Abstract
Biological media affect the physicochemical properties of cationic lipid-DNA complexes (lipoplexes) and can influence their ability to transfect cells. To develop new lipids for efficient DNA delivery, the influence of serum-containing media on the structures and properties of the resulting lipoplexes must be understood. To date, however, a clear and general picture of how serum-containing media influences the structures of lipoplexes has not been established. Some studies suggest that serum can disintegrate lipoplexes formed using certain types of cationic lipids, resulting in inhibition of transfection. Other studies have demonstrated that lipoplexes formulated from other lipids are stable in the presence of serum and able to transfect cells efficiently. In this paper, we describe the influence of serum-containing media on lipoplexes formed using the redox-active cationic lipid bis(n-ferrocenylundecyl)dimethylammonium bromide (BFDMA). This lipoplex system promotes markedly decreased levels of transgene expression in COS-7 cells as serum concentrations are increased from 0% to 2%, 5%, 10%, and 50% (v/v). To understand the cause of this decrease in transfection efficiency, we used cryogenic transmission electron microscopy (cryo-TEM) and measurements of zeta potential to characterize lipoplexes in cell culture media supplemented with 0%, 2%, 5%, 10%, and 50% serum. Cryo-TEM revealed that in serum-free media BFDMA lipoplexes form onion-like, multilamellar nanostructures. However, the presence of serum in the media caused disassociation of the intact multilamellar lipoplexes. At low serum concentrations (2% and 5%), DNA threads appeared to separate from the complex, leaving the nanostructure of the lipoplexes disrupted. At higher serum concentration (10%) disassociation increased, and bundles of multilamellae were discharged from the main multilamellar complex. In contrast, lipoplexes characterized in serum-free aqueous salt (Li2SO4) medium and in OptiMEM cell culture medium (no serum) did not exhibit significant structural changes. The zeta potentials of lipoplexes in serum-free media (salt medium and cell culture medium) were similar (e.g., approximately −35mV). Interestingly, the presence of serum caused zeta potentials to become less negative (about −20mV in OptiMEM and −10mV in Li2SO4), even though serum contains negatively charged entities that have been demonstrated to lead to more negative zeta potentials in other lipoplex systems. The combined measurements of zeta potential and cryo-TEM are consistent with the proposition that DNA threads separate from the lipoplex in the presence of serum, resulting in a decrease in the net negative charge of the surface of the lipoplex.
Keywords: lipoplex, serum, media effect, cationic lipids, transfection
Introduction
Cationic lipids are attractive as vectors for the non-viral delivery of DNA because they can interact with negatively charged DNA to form complexes (lipoplexes) that protect DNA from degradation by nucleases and facilitate internalization by cells. Despite these advantages and successes with a variety of different cationic lipid systems, many lipoplexes suffer from low transfection efficiencies, and the reasons for this remain poorly understood (1,2).
The factors that govern the transfection efficiency of cationic lipid systems can be divided into three major groups. The first one is the type (cationic, zwitterionic, etc.) and composition of the lipids used and the sizes and structures of the nucleic acid components (plasmids, oligonucleotides, etc.), both of which can influence the physicochemical properties of lipoplexes significantly. The second and third groups correspond to solution conditions (ionic strength, pH, etc.) and interactions with biological species in extracellular and intracellular environments that can also impact and change the physicochemical properties of lipoplexes in important ways (3).
Because lipoplexes can form different liquid crystalline nanostructures, including the LαC multilamellar phase and the inverted hexagonal HIIC phase (4-7), they can be extremely sensitive to changes in environmental conditions. Unfortunately, physical characterization of lipoplexes is often conducted in simple aqueous solutions of electrolyte (e.g., buffer) that do not accurately mimic the complexity of the biological media in which the lipoplexes will be used. After the lipoplexes are prepared, they are introduced to biological media such as cell culture media and blood serum that may alter their structural and physicochemical properties significantly (4-8). To predict whether a certain lipidic system is suited for gene delivery applications, it should be characterized in the relevant biological medium, or at least in the presence of serum-containing media to simulate biological conditions.
Simberg et al. (9) studied the effect of blood serum on the structural and physicochemical properties of DOTAP-based (1,2-dioleoyl-3-trimethylammonium-propane) lipoplex systems. By using cryo-TEM, zeta potential, electrical surface potential Ψ0 measurements, and FRET (fluorescence resonance energy transfer) techniques they showed that DOTAP/cholesterol lipoplexes are stable (i.e., their nanostructure is not altered) in serum, while lipoplexes formed using DOTAP and DOPE (dioleoyl phosphatidylethanolamine, which has been demonstrated to induce an inverse hexagonal phase) were unstable and disintegrated in the presence of serum. These authors suggested that serum proteins tend to adhere to the surfaces of the DOTAP/cholesterol lipoplexes and enlarge the aggregate size, rather than causing lipid-DNA disassociation.
Eliyahu et al. (10) also studied the effect of serum on DOTAP/cholesterol lipoplexes and compared it to the effect of serum on dextran-spermine polyplexes. Using FRET they showed that addition of serum increased the lipid-DNA average molecular distance (from 5.25 to 5.79 nm) and decreased the polymer-DNA average distance (from 7.66 to 6.14 nm). However, FRET efficiency experiments showed that the serum proteins did not induce significant DNA disassociation from either complex, suggesting that DNA has higher affinity than serum proteins to both cationic carriers. Their findings were in good agreement with the findings of Simberg et al. (9), showing that DOTAP/cholesterol lipoplexes preserved their shape and morphology after addition of serum, and that serum proteins did not penetrate into the vesicles, but associated only with the outer membranes.
