y, was activated at 1 µM LCA and 10 µM cholecalciferol in adenine selective media (Figure 4). These results were surprising as previous mutational analysis had shown that this residue forms an essential hydrogen bond with the 25-hydroxyl group of 1,25(OH)2D3, not present in cholecalciferol. However, the combination of both mutations in the double variant results in an increase in bulk, hence decreasing the overall volume of the ligand binding pocket. This suggests that a combination of volume changes in the pocket at residues 305 and 397 leads to the binding and activation with cholecalciferol, as seen with the H305F/H397Y mutant. With 1,25(OH)2D3, H305F/H397Y displayed activation at 10 nM in adenine selective media, similar to wild-type hVDR, H305F, and H305Y.
3.3 Testing variants in cell culture
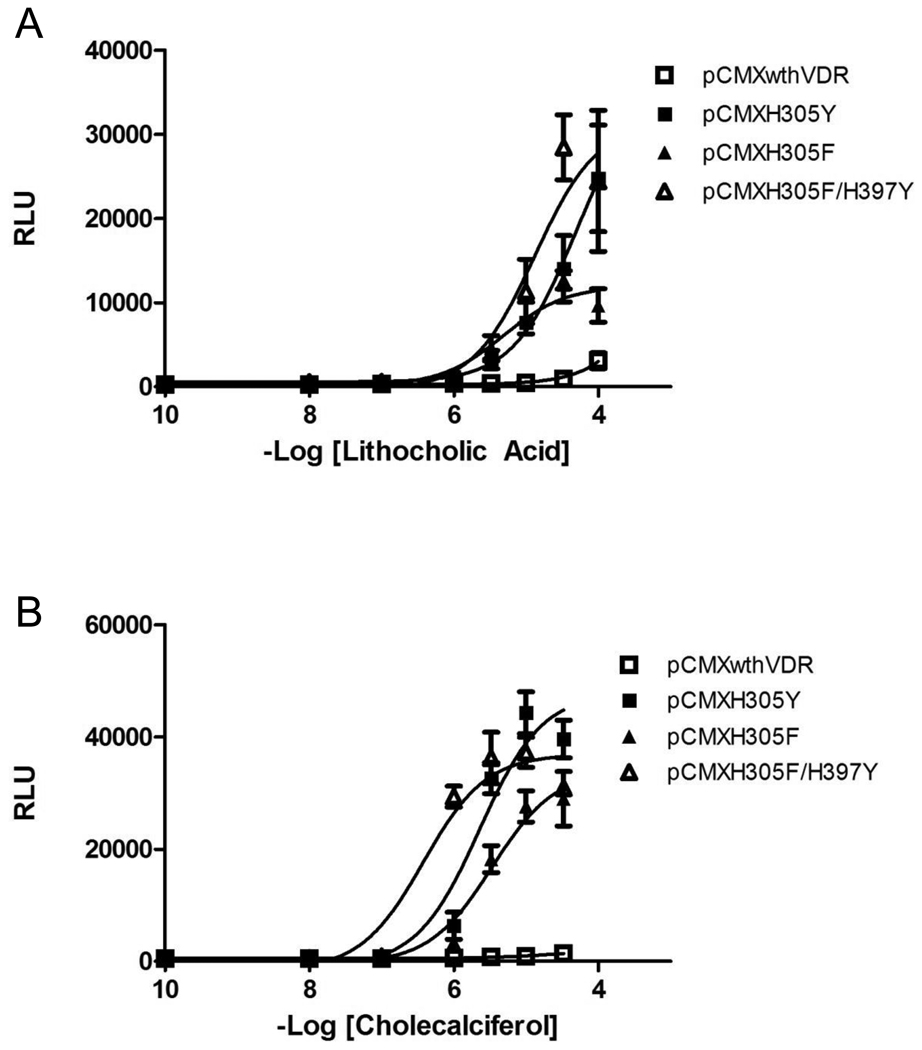
To determine whether the variants discovered with chemical complementation display the same or similar activity in mammalian cell assays, the variants were analyzed in human embryonic kidney 293T cells (HEK293T). The hVDR mutants were cloned into a mammalian expression vector (pCMX) containing a cytomegalovirus (CMV) promoter, with the Gal4 DNA binding domain fused to the variant VDR ligand binding domain (GBD:VDRLBD). This plasmid was co-transfected with a reporter plasmid containing four Gal4 response elements controlling the expression of the Renilla luciferase gene (p17*4TATAluc). As shown in Figure 5, hVDR mutants, H305F and H305Y, display activation at 3.3 µM LCA and 1 µM, respectively, in comparison to 100 µM LCA for wild-type hVDR as was the trend seen in yeast. H305F and H305Y show 40- and 74- fold activation, respectively, with LCA, whereas wild-type hVDR shows a 13-fold activation with LCA and only at the highest concentration (100 µM) of ligand (Figure 5, Table 1). The H305F/H397Y variant has a 100-fold increased sensitivity with LCA showing activation at 1 µM in comparison to 100 µM with wild-type hVDR and LCA. Additionally, H305F and H305Y display activation at 1 µM cholecalciferol resulting in a 54- and 65- fold activation with cholecalciferol, whereas no activation is observed with cholecalciferol and wild-type hVDR.
