Abstract
A combined structural and computational analysis of rabbit cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene (tBPA) has yielded insight into how the enzyme retains partial activity. Since conjugation to tBPA modifies a highly conserved active site residue, the residual activity of tBPA-labeled 2B4 observed in previous studies was puzzling. Here we describe the first crystal structures of a modified mammalian P450, which show an oxygenated metabolite of tBPA conjugated to Thr 302 of helix I. These results are consistent with previous studies that identified Thr 302 as the site of conjugation. In each structure, the core of 2B4 remains unchanged, but the arrangement of plastic regions differs. This results in one structure that is compact and closed. In this conformation, tBPA points toward helix B′, making a 31° angle with the heme plane. This conformation is in agreement with previously performed in silico experiments. However, dimerization of 2B4 in the other structure, which is caused by movement of the B/C loop and helices F through G, alters the position of tBPA. In this case, tBPA lies almost parallel to the heme plane due to the presence of helix F′ of the opposite monomer entering the active site to stabilize the dimer. However, docking experiments using this open form show that tBPA is able to rotate upward to give testosterone and 7-ethoxy-4-trifluoromethylcoumarin access to the heme, which could explain the previously observed partial activity.
Keywords: cytochrome P450 2B4, X-ray crystallography, mechanism-based inactivation, tert-butylphenylacetylene, DXMS, ligand docking
Cytochromes P450 (P450s) are a superfamily of heme monooxygenases responsible for the oxidation of numerous endogenous compounds, pharmaceuticals, xenobiotics, and environmental toxins. Inhibition of these enzymes can affect the pharmacokinetics of therapeutics and result in adverse drug-drug interactions. However, mechanism-based inactivation (MBI) of P450s presents even more serious problems. During their natural catalytic cycle, P450s can sometimes create reactive intermediates that irreversibly modify components of the protein necessary for catalysis, such as the heme or active site residues, thus rendering the enzyme unable to function (1). This may lead to the build up of other substrates of the conjugated P450, ultimately resulting in toxicity. However, MBI may also be used during therapeutic treatments when specifically designed inactivators target enzymes involved in disease progression.
Interesting mechanistic insights into P450 function have been derived from several studies of MBI of P450s 2B (2–5). This subfamily, which includes rat 2B1, rabbit 2B4, human 2B6, and dog 2B11 are known to be inactivated by a variety of agents including organosulfur compounds, arylamines, acetylenes, and furanocoumarins (1). Among these classes of compounds, acetylenes are perhaps the most highly studied. 17-α-ethynylestradiol (3), 5-phenyl-1-pentyne (6), 9-ethynylphenanthrene (7), 2-ethynylnaphthalene (8), 7-ethynylcoumarin (9), tert-butyl-1-methyl-2-propynyl ether (10, 11), and tert-butylphenylacetylene (tBPA) (11–13) have been all used to probe the topology of the active site cavity or the catalytic mechanism of P450s. Each of these compounds covalently modifies either the heme moiety or Thr 302 of P450s 2B (14).
Thr 302 lies on the I helix along the backside of the active site cavity and is highly conserved across many P450 families (although the residue number changes based on sequence). High-resolution crystal structures of the bacterial P450cam-dioxygen complex have identified a hydrogen-bonding network that includes the Thr 252 (equivalent to Thr 302) hydroxyl, molecular oxygen, and two active site water molecules (15, 16). Furthermore, residual enzymatic activity of the P450cam T252A (15) and 2B4 T302A (17) mutants and the methoxy-modified Thr 252 variant of P450cam (18) indicates that the specific role of this threonine residue is to serve as a hydrogen bond acceptor to stabilize a hydroperoxy intermediate during the catalytic cycle. Additionally, mutational studies in human 2E1 have indicated that this threonine residue is important in determining the orientation of ligands in the active site cavity (19).