In contrast to the these studies, Li et al. (11) found via FRET and ethidium bromide (EtBr) intercalation assays that for several lipidic vectors of different compositions, serum induced the disintegration of lipoplexes and resulted in DNA release. They showed that DOTAP/DOPE lipoplexes disintegrated rapidly during serum incubation, and exhibited low levels of transfection efficiency compared to DOTAP/cholesterol and DOTAP lipoplexes.
Yang and Huang (12) investigated the inhibition effect of serum on lipofection. They showed that serum has an inhibitory effect on transfection using DOTAP/DOPE, DC (3ß-[N-(N’,N’-dimethylaminoethane)-carbamoyl])-cholesterol/DOPE, and Lipofectamine lipoplex systems. However, in the DC-cholesterol and DOTAP containing systems, they found that inhibition could be overcome by optimizing the lipid-to-DNA charge ratio. To understand the reason for this serum inhibition, they removed the negatively charged proteins from the serum by DEAE-Sephacel chromatography, and the results showed that the inhibitory effect of the serum was eliminated. Therefore, one conclusion of this study was that the negatively charged proteins in the serum were responsible for the low transfection efficiency.
In spite of all these past studies, the extents to which (and the mechanisms through which) the presence of serum can lead to lipoplex disruption and disintegration remain poorly understood. In this paper we report on the influence of biological media (e.g., both serum-free and serum-containing cell culture media) on the stability of lipoplexes formulated using a novel, ferrocene-containing cationic lipid.
We have demonstrated in past studies that the redox active cationic lipid bis(n-ferrocenylundecyl)dimethylammonium bromide (BFDMA) is an effective cell transfection agent when it is used in its reduced state in a serum-free medium. Our past studies also demonstrate that the presence of serum can reduce levels of transfection significantly; for example the efficiency in media containing 10% serum was found to be lower by an order of magnitude compared to levels of transfection observed when serum-free medium is used (13-14).
This current study sought to investigate the influence of serum on the structures and properties of reduced BFDMA lipoplexes that could underlie these differences in cell transfection.
Pizzey et al. (8) characterized the complexes that BFDMA forms with plasmid DNA using cryo-TEM and small-angle neutron scattering (SANS). The lipid in its reduced state exhibited well-defined multilamellar onion-like lipoplexes, in a large range of concentrations, and lipid-to-DNA charge ratios. These past observations provide a basis from which to investigate changes in the structure of BFDMA/DNA lipoplexes that may occur upon the introduction of serum. We used cryo-TEM and measurements of zeta potential to characterize reduced BFDMA lipoplexes in (serum-free) OptiMEM cell culture medium, serum-containing OptiMEM cell culture medium, and serum-containing aqueous solutions of Li2SO4. The structural and physicochemical properties of these lipoplexes were correlated to the results of transfection experiments using the COS-7 cell-line and culture media containing increasing amounts of serum. Our results show that the presence of serum caused disassociation of DNA from the lipid-DNA complexes. As the serum concentration increased, we observed enhanced levels of DNA disassociation that correlated with decreased levels of cell transfection. We use cryo-TEM to provide levels of detail regarding the lipoplex disassociation process that have not been reported previously.
Experimental section
Materials
BFDMA was synthesized as described elsewhere (16). Adult bovine blood serum was purchased from Biological Industries Ltd. Kibbutz Beit Haemek, Israel or from Invitrogen (Carlsbad, CA). Plasmid DNA encoding enhanced green fluorescent protein (pEGFP-N1, 4.7 kb, >95% supercoiled) and firefly luciferase (pCMV-Luc, 6.2 kb) >90% supercoiled) were purchased from Elim Biopharmaceuticals, Inc., San Francisco, CA. OptiMEM cell culture medium, Dulbecco’s Modified Eagle’s Medium (DMEM), and fetal bovine serum (FBS) were purchased from Invitrogen (Carlsbad, CA). Bicinchoninic acid (BCA) protein assay kits were purchased from Pierce (Rockford, IL). Glo Lysis Buffer and Steady-Glo Luciferase Assay kits were purchased from Promega Corporation (Madison, WI). All salt solutions and buffers were prepared with deionized water (18.2 MΩ).
Preparation of Lipoplexes
Solutions of lipoplexes were prepared in the following manner: First, reduced BFDMA was dissolved in 1 mM Li2SO4 (pH=5), and the resulting solution was sonicated for about 15 minutes until all BFDMA was suspended in solution to produce a 1 mM reduced BFDMA solution. Then, to prepare lipoplexes in Li2SO4 solution, 2.9 mg/ml DNA in H2O was added to the 1mM reduced BFDMA lipid solution to produce 0.905 mM BFDMA concentration with a 1.1:1 lipid-to-DNA charge ratio. The solution of BFDMA lipoplexes in Li2SO4 salt solution was added to OptiMEM cell culture medium in a concentration that was 0.93x the original purchased concentration. The final BFDMA concentration was 0.657 mM with a 1.1:1 lipid to DNA charge ratio. Serum solution was added to the lipoplex solutions (with or without OptiMEM cell culture medium) to produce solutions containing 2%, 5%, 10%, and 50% (v/v) serum. Lipoplexes for zeta potential and transfection experiments were diluted to 40 μM and 8 μM BFDMA concentrations (respectively), and DNA concentration was adjusted to maintain the 1.1:1 lipid to DNA charge ratio. The lipoplex concentrations and the lipid-to-DNA charge ratio were chosen based on our previous studies demonstrating efficient transfection and consistency in morphology (8, 13). We note that the pH of the OptiMEM-containing solutions and the serum-containing solution was about 7.5, while the pH of the Li2SO4 containing solution was about 5.