Figure 5. Mammalian Cell Culture.
Dose response curves for the activation of wild-type VDR and VDR mutants (H305F, H305Y, and H305F/H397Y) in response to LCA (A) and cholecalciferol (B). Activity is measured in relative light units (RLU) derived from the measurement of luciferase activity and normalization against a β-galactosidase internal standard. The three variants show an EC50 value of 10 µM with LCA (A) and 3 µM with cholecalciferol for H305F and H305Y, whereas H305F/H397Y shows and EC50 of 300 nM with cholecalciferol (B) in comparison to an EC50 of 100 µM or greater with LCA (A) and cholecalciferol (B) with wild-type VDR. The 10−10 M data point indicated on the graph does not contain ligand.
Table 1.
EC50s and fold activation (FA) of wthVDR and variants
|
Lithocholic Acid HEK293T |
Cholecalciferol HEK293T |
|||
|---|---|---|---|---|
| Variant | EC50 | FA | EC50 | FA |
| Wild-Type | 100 µM | 13 + 7 | >100 µM | 3 + 1 |
| H305F | 10 µM | 40 + 14 | 3 µM | 54 + 11 |
| H305Y | 10 µM | 74 + 25 | 3 µM | 65 + 6 |
| H305F/H397Y | 10 µM | 84 + 14 | 0.3 µM | 70 + 11 |
EC50 values, which are based on activation, and fold activation represent the average of two experiments in triplicate using HEK293T cells. EC50 values were extrapolated two ways. Method one used the best fit line of the raw data. Method two used a non-linear regression from GraphPad Prism. The fold activation was calculated by taking the highest level of activation and dividing this by the basal (no ligand) activation level. The error was obtained by calculating the standard deviation: σ = Square root (Σ[(X-μ)2]).
As shown in Table 1, EC50 values, the ligand concentration where 50% of the maximal response is seen, of 10 µM for LCA and 3 µM for cholecalciferol were observed with both H305F and H305Y, whereas, EC50 values for wild-type hVDR with both LCA and cholecalciferol were 100 µM or greater. The second generation double variant, H305F/H397Y, shows an EC50 value similar to that of H305F with LCA (10 µM), but an EC50 value of 300 nM is observed with cholecalciferol, in comparison to the 3 µM EC50 value for H305F. Thus, we have engineered a variant of hVDR that is now activated by submicromolar concentrations of cholecalciferol, along with an increased sensitivity to LCA.