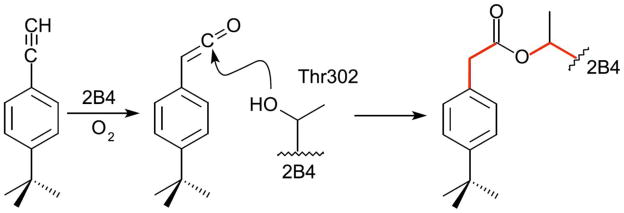
Recently, interest in the effects of tBPA modification of P450s 2B as mechanistic probes has been increasing. A proposed mechanism suggests that an oxygenated ketene metabolite of tBPA is conjugated to Thr 302 through nucleophilic attack by the hydroxyl moiety of Thr 302 to create an ester bond (Fig. 1) (12, 13). Previous studies have shown that stoichiometric covalent modification of Thr 302 in 2B4 by tBPA results in a predominantly low-spin state of the enzyme with decreased polarity of the heme pocket (13, 20). Additionally, this modification reduces catalytic activity toward benzphetamine, 7-ethoxy-4-trifluoromethylcoumarin, and testosterone by 79, 70, and 90.4%, respectively (13). Based on UV-visible and stopped-flow spectroscopy, the authors conclude that the binding of benzphetamine to the active site of tBPA-conjugated 2B4 is impaired, which is also supported by a separate resonance Raman study (20). However, the partial enzymatic activity implies that substrates are still able to access the heme and despite modification of a highly conserved active site residue, the enzyme is still catalytically competent. To further investigate these results, Zhang, et al. utilized molecular docking to probe the orientation of tBPA in the 2B4 active site (13). tBPA was found to bind above the plane of the heme with the terminal alkyne carbon 3.65 Å from Thr 302.
Figure 1.
Proposed mechanism for tBPA modification of 2B4. During catalytic turnover, tBPA is oxygenated to create a ketene intermediate. Nucleophilic attack by the hydroxyl group of Thr 302 forms an ester bond between 2B4 and the inhibitor. Selected bonds (red lines) indicate points of rotation to allow tBPA to change conformation once bound to the enzyme.
Despite this growing wealth of experimental data, several questions remained unanswered. First, in silico docking methods can only predict tBPA positions based on the conformation of the starting model and do not take into account changes to the protein backbone. Furthermore, unoxidized tBPA was used for docking and not the proposed oxygenated metabolite or estertified conjugate. It was unclear how the Thr 302-tBPA conjugate would orient itself in the active site and if the modification would affect the tertiary structure of 2B4. Other questions also remained about the volume of the active site void once the enzyme was modified, which presumably was still large enough to accommodate some substrates for catalysis. It was also unknown how neither tBPA binding near the heme, nor the additional, albeit hindered, binding of benzphetamine did not affect the spin state of 2B4 (13, 20). To answer these questions, we turned to X-ray crystallography and deuterium exchange mass spectrometry (DXMS) to determine and analyze the first structures of a modified mammalian P450, the 2B4-tBPA adduct. Additionally, we utilized ligand docking to propose methods by which the modified enzyme is still able to retain partial catalytic activity.
Materials and Methods
Materials
5-Cyclo-hexylpentyl-®-D-maltoside (Cymal-5) was from Anatrace (Maumee, OH). CM-sepharose resin was from Bio-Rad Laboratories (Hercules, CA). C41(DE3) cells were from Avidis (Saint Beauzire, France). Nickel-nitrilotriacetic acid (Ni-NTA) affinity resin, dilauroylphosphatidylcholine (DLPC), catalase, NADPH, and tBPA were from Sigma-Aldrich Inc. (St. Louis, MO). Trifluoroacetic acid (TFA) was from Pierce Chemicals (Rockford, IL). IndexHT and PEGRx 1 high throughput crystal screening kits were from Hampton Research (Alisa Viejo, CA). The synthesis of the facial amphiphile 3α,7α,12α-tris[(β-D-maltopyranosyl)ethyloxy]cholane (232-chol) was performed essentially according to a previous method (21).
Over-expression and purification of 2B4, NADPH-dependent cytochrome P450 reductase, and cytochrome b5
CYP2B4dH (H226Y), an N-terminal truncated form of CYP2B4 with an internal mutation and C-terminal His-tag (from hereon referred to as 2B4), was over-expressed in Escherichia coli C41(DE3) cells and purified with a Ni-NTA affinity column followed by a CM-sepharose cation exchange column as described previously (22). NADPH-dependent cytochrome P450 reductase (CPR) and cytochrome b5 (b5) were over-expressed and purified as described previously (23).
Preparations of tBPA-modified 2B4 for crystallization
Chemical modification of 2B4 by tBPA was achieved in a reconstituted system under steady-state turnover conditions. In a typical reaction, 2B4 (1 μmole), CPR (0.5 μmole), and b5 (3 μmoles) were reconstituted in the presence of DLPC (60 μmoles) for 60 min at 4 °C. The mixture was then diluted from approximately 50–80 ml to 250 ml with 50 mM potassium phosphate (pH 7.4 at 4 °C). tBPA and catalase were added into the solution at final concentrations of 20 μM and 300 units/ml respectively. Because of the large volume (~250 ml), the solution was incubated at 30 °C for 10 min to reach thermal equilibrium. The reaction was then initiated by the addition of 1 mM NADPH. It was estimated that the reaction to covalently modify 2B4 by tBPA should reach completion in approximately 20 min based on a t1/2 of 2.8 min as previously reported (13). Therefore, the reaction mixture was placed in ice-cold water bath after a 20 min incubation at 30 °C, and an aliquot (20 μL) of the reaction mixture was used to confirm the chemical modification.