Transfection and Gene Expression Assays
Transfection experiments were conducted using COS-7 cells, which were grown in clear (for pEGFP-N1 expression) or opaque (for pCMV-Luc expression) polystyrene 96-well culture plates. The initial seeding densities of the cells were 15000 cells/well in 200 μl of growth medium (90% Dulbecco’s modified Eagle’s medium, 10% fetal bovine serum, 100 units/ml Penicillin, and Streptomycin 100 μg/ml). The cells were incubated at 37 °C for 24 hours and transfection experiments were conducted at approximately 80% confluence. The culture medium was replaced by 200 μl OptiMEM with or without added serum, followed by addition of 50 μl of a previously-prepared lipoplex solution. After the addition of lipoplexes, the cells were incubated for 4 hours at 37°C, after which time lipoplex-containing medium was aspirated from all wells and replaced with 200 μl of fresh serum-containing medium. Samples were incubated for additional 48 hours, and then gene expression was characterized. EGFP-N1 expression was characterized using phase-contrast and fluorescence microscopy (Olympus IX70 fluorescence microscope with MetaVue v.7.1.2.0 Software package). Luciferase expression was determined using a commercially available luminescence based luciferase assay kit according to the manufacturer’s protocol. Samples were compared with signals from control wells and/or normalized against total cell protein in each respective well, using a commercially available bicinchoninic acid (BCA) assay kit (Pierce).
Cryogenic Transmission Electron Microscopy
Vitrified specimens of lipoplex solutions were prepared in the following manner. A drop of a lipoplex solution was applied to a perforated carbon film supported on a TEM copper grid in a controlled environment vitrification system (CEVS). The CEVS is a closed chamber, which allows maintaining a constant temperature (25 °C for our experiments) and 100% relative humidity to prevent evaporation of the solution from the grid. Then, the drop was blotted to create a thin film (< 300 nm), and immediately plunged into liquid ethane at its freezing point (~ −183°C). Before cryo-TEM imaging, the vitrified specimens were stored in clean liquid nitrogen. The specimens were transferred to a Gatan 626 or to an oxford CT-3500 cooling holder via their “transfer station”, and equilibrated in the microscope at about −178 °C. The samples were observed in a Philips CM120 or in a FEI T12 G2 transmission electron microscope, operated in 120 kV, low-dose mode to minimize electron-beam radiation damage. We used a Gatan MultiScan 791 or a Gatan US 1000 high-resolution cooled-CCD camera to record the images at a nominal underfocus of 1-2 μm, using the Digital-Micrograph software package.
ζ- potential measurements
5 ml of lipoplex solution was injected to a Zetasizer 3000HS (Malvern Instruments, Worcestershire, UK) instrument. Measurements were made using an applied voltage of 150 V at ambient temperature. The Henry equation was used to calculate ζ-potentials from measurements of electrophoretic mobility (17). For every solution five measurements were made; the viscosity was assumed to be the same as water.
Results
Characterization of BFDMA-Mediated Transfection in the Presence of Serum.
Gene expression levels were determined in OptiMEM cell culture medium containing 0%, 2%, 5%, 10%, and 50% serum. Levels of expression of pEGFP and luciferase are reported in Figures 2 and 3, respectively. Figure 2 shows fluorescence micrographs of EGFP expression and permits identification of qualitative trends. At low (0% and 2%) serum concentrations (Fig. 2A-B), we observed high levels of expression; the difference in the levels of expression under these two conditions is not generally apparent in these data sets. However, at higher serum concentrations (5%, 10%, 50%, Fig 9C-E), the EGFP expression levels decrease, with the presence of 10% serum concentration leading to very low expression levels. At 50% serum concentration (Fig. 2E), no expression is visible. Additional transfection experiments using plasmid DNA encoding luciferase were conducted to quantify differences in these levels of gene expression. Figure 3 shows that transfection efficiency decreased with increased serum concentration. The results show that the presence of 10% serum resulted in a one order of magnitude decrease in the gene expression level relative to controls performed in the absence of serum. However, the expression levels at low serum concentrations (0%, 2%, and 5%) are still on the order of 107 RLU/mg. At 50% serum concentration the expression levels are very low, about 104 RLU/mg.
Figure 2.
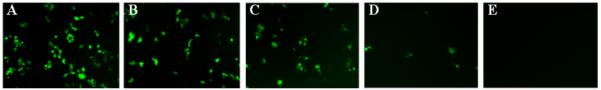
Representative fluorescence micrographs (100× magnification; 1194 μm × 895 μm) showing relative levels of EGFP expression in a confluent layer of COS-7 cells 48 hours after treatment with lipoplexes formed from reduced BFDMA. A-E) Expression levels mediated in cell culture media supplemented with 0%, 2%, 5%, 10%, and 50% serum, respectively.