3.4 Docking of hVDR mutants
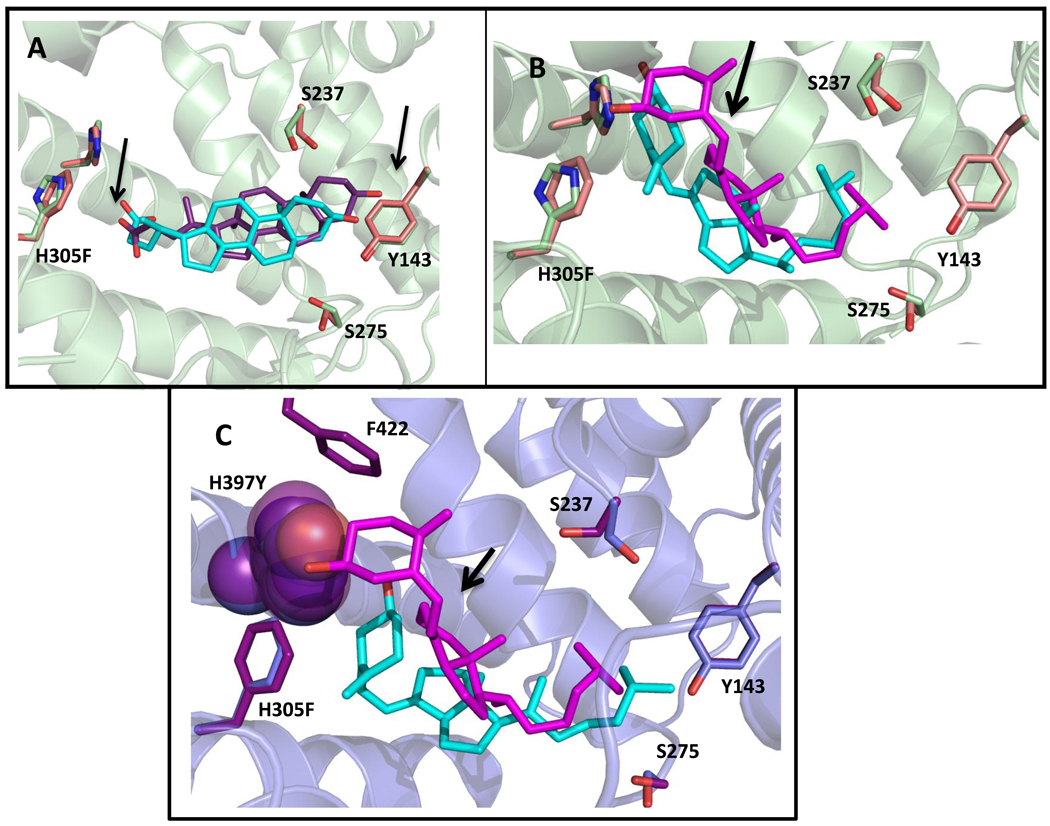
To visualize the impact of these mutations on ligand binding through in silico methods, LCA and cholecalciferol were docked into wild-type hVDR, H305F, and H305F/H397Y using AutoDock Vina (Figure 6) [35]. When comparing the superimposition of the docked structures of wild-type hVDR and H305F bound to lithocholic acid (Figure 6A), a slight shift in the ring system of the LCA towards Y143 and a small repositioning of the carboxyl group of LCA is observed. The bulkier residues, phenylalanine or tyrosine, in the place of histidine lead to an increase in volume at position 305. This increase seems to contribute to the repositioning and shifting of the ligand, resulting in more efficient interactions between residues in the pocket and LCA, specifically residues L227, L414, and V418 as previously mentioned by Mizwicki et al.; thus, leading to the higher sensitivity with this ligand [40].
Figure 6. Results of docking lithocholic acid and cholecalciferol to wild-type hVDR and variants.