Analyses of the tBPA-modified 2B4 protein by ESI-LC/MS
Covalent modification of 2B4 by tBPA was confirmed by analyses of the protein mass of 2B4 as the covalent modification led to an increase in the mass of 2B4 by 174 Da as previously reported (13). The protein mass of the tBPA-modified 2B4 was determined on a LCQ ion-trap mass spectrometer (ThermoScientific, Waltham, MA) as previously described (13). The aliquot of 20 μL of the reaction mixture was applied to a reverse-phase C3 column (2×150 mm, 5 μm) (Agilent Technologies, Santa Clara, CA). 2B4 was separated from the other reaction components with a binary solvent system consisting of 0.1% TFA in water (Solvent A) and 0.1% TFA in acetonitrile (Solvent B) using the following gradient: 30% B for 5 min, linearly increased to 90% B in 20 min, and held at 90% B for 30 min. The flow rate was 0.25 ml/min. The molecular masses of the control and tBPA-modified 2B4 were determined by deconvolution of the apo protein charge envelopes using the Bio-works software (Thermo Scientific, Waltham, MA).
Once the stoichiometric chemical modification of 2B4 was confirmed (Suppl. Fig. S1), 0.5 M NaCl and 4.8 mM Cymal-5 were added into the ice-cold reaction mixture to completely terminate the reaction. The tBPA-modified 2B4 was then purified to homogeneity from the reaction as described previously (22). The purified tBPA-modified 2B4 was then stored on ice to be used for crystallization.
Crystallization and data collection
The tBPA-modified protein was diluted to 437 μM in 50 mM potassium phosphate (pH 7.4 at 4 °C), 500 mM NaCl, 500 mM sucrose, 1 mM EDTA, and 0.2 mM DTT. Cymal-5 and 232-chol were added to this solution to final concentrations of 4.8 mM and 0.028% (w/v), respectively. Initial crystallization attempts in the absence of 232-chol did not produce any crystals of 2B4-tBPA. The protein and detergents were allowed to equilibrate for approximately 20 min before mixing with crystallization reagents.
For the open crystal structure, screening was performed by sitting drop vapor diffusion using the PEGRx 1 high throughput kit by mixing equal volumes of protein solution and well solution. Drops were equilibrated against the well solutions at 18 °C. Crystals suitable for X-ray diffraction appeared over the course of approximately three weeks in drops containing 0.1 M HEPES (pH 7.5) and 12% (w/v) PEG 3350. Crystals were briefly transferred to a solution of mother liquor supplemented with 335 mM sucrose before flash freezing in liquid nitrogen. 150° of data were collected using 1° oscillations and 20 s exposures at 100 K on a Quantum CCD detector (Area Detector Systems Corp., Poway, CA) at BL 11-1 of the Stanford Synchrotron Radiation Lightsource (SSRL) (Stanford, CA). The data were processed to 3.0 Å using iMOSFLM (24) and SCALA (25).
Screening, crystal incubation, and cryoprotection for the closed structure crystals were done essentially as above, substituting the IndexHT kit for PEGRx 1. Crystals suitable for X-ray diffraction appeared over the course of one month and grew in 0.1 M magnesium formate dihydrate and 15% (w/v) PEG 3350. 90° of data were collected using 1° oscillations and 20 s exposures on the same detector as above. The data were processed to 3.5 Å using iMOSFLM (24) and SCALA (25).
Structure determination and refinement
Both tBPA-conjugated 2B4 structures were determined by molecular replacement using the previously determined 2B4-4-(4-chlorophenyl)imidazole (4-CPI) complex (PDB ID 1SUO) (with the inhibitor molecular removed from the coordinates) in Phaser (26). The closed structure solution was found in space group P212121, containing 52.3% solvent assuming eight molecules per asymmetric unit. The initial model was first subjected to a simulated annealing step followed by restrained refinement in PHENIX (27) using non-crystallographic symmetry restraints to remove model bias. Model building was performed in COOT (28) using both 2Fo−Fc and Fo−Fc electron density maps contoured to 1-σ and 3-σ, respectively. The model was modified to fit the electron density and refined in an iterative manner until a final R-factor of 20.6% and an Rfree of 28.5% were reached. Non-crystallographic symmetry restraints were slowly released during the refinement process.