Figure 3.
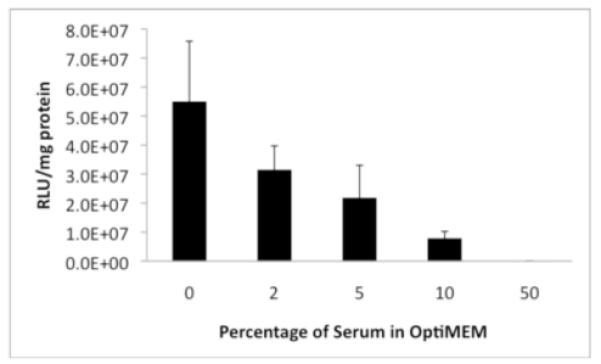
Luciferase expression in COS-7 cells (expressed as relative light units normalized to total cell protein) mediated by lipoplexes formed from reduced BFDMA in the absence or presence of serum. Percentages of serum (v/v) correspond to conditions used to generate the data in Figure 2.
Characterization of Lipoplexes Using Cryo-TEM
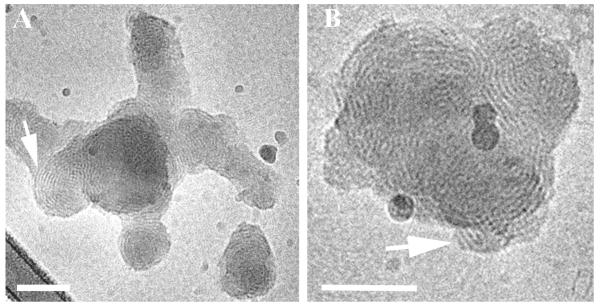
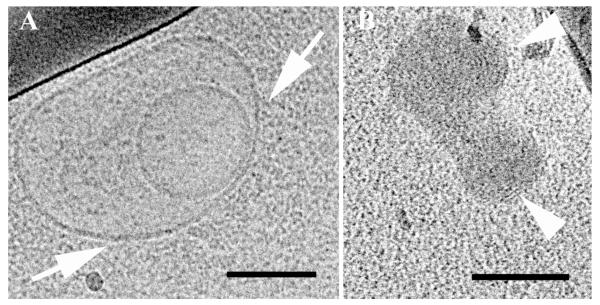
Reduced BFDMA lipoplexes were prepared in an aqueous Li2SO4 salt solution (pH = 5). The lipoplexes exhibited multilamellar nanostructures as reported previously by Pizzey et al. (8). The majority of the nanostructures were onion-like multilamellar aggregates where the DNA is sandwiched between the lipid bilayers (Fig. 4A; arrows).
Figure 4.
Cryo-TEM micrographs of reduced BFDMA lipid-DNA complexes in an aqueous Li2SO4 solution, showing multilamellar nanostructures. The arrows point at multilamellar onion-like aggregates. Bars represent 100 nm.
Figure 5 represents a cryo-TEM micrograph of the pEGFP-N1 plasmid DNA in water. The GFP plasmids, due to their supercoiled nature (more than 95% supercoiled) exhibited thread-like structures (not circular), which are thicker than the 2 nm diameter expected for a double stranded DNA.
Figure 5.
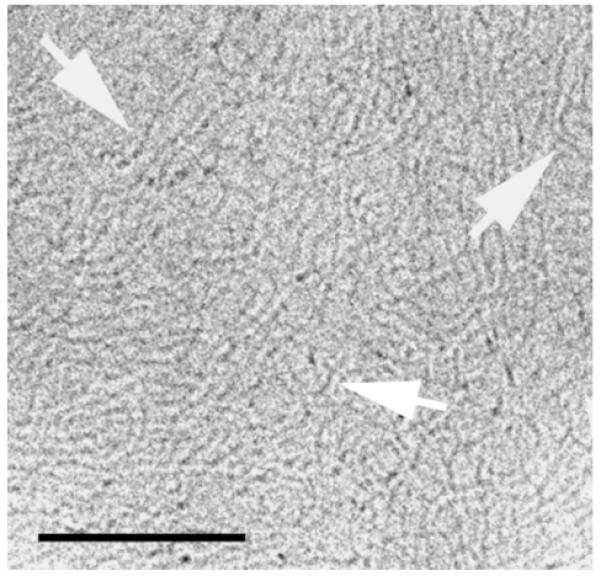
Cryo-TEM image of 2.9mg/ml pEGFP-N1 plasmid DNA (4.7kbps) > 95% supercoiled, in water. The arrows point at thread-like structures that are formed by the DNA plasmids. Bar represents 100 nm.
To mimic the conditions under which transfection is performed in vitro and thereby observe the effect of biological media on the nanostructure of the lipoplexes, we characterized reduced BFDMA lipoplexes in OptiMEM cell culture medium, serum-containing OptiMEM cell culture medium, and serum-containing aqueous salt solution.