Wild-type docked structures are green (residues) and cyan (ligand). Variant docked structures are pink/blue (residues) and purple/pink (ligand). In A, as shown with the arrows, an upward shift in the pocket and a change in position of the carboxyl of LCA, places the carboxyl group into closer proximity to the phenylalanine, which may be responsible for the increased activity of H305F with LCA. In B, cholecalciferol is flipped 180° from the conformation seen for 1,25(OH)2D3 in the wild-type crystal structure (Figure 3). This flipped conformation is also observed when 3-keto lithocholic acid is docked to hVDR [22]. The arrow shows a slight shift of the cholecalciferol bound to H305F (pink) when compared to wild-type (cyan). C shows a spacefill of H397 (blue) and H397Y (purple) in order to show the difference in volumes leading to a drastic shift of the cholecalciferol to a more bowl-shaped conformation in the pocket in H305F/H397Y (cyan) in comparison to H305F (pink). The distance between the hydroxyl group of cholecalciferol and the hydrogen bonding atom on residue 397 (H and Y) is annotated. The binding energies are as follows: wild-type hVDR/LCA (−9.7 kcal/mol), H305F/LCA (−9.6 kcal/mol), wild-type hVDR/cholecalciferol (−10.4 kcal/mol), H305F/cholecalciferol (−10.6 kcal/mol), and H305F/H397Y/cholecalciferol (−10.2 kcal/mol).
In the superimposition of wild-type hVDR and H305F with cholecalciferol, a shift upward is observed with cholecalciferol docked to H305F (Figure 6B). Cholecalciferol, in the docked structures, shows a 180° flip in conformation in the binding pocket when compared to the crystal structure of wild-type hVDR and 1,25(OH)2D3. Previously Yamada et al. observed a 180° shift with 3-ketolithocholic acid in the hVDR pocket, similar to the observations with cholecalciferol (Figure 6B) [22]. Also, in silico structures with cholecalciferol bound to wild-type and H305F (Figure 6B), display a bent/kinked conformation of the ligand, whereas the LCA docked structures have a more linear, bowl-shaped conformation. The crystal structure of hVDR with 1,25(OH)2D3, and previous work with different hVDR agonists suggests that a bowl-shaped conformation of the ligand is preferred [19–21]. Therefore, the bent/kinked conformation with cholecalciferol and wild-type hVDR may be leading to a non-optimal conformation by not forming key contacts with R274, Y143, and S278 (Figure 6B, Figure 3A). In the case of the double variant H305F/H397Y, the combination of both mutations causes a drastic shift in the cholecalciferol when compared to H305F. As seen in Figure 6C, the cholecalciferol is in a bowl-shaped conformation, resembling the conformation of 1,25(OH)2D3 and the wild-type receptor. This shift observed with cholecalciferol with the double variant is most likely due to the increased bulk at 397 due to the presence of the tyrosine instead of a histidine, leading to an increase in the bulk at that end of the pocket. This increase in bulk perhaps leads more favorable hydrophobic van der Waals interactions between residue 397 and F422, an important residue in the AF-2 domain of hVDR. This may be leading to a stabilizing effect, such as tighter co-activator binding, as shown previously [27, 43].
4. Discussion
Unique characteristics of the ligand binding domains of different nuclear receptors contribute to the variety of ligand/receptor pairs, leading to their different roles in various regulatory pathways. Several factors, such as the volume, shape, and polarity of the ligands and residues that are present in the pocket contribute to specific interactions that affect ligand/nuclear receptor binding [1, 2]. As a member of the nuclear receptor superfamily, VDR is involved in a number of biological processes and binds a variety of ligands. A disruption of the critical network of interactions between the ligand and the receptor has been connected to disease, such as in the case of a point mutation, H305Q, causing Type II Rickets, leading to a 10-fold decrease in activation with 1,25(OH)2D3 [16]. To further understand the specific molecular interactions required for binding and activation of hVDR, a library was designed challenging the mutational tolerance of hVDR’s binding pocket.
Initially, a designed library for hVDR included mutations at seven key residues, based on the crystal structure of hVDR with 1,25(OH)2D3, sequence alignments with VDR orthologs, and previous mutational studies [17, 18]. Through the rationally designed library, variants H305F and H305Y were discovered, displaying a 100-fold increase in sensitivity to LCA over wild-type hVDR in both yeast and in mammalian cell culture, as well as displaying growth at 10 nM 1,25(OH)2D3 as is also seen with wild-type hVDR. An EC50 value of 10 µM LCA was observed as shown in Figure 5A. It has been shown that changing residue 305 from histidine to glutamine causes a decrease in the affinity of the receptor for 1,25(OH)2D3. However, a phenylalanine or tyrosine at the same position have been shown to have no affect on binding and activation in response to 1,25(OH)2D3, but does lead to increased sensitivity to LCA (Figure 4A, 4B) [27, 40].