A partial solution was found for the open structure in space group C121, containing 67.5% solvent assuming four molecules per asymmetric unit. Despite several attempts to find a complete solution using various other programs, the solution found in PHASER containing three molecules was the best (or at least equivalent to others). Based on differences observed between the initial model and electron density maps, residues 100–120 and 200–230 were deleted prior to modeling and refinement. Density for the fourth molecule was evident upon inspection of the electron density maps. This last copy of the protein was fit manually using the planar density for the heme group as a guide point. The model containing all four monomers was modeled and refined as described above until an R-factor of 24.2% and an Rfree of 28.8% were reached. Structure refinement statistics are summarized in Table 1.
Table 1.
X-ray Data Collection & Refinement Statistics
| Crystal Data | ||
| Complex | Open | Closed |
| Space Group | C121 | P212121 |
| Unit Cell | ||
| a, b, c (Å) | 193.9, 153.7, 129.7 | 134.4, 144.5, 229.3 |
| α, β, γ (°) | 90, 122.2, 90 | 90, 90, 90 |
| Molecules per A.U. | 4 | 8 |
| Data Collection | ||
| X-ray source | SSRL BL 11-1 | SSRL BL 11-1 |
| Wavelength (Å) | 0.98 | 0.98 |
| Resolution Range (Å) | 53.43 – 3.00 | 98.42 - 3.50 |
| Total Observations | 203,162 | 163,147 |
| Unique Observations (F>0) | 63,932 | 51,302 |
| Completeness (%)a | 99.4 (99.4) | 90.2 (90.2) |
| Redundancya | 3.2 (3.1) | 3.2 (3.2) |
| I/σa | 7.5 (1.7) | 6.5 (2.2) |
| Rmerge (%)a,b | 9.2 (58.6) | 15.6 (57.7) |
| Refinement Statistics | ||
| R-factor (%)c | 24.20 | 20.58 |
| Rfree (%)c | 28.76 | 28.49 |
| RMS bond lengths (Å) | 0.008 | 0.004 |
| RMS bond angles (°) | 1.351 | 0.889 |
| Number of Atoms | ||
| Proteind | 14,020 (101.4) | 28,357 (74.6) |
| Hemed | 172 (76.8) | 344 (62.6) |
| tBPAd | 52 (79.1) | 104 (62.2) |
| Cymal-5d | 160 (111.6) | NA |
| Sucrosed | 92 (105.0) | NA |
| Waterd | NA | 8 (43.3) |
Values for the highest resolution shell are in parentheses.
Rmerge = [ΣhΣi|Ih−Ihi|/ΣhΣiIhi] where Ih is the mean of Ihi observations of reflection h.
R-factor & Rfree = Σ||Fobs|−|Fcalc||/Σ|Fobs| × 100 for 95% of the recorded data (R-factor) and 5% of data (Rfree).
Average B-factors (Å2) are in parentheses.
Ligand docking
Docking of 7-ethyoxy-4-trifluoromethylcoumarin (7-EFC) and testosterone into the open structure of tBPA-labeled 2B4 was performed using AutoDock Vina version 1.1.1 (29). Residues not modeled into the experimental electron density were added using COOT (28), and Cymal-5 molecules were removed from the file. Thr 302 was manually linked to tBPA to define the adduct and an AutoDockTools4 script added all hydrogen atoms to the protein (30). Gasteiger charges were utilized for small molecules. Heme charges were modified by a separate script using previously reported values (31). Using AutoDockTools4, generation of a flexible receptor file containing Thr 302 and the linked tBPA allowed the adduct to move freely during docking runs; the remainder of the protein was contained in a rigid receptor file. The docking experiment for each substrate included 100 events with a box size of 25 Å 25 Å 25 Å centered 10 Å above the heme iron.
Dynamic light scattering (DLS) of tBPA-modified 2B4
DLS was used to determine the aggregation state of tBPA-conjugated 2B4 in solution. Buffer conditions were 50 mM potassium phosphate (pH 7.4 at 4 °C), 500 mM NaCl, 20% (v/v) glycerol, 1 mM EDTA, 0.2 mM DTT and 1 mM Cymal-5. For a positive control, bacterial cytochrome P450 eryF (P450eryF) was used, since it is known to be monomeric in solution. DLS experiments were performed on a Zetasizer Nano-ZS (Malvern Instruments, Worcestershire, UK) instrument. The results were analyzed using the Zetasizer software, assuming a viscosity and refractive index of 20% glycerol and 80% water to approximate the solution conditions.