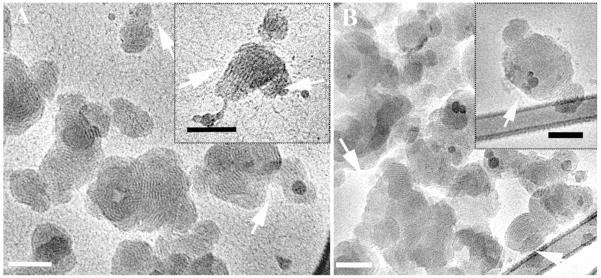
While the effect of OptiMEM cell culture medium on the lipoplexes was modest, the lipoplexes did disassociate to some extent. The process is shown in Figure 6A, indicated by some disassociation of the onion-like complexes releasing DNA threads, leaving the aggregate distorted (inset in Figure 6A). This process may occur due to the different ionic components of the OptiMEM cell culture medium. However, their effect on the morphology of the complexes is not dominant, and most of the multilamellar complexes are preserved (Figure 6B).
Figure 6.
Cryo-TEM micrographs of reduced BFDMA lipid-DNA complexes in OptiMEM cell culture medium. The arrows in 6A point at thread-like structures disassociated from the lipid-DNA complexes. Arrows in 6B point at intact complexes (no disassociation features). The bars represent 100 nm.
To understand the effect of blood serum on the nanostructures of the lipoplexes we prepared solutions of the lipoplexes in OptiMEM containing 2%, 5%, 10%, and 50% bovine blood serum. The results of the solution containing 5% serum showed similar behavior to the solution containing 2% serum, and therefore are not presented here.
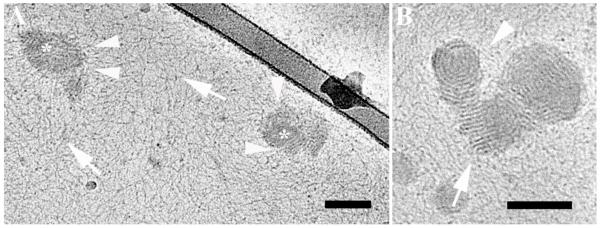
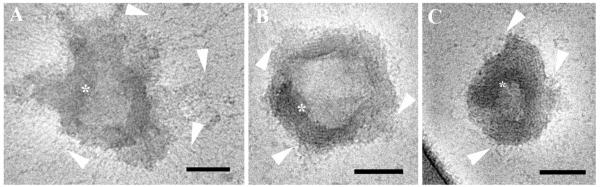
At 2% serum concentration, we observed DNA threads released from the onion-like aggregates (Figure 7). In Figure 7A, the arrowheads point at disassociating, distorted multilamellar aggregates, and the arrows point at free DNA threads, probably disassociated from the complexes. Figure 7B shows at higher magnification a multilamellar aggregate in the process of disassociation.
Figure 7.
Cryo-TEM images of reduced BFDMA-DNA complexes in OptiMEM cell culture and 2% (v/v) serum. A) The arrowheads point at thread-like structures disassociated from the edges of the lipid-DNA complexes. Arrows indicate free DNA threads, and asterisks decorate multilamellar complexes in the midst of disassociation process. B) The arrow points at the edge of a multilamellar complex, where the DNA threads are released into the solution. The arrowhead points at “leftovers” of the roundish onion-like lipoplex. The bars represent 100 nm.
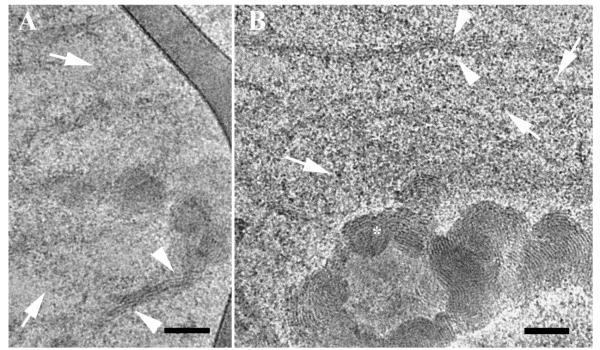
At higher serum concentrations, the background in each image was grainier due to the high density of serum proteins (arrows in Figure 8). Figure 8 shows representative nanostructures observed in media containing 10% serum. Here, too, we observed disassociation of lipoplexes. However, the disassociation process under these conditions was more severe. In 2% serum, the disassociation occurs due to the release of DNA threads from the confined zone between two adjacent bilayers. At 10% serum, the DNA threads are not visible due to the high density of serum proteins. However, disassociation of the lipoplexes is evident, and it appears to involve the simultaneous rupture of several lamellae (arrowheads in Figures 8A and 8B).
Figure 8.
Cryo-TEM micrograph of reduced BFDMA-DNA complexes in OptiMEM cell culture medium and 10% (volume) serum. The arrowheads in 8A and 8B point at lamellae bundles that are released from lipoplex aggregates. The arrows point at serum proteins in the background. The bars represent 100 nm.
At higher serum concentrations (e.g., 50% serum), the multilamellar aggregates were almost completely absent. The few aggregates that we did observe were rather small (indicating high extent of disassociation), and their multilamellar nature was not preserved. Figure 9B shows a representative example of such a distorted lipoplex. The arrowheads point at the edges of the aggregate, where traces of lamellae can be seen. In addition, we observed mostly empty liposomes which are represented in Figure 9A. Figure 9A shows that serum proteins tend to surround the BFDMA vesicle, but do not penetrate nor disassociate it.
Figure 9.