In silico docking showed that the mutations, H305F and H305Y, cause a shift in the position of LCA in the ligand binding pocket. We hypothesize that this ligand shift may be due to the change in volume of the side chains of tyrosine or phenylalanine, compared to histidine. The decrease in the overall volume of the pocket may possibly be leading to closer interactions between LCA and the residues in the pocket, not only by the upward shift of LCA but also the repositioning of the carboxyl group of LCA [40, 41]. The lack of potential hydrogen bonding for H305 in the H305F variant indicates that polarity at this position may not be as crucial for activation as a modification in the shape and/or volume of the pocket seems to be.
When H305F was subjected to random mutagenesis using error-prone PCR in an attempt to discover a variant capable of binding cholecalciferol, a double variant, H305F/H397Y, displayed LCA activation profiles similar to those of H305F but a ∼10-fold increased sensitivity towards cholecalciferol, displaying an EC50 of 300 nM in mammalian cells (Figure 4, Figure 5). In silico docking of cholecalciferol to the H305F variant using AutoDock Vina displays a similar conformation to that of cholecalciferol docked to the wild-type receptor (Figure 6B) [35]. However, the increased bulk of the phenylalanine is most likely leading to increased contacts cholecalciferol and therefore displaying ligand-activation in mammalian cells. In the double variant, when comparing the spacefill of histidine and tyrosine at 397, the bulkier tyrosine results in a shift to a more bowl-shaped conformation of the cholecalciferol in the pocket, perhaps allowing for more favorable van der Waals interactions between cholecalciferol and the surrounding residues. In the docked structures, the hydroxyl group of cholecalciferol does not seem to be within hydrogen bonding distance of the histidine (6.09 Å) or tyrosine (6.59 Å) of the variants. However, this could be an artifact of the docking program as the protein is held rigid during docking; therefore, the residues may be in a different conformation and may be able to form a hydrogen bond with cholecalciferol. In addition, water molecules are known to form hydrogen bonds with ligands, such as in the case of rat VDR [22, 44]. Thus, a water molecule may be present in the ligand binding pocket capable of forming a hydrogen bond with H397Y and cholecalciferol [22]. As previously mentioned, H397Y also leads to increased favorable van der Waals interactions with F422, possibly leading to tighter co-activator binding and improved activation [27, 43].
In conclusion, more insight into the relationship and crucial interactions between hVDR and various ligands has been determined. Single point mutations at position 305, H305F and H305Y, result in an increased sensitivity with LCA and cholecalciferol. A hVDR mutant H305F/H397Y, is able to bind cholecalciferol at submicromolar concentrations in mammalian cells, whereas wild-type hVDR does not display activation with this ligand. The addition of the H397Y mutation in combination with H305F, introduces a modification in the volume of the pocket, leading to activation with cholecalciferol. Future work, including in vitro binding studies, as well as additional mutational analysis, to determine if the tyrosine is necessary for maximal activation with cholecalciferol will be performed. This will allow a more comprehensive understanding of the dynamic interactions between the wild-type hVDR and these variants with their potential ligands.
Acknowledgements
We would like to acknowledge Hally Shaffer for considerable help with Pymol (DeLano Scientific LLC, USA). We would also like to offer many thanks to the members of our research group for their help with the preparation of this manuscript. This work was supported by the National Institute of Health (Grant # 5R01AI064817-02).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Amanda M. Ousley, Email: gth783d@mail.gatech.edu.
Hilda S. Castillo, Email: hilda.castillo@chemistry.gatech.edu.
Anna Duraj-Thatte, Email: annaduraj@gatech.edu.
Donald F. Doyle, Email: donald.doyle@chemistry.gatech.edu.
Bahareh Azizi, Email: bahareh.azizi@chemistry.gatech.edu.