Deuterium exchange analysis
Optimization of the 2B4 fragmentation conditions for DXMS experiments was previously completed, and the preparation of deuterated samples and subsequent DXMS analysis was carried out as previously described (32). Briefly, 5 μl of protein sample (190 μM, unlabeled or labeled 2B4) was combined with 15 μl of deuterated CM elution buffer (50 mM potassium phosphate (pH 7.4 4 °C), 500 mM NaCl, 20% glycerol (v/v), 1 mM EDTA, 0.2 mM DTT, and 1 mM Cymal-5 in D2O. After incubation at 0 °C for 10, 100, 1000, 10,000, and 100,000 s, the reaction was quenched by addition of 30 μl of ice-cold quench solution (1.6 M guanidine hydrochloride in 0.8% (v/v) formic acid, pH 2.2 – 2.5). The samples were then transferred to ice-cooled autosampler vials, frozen on dry ice, and stored at −80 °C. Frozen samples were transferred to the dry ice-containing sample basin of the cryogenic autosampler module of the DXMS apparatus. Samples were thawed on ice and immediately passed over a protease column (66 μl bed volume) filled with porcine pepsin at a flow rate of 100 μl min−1 with 0.05% TFA for a 40 s digestion. Proteolytic products were directly collected by a C18 column (Vydac cat# 218MS5105, W. R. Grace and Co., Columbia, MD) and then eluted with a linear gradient of 0.046% (v/v) TFA, 6.4% (v/v) acetonitrile to 0.03% (v/v) TFA, 38.4% (v/v) acetonitrile for 30 min. The column effluent was analyzed on an LCQ Classic electrospray ion trap-type mass spectrometer (Thermo Finnigan, Inc., San Jose, CA) and an electrospray Q-TOF mass spectrometer (Micromass, Manchester, England), and data acquisition in either data-dependent tandem mass spectrometric mode or MS1 profile mode. Determination of pepsin-generated peptides from the resulting MS/MS data sets was facilitated through the use of SEQUEST (Thermo Finnigan, Inc., San Jose, CA). This set of peptides was then further examined by specialized software, DXMS Explorer (Sierra Analytics Inc., Modesto, CA), and all data processing was the same as previously described (25). Corrections for back-exchange were determined via the methods of Zhang and Smith (29).
Protein figures
All protein figures were generated using PyMOL (33).
Results
Closed 2B4-tBPA structure
The protein backbone of the closed tBPA-conjugated 2B4 structure is very similar to those of the previously determined 2B4 complexes bound to 4-CPI (34), 1-(4-chlorophenyl)imidazole (1-CPI) (35), ticlopidine, or clopidogrel (22) as well as the closed 2B4 ligand free structure (32), with an average RMSD of 0.62 Å (Suppl. Fig. S2). In the active site cavity, a strong tube of electron density was found protruding from the hydroxyl oxygen (OG1) of Thr 302 (Fig. 2A) that is consistent with the size and shape of the previously proposed 2B4-tBPA conjugate (Fig. 1) (13). During the labeling of 2B4 with tBPA, an oxygenated metabolite of the inactivator forms an ester bond with OG1 of Thr 302 (Fig. 1). The ligand was modeled to orient the carbonyl oxygen of the ester toward the heme iron where it can interact with a water molecule that was observed in the electron density (Fig. 2A). In this orientation, tBPA points toward the B′ helix, making a 31° angle with respect to the plane of the heme and a 42° angle with helix I. This orientation is in agreement with previously performed in silico experiments that dock unmetabolized tBPA in the 2B1 and 2B4 active sites (12, 13). The amino acid side chains making contact (defined as those within a 5 Å radius) with tBPA in this orientation are Val 104, Phe 108, Ile 114, Phe 115, Phe 206, Ile 209, Phe 297, Ala 298, Thr 302, Ile 363, Val 367, and Val 477 (Fig. 2A).
Figure 2.