Cryo-TEM micrograph of reduced BFDMA lipid-DNA complexes in OptiMEM cell culture and 50% serum (volume). The background looks grainy due to high concentration of serum proteins. A) The arrows point at the edge of a lipid vesicle, where serum proteins surround the outer surface of the lipid bilayers. B) The arrowheads show a distorted multilamellar aggregate. The bars represent 100 nm.
We next sought to determine whether the presence of the cell culture medium itself contributed to the disassociation of the lipid-DNA complexes. Therefore, we prepared solutions of the lipoplexes in 2%, 5%, and 10% serum in Li2SO4 salt solution. In these samples, the disassociation extent was similar to the samples containing cell culture medium and serum, at the same serum concentration. Since we did not observe any difference in disassociation from the samples containing cell culture medium and serum, we show only representative results of the sample containing 5% serum (Fig. 10).
Figure 10.
Cryo-TEM micrograph of reduced BFDMA lipid-DNA complexes 5% serum (v/v; no cell culture OptiMEM medium). The arrowheads show DNA threads disassociate from the lipoplex aggregate (asterisks). A) High extent of disassociation (such as in Figure 7A) B) Moderate extent of disassociation. C) Low extent of disassociation. The bars represent 100 nm.
Figure 10 shows lipid-DNA complexes during disassociation. In Figure 10A, the complex is distorted, DNA threads are released from the complex, and the multilamellar nature is not preserved. In Figure 10B the extent of disassociation is moderate because we see just a few DNA threads being released from the aggregate; some of the original nanostructure is preserved. The complex in Figure 10C is almost intact, and only a few DNA threads are released from it. However, the aggregate is small and was probably detached from a larger lipoplex.
ζ- Potential Measurements
Zeta potential measurements of reduced BFDMA lipoplexes in Li2SO4 and in OptiMEM media are summarized in Table 1. Table 1 shows that the zeta potential of the cationic reduced BFDMA lipid-only solution in Li2SO4 was positive, and complexes of reduced BFDMA lipid with plasmid DNA at 1.1:1 lipid to DNA charge ratio exhibited zeta potentials that were negative in both Li2SO4 and in OptiMEM media. Also, the zeta potential values of lipoplexes in OptiMEM medium and in Li2SO4 aqueous medium without serum were comparable (approximately −35 mV). Interestingly, however, the presence of serum in both media caused the zeta potential to be less negative. For example, in the OptiMEM cell culture at 5%, 10% and 50% serum concentrations, the zeta potential increased (became less negative) to values of approximately −20 mV (in the salt solution, values increased to approximately −10 mV).
Table 1.
Zeta potential measurements of reduced BFDMA solutions.
| Sample | 1mM Li2SO4 (mV) | OptiMEM (mV) |
|---|---|---|
| Reduced BFDMA lipid | 29.3 ± 1.1 | − |
| DNA only | −1.0 ± 2.7 | − |
| Reduced BFDMA lipoplexes, 0% serum |
−35.1 ± 1.5 | −33.1 ± 2.0 |
| Reduced BFDMA lipoplexes, 2% serum |
−15.1 ± 0.4 | −24.9 ± 1.6a |
| Reduced BFDMA lipoplexes, 5% serum |
−13.3 ± 0.3 | −20.8 ± 1.2 |
| Reduced BFDMA lipoplexes, 10% serum |
−9.5 ± 0.4 | −18.5 ± 0.7 |
| Reduced BFDMA lipoplexes, 50% serum |
−9.5 ± 1.1 | −20 ± 1.9 |
Discussion
Our results demonstrate that lipoplexes prepared using reduced BFDMA disassociate in the presence of biological media, particularly when serum proteins are added to cell culture media in which they are suspended. Lipoplexes in aqueous salt media (in Li2SO4) are multilamellar, roundish, and intact (Fig. 4). Addition of the lipoplex solution to OptiMEM cell culture medium does not alter the lipoplex structure (Fig. 6) or the zeta potential value (around - 35 mV for lipoplexes both in Li2SO4 and in OptiMEM). However, the addition of serum to these solutions caused significant changes in the physicochemical properties of the lipoplex aggregates, as determined by cryo-TEM and zeta potential measurements. Cryo-TEM revealed levels of lipoplex disassociation that varied with the level of serum added. At low serum concentrations (2% and 5%), both in Li2SO4 and OptiMEM media, we observed DNA thread-like structures that were released from the onion-like lipoplex aggregate (Fig. 7 and 10). At higher serum concentrations (10% and 50%), lipoplex disassociation was more substantial. At 10% serum concentration (Figure 8), we observed bundles of multilamellae that disassociate from multilamellar lipoplexes. Because of the higher serum protein concentration (the grainy background in Fig. 8), the proteins may interact with the lipoplex, and tear away several lamellae from a larger multilamellar aggregate (this may explain the lamellae bundles we observed in Figure 8).
At 50% serum concentration (Fig. 9), the presence of the lipoplex aggregates was rare, and the aggregates that we did observe seemed to be distorted (Fig. 9B), indicating that most of the lipid-DNA complexes have already disassociated at such serum concentrations. In addition, we observed that serum proteins do not disintegrate the lipid bilayers because they do not penetrate to the inner region of the vesicle, but rather attach to the outer surface of it. That serum proteins do not penetrate nor disassociate the lipid bilayers suggests that the complex disassociation occurs due to DNA release, rather than liposome bilayers disintegration. Therefore, the lipoplexes disassociation may occur due to attachment of serum proteins on the positively charged lipids membrane, and by neutralizing the charge, the DNA is released from the complex (i.e., effectively, an ion exchange process on the surface of the lipoplex). In this physical scenario, the serum proteins and DNA are competing for the surface of the lipid bilayers.