References
- 1.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The Nuclear Receptor Superfamily - The Second Decade Cell. 1995;83(6):835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Egea PF, Mitschler A, Rochel N, Ruff M, Chambon P, Moras D. Crystal structure of the human RXR alpha ligand-binding domain bound to its natural ligand: 9-cis retinoic acid. Embo J. 2000;19(11):2592–2601. doi: 10.1093/emboj/19.11.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khorasanizadeh S, Rastinejad F. Nuclear-receptor interactions on DNA-response elements. EMBO J. 2001;26(6):384–390. doi: 10.1016/s0968-0004(01)01800-x. [DOI] [PubMed] [Google Scholar]
- 4.Ottow E, Weinmann H, editors. Nuclear Receptors as Drug Targets. Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; 2008. [Google Scholar]
- 5.Schwimmer LJ, Rohatgi P, Azizi B, Seley KL, Doyle DF. Creation and discovery of ligand-receptor pairs for transcriptional control with small molecules. Proc. Natl. Acad. Sci. U. S. A. 2004;101(41):14707–14712. doi: 10.1073/pnas.0400884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang JC. Finding Primary Targets of Transcriptional Regulators. Cell Cycle. 2005;4(3):356–358. doi: 10.4161/cc.4.3.1521. [DOI] [PubMed] [Google Scholar]
- 7.Azizi B, Chang EI, Doyle DF. Chemical complementation: small-molecule-based genetic selection in yeast. Biochem. Biophys. Res. Commun. 2003;306(3):774–780. doi: 10.1016/s0006-291x(03)01039-8. [DOI] [PubMed] [Google Scholar]
- 8.Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr. Opin. Cell Biol. 1998;10(3):384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 9.Zhang ZD, Burch PE, Cooney AJ, Lanz RB, Pereira FA, Wu JQ, Gibbs RA, Weinstock G, Wheeler DA. Genomic Analysis of the Nuclear Receptor Family: New Insights Into Structure, Regulation, and Evolution From the Rat Genome. Genome Res. 2004;14(4):580–590. doi: 10.1101/gr.2160004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hourai S, Rodrigues LC, Antony P, Reina-San-Martin B, Ciesielski F, Magnier BC, Schoonjans K, Mourino A, Rochel N, Moras D. Structure-Based Design of a Superagonist Ligand for the Vitamin D Nuclear Receptor. Chem. Biol. 2008;15(4):383–392. doi: 10.1016/j.chembiol.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 11.Rochel N, Wurtz JM, Mitschler A, Klaholz B, Moras D. The Crystal Structure of the Nuclear Receptor for Vitamin D Bound to Its Natural Ligand. Mol. Cell. 2000;5(1):173–179. doi: 10.1016/s1097-2765(00)80413-x. [DOI] [PubMed] [Google Scholar]
- 12.Vaisanen S, Ryhanen S, Saarela JTA, Maenpaa PH. Structure-function studies of new C-20 epimer pairs of vitamin D3 analogs. Eur. J. Biochem. 1999;261(3):706–713. doi: 10.1046/j.1432-1327.1999.00318.x. [DOI] [PubMed] [Google Scholar]
- 13.Nayeri S, Kahlen JP, Carlberg C. The high affinity ligand binding conformation of the nuclear 1,25-dihydroxyvitamin D3 receptor is functionally linked to the transactivation domain 2 (AF-2) Nucleic Acids Res. 1996;24(22):4513–4518. doi: 10.1093/nar/24.22.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jurutka PW, Thompson PD, Whitfield GK, Eichhorst KR, Hall N, Dominguez CE, Hsieh JC, Haussler CA, Haussler MR. Molecular and Functional Comparison of 1,25-Dihydroxyvitamin D3 and the Novel Vitamin D Receptor Ligand, Lithocholic Acid, in Activating Transcription of Cytochrome P450 3A4. J. Cell. Biochem. 2005;94(5):917–943. doi: 10.1002/jcb.20359. [DOI] [PubMed] [Google Scholar]
- 15.Jones G, Strugnell SA, DeLuca HF. Current Understanding of the Molecular Actions of Vitamin D. Physiol. Rev. 1998;78(4):1193–1231. doi: 10.1152/physrev.1998.78.4.1193. [DOI] [PubMed] [Google Scholar]