Ribbon and stick diagrams showing the active sites of both tBPA-modified 2B4 conformations. Residues found within 5 Å of tBPA make up the inactivator binding sites. In gray mesh, unbiased Fo − Fc simulated annealing omit maps contoured at 3-σ surround the Thr302-tBPA conjugates. In all three panels, the A chain of each structure is shown, which is representative of the other protein chains in the asymmetric unit. A) The active site of the closed structure surrounds tBPA with Val 104, Phe 108, Ile 114, Phe 115, Phe 206, Ile 209, Phe 297, Ala 298, Glu 301, Thr 302, and Ile 363. There is also density for a water molecule, which is also shown. Heme and tBPA are depicted as red and gray sticks, respectively and the protein is colored blue. B) Dimerization results in a different active site composition. tBPA makes contact with Arg 98, Trp121, Ser 294, Ala 298, Thr 302, Ile 363, and Val 367. From the other monomer, Val 216, Phe 217, Phe 220, Pro 221 also interact with tBPA. Heme and tBPA are colored as in A) and one half of the protein dimer is colored in green, while its symmetrical partner is in yellow. C) An overlay of the two protein conformations highlights the difference in binding modes for the Thr 302-tBPA conjugate.
Open 2B4-tBPA structure
Crystals grown in a second condition were also analyzed by X-ray diffraction and yielded a structure different from that of closed tBPA-modified 2B4. Significant portions of the protein backbone had to be rebuilt during modeling of the second tBPA 2B4 adduct structure due to differences compared to the molecular replacement solution. The B/C loop, F-G cassette, C-terminus of the helix H, and N-terminus of helix I of the solution, all corresponding to known plastic regions (36), did not fit the density of the initial map. Upon rebuilding to fit the electron density, the protein was found to dimerize by burying the largely hydrophobic F′ helix of one chain in the active site of the opposite monomer (Suppl. Fig. S3), similarly to the previously determined 1-phenylbenzyl imidazole (1-PBI) (37) and bifonazole (36) complexes of 2B4. In this conformation, the B/C loop extends upward and slightly away from the active site. Additionally, after an approximately 180° turnaround in the B/C loop, a longer than usual B′ helix, was observed to span from Pro 106 to Ala 116 before connecting to the C helix. The C helix was found in a conformation more similar to the closed structures than to those of the two other dimerized forms of 2B4. The F-G cassette also differed significantly from the initial model. The F helix begins to unwind at Phe 203 and extends toward its dimer pair. Stretching from Phe 212 to Phe 220, the F′ helix of one chain packs against the tBPA molecule and a small portion of the I helix of the other chain. Once the F′ helix terminates, the backbone meanders back toward the G helix. As observed in other the 2B4 dimers, the G helix is angled upward, away from the I helix, to allow for the F′ helix of the other monomer to enter the active site cavity. Furthermore, the H and I helices alter their conformations as well. The H helix is a half turn shorter than in the closed structures and turns slightly upward at its C-terminus, rising toward the I helix. The I helix of this conformation begins at Pro 279 and is one and a half turns longer at its N-terminus than the closed forms of 2B4. It also bends upward toward the G helix.
As in the closed structure, electron density connected to OG1 of Thr 302 is consistent with the size and shape of the proposed 2B4-tBPA conjugate (Fig. 2B). However, in contrast to the closed structure, there is no evidence in the electron density maps for a water molecule above the plane of the heme. The orientation of tBPA in the open structure points the inactivator toward the N-terminus of the C helix, where it lies almost parallel to the heme plane, but maintains the angle with respect to the I helix observed in the closed structure. Residues that were found within 5 Å of tBPA are Arg 98, Trp 121, Ser 294, Ala 298, Thr 302, Ile 363, and Val 367. Because of the dimerization, Val 216, Phe 217, Phe 220, and Pro 221 from the F′ helix of the other monomer also contact tBPA (Fig. 2B).
Several whole or partial Cymal-5 molecules as well as four sucrose molecules (one per chain) were also observed in the electron density and modeled.
DLS reveals that the tBPA-modified 2B4 is a monomer
To determine which conformation observed by X-ray crystallography occurred in solution, DLS was used to determine the aggregation state of the modified enzyme. DLS results of tBPA-conjugated 2B4 and P450eryF are summarized in Table 2. The hydrodynamic radii and molecular weights of the aggregate are shown in the first two columns. Because of estimations made to calculate these values, they should be considered approximate. The third column shows the molecular weights of the monomer calculated from the amino acid sequence. The ratio of the second and third columns is shown in the last column to compare the aggregation states of monomeric P450eryF to tBPA-conjugated 2B4. The ratio of 2B4-tBPA was similar to monomeric P450eryF, suggesting that the modified 2B4 is also a monomer under these conditions. Previous results using analytical ultracentrifugation of (38) have shown that unlabeled 2B4 is monomeric under similar conditions. Additionally, results from DLS experiments using unlabeled 2B4 under these monomeric conditions (unpublished results) were consistent with those observed with tBPA-labeled enzyme.