Zeta potential results support the fact that serum causes structural changes. Blood serum contains mostly negatively charged entities (albumin). Thus, zeta potential is expected to become more negative upon introduction of positively charged lipoplexes to serum-containing media. Simberg et al. (9) and Eliyahu et al. (10) showed that for DOTAP-containing lipoplexes, zeta potential becomes more negative in the presence of serum, suggesting that serum proteins tend to coat the surface of the lipoplex and induce aggregation. In the BFDMA lipoplex system, that is not the case; instead of turning the zeta potential more negative, the presence of serum causes the zeta potential to become less negative.
The combination of zeta potential measurements (Table 1) with cryo-TEM suggests that serum proteins cause DNA disassociation from the lipid-DNA complex. Serum proteins compete for the surface of the positively charged lipid bilayers, resulting in the dissociation of DNA from the complexes. Because of DNA release, there are less negatively charged entities associated in the complex, and therefore the zeta potential becomes less negative. Even if serum proteins replace the DNA, their charge density is lower than that of the DNA plasmids, thus causing the zeta potential to be less negative, but still in the negative charge zone.
Transfection experiments and gene expression results show that levels of transfection mediated by reduced BFDMA lipoplexes are significantly lower in the presence of serum. As shown in Figs. 2 and 3, levels of both EGFP and luciferase expression decrease as serum concentrations are increased. These decreases in expression levels correlate with changes in the physicochemical properties of the lipoplexes in the presence of serum that are revealed by our cryo-TEM and zeta potential measurements. Although it is difficult to determine the specific reasons for lower transfection on the basis of these current measurements, these large changes in structure and charge could potentially lead to changes in the extent to which DNA is internalized by cells (and thereby lead to lower overall levels of cell transfection). All our findings show the ability of serum to disassociate the lipid-DNA complex. This is in contrast to other past reports, in which suggested that DNA is not released from lipoplexes based on other cationic lipids (9-10, 18-20). Furthermore, Simberg et al. (9) showed by cryo-TEM that serum proteins do not penetrate or disassociate DOTAP/cholesterol-based lipoplexes, but rather adhere to the surface of the lipoplexes without causing them to disintegrate or undergo other large changes in structure. Those findings partly resemble ours. We also demonstrated in Figure 9A that serum proteins do not penetrate the lipids membrane. However, their results suggest that DNA does not disassociate from the lipoplex in the presence of serum. In our work, we observed DNA disassociation from lipoplexes in the presence of serum.
One wonders why the presence of blood serum causes DNA release from the cationic BFDMA lipid-DNA complexes, while other cationic-based lipoplexes stay intact. Although the multilamellar morphology of lipoplexes based on reduced BFDMA is quite ordinary and resembles “standard” lipoplexes based on cationic lipids, the structure of this lipid is different in several ways from lipids investigated in the past studies noted above. Standard cationic lipids used for gene delivery are composed of a cationic polar head group, and two hydrocarbon tails. Similarly, reduced BFDMA lipid is composed of a cationic head group and double hydrocarbon tail. However, reduced BFDMA contains, at the end of each hydrocarbon tail, a ferrocene group. We note that past studies (21) of monolayers of ferrocene-containing surfactants concluded that the ferrocene group changes substantially the conformation of the surfactant molecules at the surface of water. Similarly, the ferrocene groups of BFDMA may perturb the packing of the lipid within the bilayer structure, making it less stable, resulting in weaker electrostatic interactions between the lipid bilayers and the DNA. When serum is added, negatively charged proteins might interact with the positively charged lipid membrane and neutralize it. When some parts of the lipid membrane are neutralized, the interactions with the DNA are weakened and DNA may be released. Another secondary effect that may weaken the lipid-DNA interactions is the hydrophobic interactions between the hydrophobic domains of the serum proteins and the hydrophobic segments of the BFDMA lipids. In standard cationic lipids, the lipids bilayers may be stable and be packed better (because their hydrophobic tails contain only hydrocarbon chains), allowing stronger electrostatic interactions with the DNA.
Conclusions
We have demonstrated that lipoplexes formed using reduced BFDMA, when exposed to biological media, can undergo significant disassociation. The disassociation process and its extent depend on the type of the medium and its concentration. OptiMEM cell culture medium induced some lipoplex disassociation, but did not affect the zeta potential. In contrast, the addition of serum caused lipoplexes to disassociate, resulting in structural changes and changes in zeta potential. We used cryo-TEM to characterize how the lipoplexes disassemble in the presence of serum and correlated these observations to transfection efficiency, which demonstrated that as serum concentration increased, transfection levels decreased.
Figure 1.