- 16.Malloy PJ, Eccleshall TR, Gross C, VanMaldergem L, Bouillon R, Feldman D. Hereditary vitamin D resistant rickets caused by a novel mutation in the vitamin D receptor that results in decreased affinity for hormone and cellular hyporesponsiveness. J. Clin. Invest. 1997;99(2):297–304. doi: 10.1172/JCI119158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada S, Yamamoto K, Masuno H. Structure-Function Analysis of Vitamin D and VDR Model. Curr. Pharm. Design. 2000;6(7):733–748. doi: 10.2174/1381612003400353. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto K, Masuno H, Choi M, Nakashima K, Taga T, Ooizumi H, Umesono K, Sicinska W, VanHooke J, DeLuca HF, Yamada S. Three-dimensional modeling of and ligand docking to vitamin D receptor ligand binding domain. Proc. Natl. Acad. Sci. U. S. A. 2000;97(4):1467–1472. doi: 10.1073/pnas.020522697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tocchini-Valentini G, Rochel N, Wurtz JM, Mitschler A, Moras D. Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands. Proc. Natl. Acad. Sci. U. S. A. 2001;98(10):5491–5496. doi: 10.1073/pnas.091018698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tocchini-Valentini G, Rochel N, Wurtz JM, Moras D. Crystal structures of the vitamin D nuclear receptor liganded with the vitamin D side chain analogues calcipotriol and seocalcitol, receptor agonists of clinical importance. Insights into a structural basis for the switching of calcipotriol to a receptor antagonist by further side chain modification. J. Med. Chem. 2004;47(8):1956–1961. doi: 10.1021/jm0310582. [DOI] [PubMed] [Google Scholar]
- 21.Hourai S, Fujishima T, Kittaka A, Suhara Y, Takayama H, Rochel N, Moras D. Probing a water channel near the A-ring of receptor-bound 1 alpha,25-dihydroxyvitamin D3 with selected 2 alpha-substituted analogues. J. Med. Chem. 2006;49(17):5199–5205. doi: 10.1021/jm0604070. [DOI] [PubMed] [Google Scholar]
- 22.Yamada S, Yamamoto K. Ligand recognition by vitamin D receptor: Total alanine scanning mutational analysis of the residues lining the ligand binding pocket of vitamin D receptor. Curr. Top. Med. Chem. 2006;6(12):1255–1265. doi: 10.2174/156802606777864881. [DOI] [PubMed] [Google Scholar]
- 23.Nakajima S, Hsieh JC, Jurutka PW, Galligan MA, Haussler CA, Whitfield GK, Haussler MR. Examination of the Potential Functional Role of Conserved Cysteine Residues in the Hormone Binding Domain of the Human 1,25-dihydroxyvitamin D-3 Receptor. J. Biol. Chem. 1996;271(9):5143–5149. [PubMed] [Google Scholar]
- 24.Solomon C, Macoritto M, Gao XL, White JH, Kremer R. The unique tryptophan residue of the vitamin D receptor is critical for ligand binding and transcriptional activation. J. Bone Miner. Res. 2001;16(1):39–45. doi: 10.1359/jbmr.2001.16.1.39. [DOI] [PubMed] [Google Scholar]
- 25.Reddy MD, Stoynova L, Acevedo A, Collins ED. Residues of the human nuclear vitamin D receptor that form hydrogen bonding interactions with the three hydroxyl groups of 1alpha,25-dihydroxyvitamin D3. J Steroid Biochem Mol Biol. 2007;103(3–5):347–351. doi: 10.1016/j.jsbmb.2006.12.045. [DOI] [PubMed] [Google Scholar]
- 26.Swamy N, Xu WR, Paz N, Hsieh JC, Haussler MR, Maalouf GJ, Mohr SC, Ray R. Molecular Modeling, Affinity Labeling, and Site-Directed Mutagenesis Define the Key Points of Interaction between the Ligand-Binding Domain of the Vitamin D Nuclear Receptor and 1 alpha,25-Dihydroxyvitamin D3. Biochemistry. 2000;39(40):12162–12171. doi: 10.1021/bi0002131. [DOI] [PubMed] [Google Scholar]