Table 2.
Analysis of DLS
| Hydrodynamic Radius (nm)a | Molecular weight of the Aggregate (kDa)a | Molecular weight of the Monomer (kDa)b | Ratioc | |
|---|---|---|---|---|
| 2B4-tBPA | 4.01 | 86.8 | 54 | 1.6 |
| P450eryF | 3.60 | 67.2 | 45 | 1.5 |
Hydrodynamic radii and the calculated molecular weight were determined assuming a viscosity and refractive index.
The molecular weight were calculated using the amino acid sequences of truncated 2B4 (34) and P450eryF (46).
The ratio between the calculated MW of the aggregate and the theoretical MW of the monomer.
Deuterium incorporation shows differential solvent exposure upon covalent labeling of 2B4
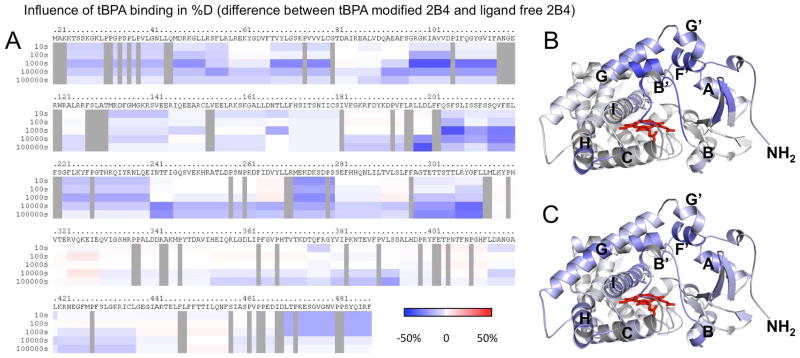
To give further insight into the conformation of tBPA-modified 2B4 in solution, we used DXMS, which correlates amide proton-deuterium exchange and solvent accessibility. Covalent labeling of the protein by tBPA caused slowing of the deuteration rate across the plastic regions of the enzyme as seen previously in the enzyme when bound to 4-CPI (Fig. 3A) (32). The greatest difference in deuteration rates across the B′- and C-helices for tBPA labeled protein and protein in the presence of 4-CPI compared to enzyme alone is at 1000 s. Across the F-G cassette, 2B4 in the presence of 4-CPI shows the greatest difference in deuteration rates at 10000 s, but in tBPA labeled protein the greatest difference is at 1000 s. This difference in deuteration rates could be explained by the exchange of 4-CPI between bound and unbound states, while tBPA is covalently attached to the protein, not forming an “unbound” state. Furthermore, in addition to changes in deuteration rates across the plastic regions of 2B4, tBPA labeled protein also exhibits slowed deuteration rates across the A′, A, E, H, and I helices, the H/I Loop, and β-strands β1-1, β1–2, β1–3, β1–4, β1–5, β2–1, and β2–2 (Suppl. Fig. S2). Furthermore, the changes in deuteration rates between ligand-free and tBPA-labeled protein appear to be biphasic across time. Labeling causes immediate slowing across the N- and C-termini, the A′, B′, C, F, F′, G′, and H helices, the N-terminal end of the G helix, the C-terminal end of the I helix, and the C/D and H/I Loops (Fig. 3B). These portions of the protein show continued differences through the 1000s time point. After that time, the A and E helices, the C-terminal end of the G helix, and the β-strands show varying levels of differences in deuteration rates between the unlabeled and labeled protein conditions (Fig. 3C).
Figure 3.
Effects of labeling on hydrogen-deuterium exchange of 2B4. A) Difference in deuteration level of ligand-free 2B4 and tBPA-labeled 2B4 is shown with slowing in blue and acceleration in red and darker colors indicating greater magnitude of change, as indicated by the color bar. Each bar under the primary sequence is divided into rows corresponding to each time point from 10 to 100,000 s (top to bottom). B) Difference in deuteration levels after 1000 s mapped onto the closed tBPA-modified 2B4 structure. C) Difference in deuteration levels after 10,000 s mapped onto closed tBPA-modified 2B4 structure.