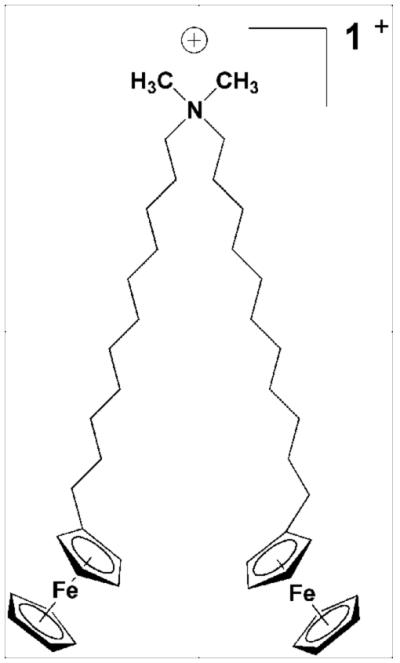
Molecular structure of BFDMA (15).
Acknowledgment
Financial support was provided by the Technion Russell Berrie Nanotechnology Institute (RBNI), and the Technion Soft Matter Electron Microscopy Laboratory. In addition, support from the US National Science Foundation (CBET 075921) and the National Institutes of Health (1 R21 EB006168) is gratefully acknowledged.
References
- (1).Verma IM, Weitzman D. Annu. Rev. Biochem. 2005;74:711–738. doi: 10.1146/annurev.biochem.74.050304.091637. [DOI] [PubMed] [Google Scholar]
- (2).Patil SD, Rhodes DG, Burgess DJ. The AAPS Journal. 2005;07(01):E61–E77. doi: 10.1208/aapsj070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Boktov J, Hirsch-Lerner D, Barenholz Y. J. Gene. Med. 2007;9:884–893. doi: 10.1002/jgm.1079. [DOI] [PubMed] [Google Scholar]
- (4).Koltover I, Salditt T, Radler JO, Safinya CR. Science. 1998;281:78–81. doi: 10.1126/science.281.5373.78. [DOI] [PubMed] [Google Scholar]
- (5).Koltover I, Salditt T, Safinya CR. Biophys J. 1999;77(2):915–924. doi: 10.1016/S0006-3495(99)76942-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Safinya CR, Ewert K, Ahmad A, Evans HM, Raviv U, Needleman DJ, Lin AJ, Slack NL, George C, Samuel CE. Phil. Trans. R. Soc A. 2006;364:2573–2596. doi: 10.1098/rsta.2006.1841. [DOI] [PubMed] [Google Scholar]
- (7).Weisman S, Hirsch-Lerner D, Barenholz Y, Talmon Y. Biophys J. 2004;87:609–614. doi: 10.1529/biophysj.103.033480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Pizzey CL, Jewell CM, Hays ME, Lynn DM, Abbott NL, Kondo Y, Golan S, Talmon Y. J. Phys. Chem. B. 2008;112:5849–5857. doi: 10.1021/jp7103903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Simberg D, Weisman S, Talmon Y, Faerman A, Shoshani T, Barenholz Y. J. Biol. Chem. 2003;278(41):39858–39865. doi: 10.1074/jbc.M302232200. [DOI] [PubMed] [Google Scholar]
- (10).Eliyahu H, Joseph A, Schillemans JP, Azzam T, Domb AJ, Barenholz Y. Biomaterials. 2007;28:2339–2349. doi: 10.1016/j.biomaterials.2006.09.001. [DOI] [PubMed] [Google Scholar]
- (11).Li S, Tseng W-C, Stolts DB, Wu S-P, Watkins SC, Huang L. Gene Therapy. 1999;6:585–594. doi: 10.1038/sj.gt.3300865. [DOI] [PubMed] [Google Scholar]
- (12).Yang J-P, Huang L. Gene Therapy. 1997;4:950–960. doi: 10.1038/sj.gt.3300485. [DOI] [PubMed] [Google Scholar]
- (13).Jewell CJ, Hays ME, Kondo Y, Abbott NL, Lynn DM. J. Controlled release. 2006;112:129–138. doi: 10.1016/j.jconrel.2006.01.014. [DOI] [PubMed] [Google Scholar]
- (14).Abbott NL, Jewell CJ, Hays ME, Kondo Y, Lynn DM. J. Am. Chem. Soc. 2005;127:11576–11577. doi: 10.1021/ja054038t. [DOI] [PubMed] [Google Scholar]
- (15).Kakizawa Y, Sakai H, Nishiyama K, Abe M. Langmuir. 1996;12:921–924. [Google Scholar]
- (16).Yoshino N, Shoji H, Kondo Y, Kakizawa Y, Sakai H, Abe MJ. Jpn. Oil Chem. Soc. 1996;45:769. [Google Scholar]
- (17).Hunter RJ. Zeta Potential in Colloid Science. Academic Press; New York: 1981. [Google Scholar]
- (18).Marchini C, Montani M, Amici A, Amenitsch H, Marianecci C, Pozzi D, Caracciolo G. Langmuir. 2009;25:3013–3021. doi: 10.1021/la8033726. [DOI] [PubMed] [Google Scholar]
- (19).Gao X, Kim K-S, Liu D. The AAPS Journal. 2007;9(1):E92–E104. doi: 10.1208/aapsj0901009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Madeira C, Loura LMS, Prieto M, Fedorov A, Aires-Barros MR. BMC Biotechnology. 2008;8(20) doi: 10.1186/1472-6750-8-20. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Gallardo BS, Metcalfe KL, Abbott NL. Langmuir. 1996;12:4116–4124. [Google Scholar]