- 27.Mizwicki MT, Bula CM, Bishop JE, Norman AW. New insights into Vitamin D sterol-VDR proteolysis, allostery, structure-function from the perspective of a conformational ensemble model. J Steroid Biochem Mol Biol. 2007;103(3–5):243–262. doi: 10.1016/j.jsbmb.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higuchi R, Krummel B, Saiki RK. A General-Method of Invtro Preparation and Specific Mutagenesis of DNA Fragments - Study of Protein and DNA Interactions. Nucleic Acids Res. 1988;16(15):7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gietz RD, Woods RA. Guide to Yeast Genetics and Molecular and Cell Biology, Pt B. San Diego: Academic Press Inc; 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method; pp. 87–96. [Google Scholar]
- 30.Taylor JL, Rohatgi P, Spencer HT, Doyle DF, Azizi B. Characterization of a molecular switch system that regulates gene expression in mammalian cells through a small molecule. BMC Biotechnol. 10:12. doi: 10.1186/1472-6750-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sali A, Blundell TL. Comparative Protein Modeling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 32.Damborsky J, Prokop M, Koca J. TRITON: graphic software for rational engineering of enzymes. Trends Biochem.Sci. 2001;26(1):71–73. doi: 10.1016/s0968-0004(00)01708-4. [DOI] [PubMed] [Google Scholar]
- 33.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Sanner MF. Python: A programming language for software integration and development. J. Mol. Graph. 1999;17(1):57–61. [PubMed] [Google Scholar]
- 35.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2009:456–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baker K, Bleczinski C, Lin H, Salazar-Jimenez G, Sengupta D, Krane S, Cornish VW. Chemical complementation: a reaction-independent genetic assay for enzyme catalysis. Proc Natl Acad Sci U S A. 2002;99(26):16537–16542. doi: 10.1073/pnas.262420099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144(4):1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Struhl K, Davis RW. Production of a Functional Eukaryotic Enzyme in Escherichia-coli -Cloning and Expression of Yeast Structural Gene for Imidazoleglycerolphosphate Dehydratase (HIS3) Proc. Natl. Acad. Sci. U. S. A. 1977;74(12):5255–5259. doi: 10.1073/pnas.74.12.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruau D, Duarte J, Ourjdal T, Perriere G, Laudet V, Robinson-Rechavi M. Update of NUREBASE: nuclear hormone receptor functional genomics. Nucleic Acids Res. 2004;32:D165–D167. doi: 10.1093/nar/gkh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizwicki MT, Bula CM, Mahinthichaichan P, Henry HL, Ishizuka S, Norman AW. On the mechanism underlying (23S)-25-dehydro-1alpha(OH)-vitamin D3-26,23-lactone antagonism of hVDRwt gene activation and its switch to a superagonist. J Biol Chem. 2009;284(52):36292–36301. doi: 10.1074/jbc.M109.042069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reichert J, Suhnel J. The IMB Jena Image Library of Biological Macromolecules: 2002 update. Nucleic Acids Res. 2002;30(1):253–254. doi: 10.1093/nar/30.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bloom JD, Arnold FH. In the light of directed evolution: Pathways of adaptive protein evolution. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9995–10000. doi: 10.1073/pnas.0901522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carlberg C. Molecular basis of the selective activity of vitamin D analogues. J Cell Biochem. 2003;88(2):274–281. doi: 10.1002/jcb.10337. [DOI] [PubMed] [Google Scholar]
- 44.Vanhooke JL, Benning MM, Bauer CB, Pike JW, DeLuca HF. Molecular structure of the rat vitamin D receptor ligand binding domain complexed with 2-carbon-substituted vitamin D3 hormone analogues and a LXXLL-containing coactivator peptide. Biochemistry. 2004;43(14):4101–4110. doi: 10.1021/bi036056y. [DOI] [PubMed] [Google Scholar]