Ligand Docking
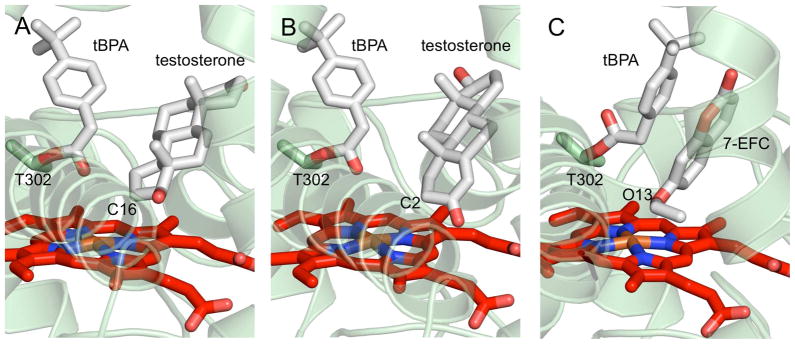
In order to explore the structural basis for the previously reported partial activity of tBPA-modified 2B4 toward several substrates (13), we utilized computer aided docking of 7-EFC or testosterone into the active site of the open structure of 2B4-tBPA. These experiments resulted in multiple complexes with similar energy between −5.6 and −7.2 kcal mol−1 for 7-EFC (100 modeling events) and between −6.8 and −9.0 kcal mol−1 for testosterone (99 modeling events). In both cases, a large number of models showed movement of tBPA away from the heme regardless of substrate proximity to the heme, 44 for 7-EFC and 52 for testosterone. The substrates were docked within 5 Å of the heme iron in seventeen poses with 7-EFC and eleven poses with testosterone. For testosterone, the substrate was within 5 Å of the heme iron in eleven poses, and C16 was positioned within 5 Å of the heme iron (~4 Å) in five poses (Fig. 4A). C2 was with 5 Å of the heme iron in three poses (Fig. 4B). In the case of 7-EFC, four models showed O13 within 5 Å of the heme iron (Fig. 4C); if the cutoff is expanded to 6Å, nine models are included.
Figure 4.
Ribbon and stick depictions of results of ligand docking into the open tBPA-modified 2B4 structure. These experiments show that a flexible Thr 302-tBPA adduct is able to rotate away from the heme to give testosterone or 7-EFC access to the heme iron. Known points of oxidation were found within 5 Å of the heme iron in several poses. These were A) carbon 16 of testosterone in five poses, B) carbon 2 of testosterone in three poses, and C) oxygen 13 of 7-EFC in four poses.
Discussion
Nearly thirty years have elapsed since the first description of covalent modification of the protein moiety of a P450 by an inactivator (39). Despite the interest in MBI as a probe of P450 function and the long list of P450 structures available in the PDB, we now present the first two structures of a mammalian P450 with a bound adduct. The two 2B4 conformations reported here highlight the pronounced flexibility of P450s 2B, where several different enzyme conformations have been previously observed by X-ray crystallography in the presence of a variety of ligands. This is the first time that two crystal forms of the same mammalian P450 complex have been reported in two distinct protein conformations.
Comparison of the open (dimeric) and closed (monomeric) forms of tBPA-modified 2B4 shows that the enzyme retains its previously observed plasticity (34, 36, 37) in the presence of the covalent label and reveals movement of tBPA while attached to Thr 302 in the active site. A previous in silico study utilized another closed form of 2B4 (the 4-CPI complex) and docked unmetabolized tBPA that was not linked to Thr 302. Analysis of those results showed that tBPA binds above the heme plane in the active site of 2B4 while making contact with several residues found within a 5 Å radius. These are Ile 101, Val 104, Ile 114, Phe 115, Ile 209, Phe 297, Ala 298, Glu 301, Thr 302, Ile 363, Gly 366, Val 367, Pro 368, Val 477 (13). Not surprisingly, that analysis most closely agrees with the results of the closed tBPA-modified 2B4 structure reported here. In the closed crystal structure, tBPA makes a smaller angle with the heme plane compared to that in the previous docking study, which yields a slightly different list of contact residues. Val 104, Phe 108, Ile 114, Phe 115, Phe 206, Ile 209, Phe 297, Ala 298, Glu 301, Thr 302, Ile 363, Val 367, and Val 477 were all found within 5 Å of bound tBPA. The orientation of tBPA in the closed crystal structure also allows for an interaction between the carbonyl oxygen and a water molecule bound to the heme iron. The presence of this water molecule bound to the heme iron is supported by previously reported low-spin resonance Raman (20) and EPR spectra (40).
The conformations of the majority of active site residues in this closed form of 2B4-tBPA are unchanged when compared with other closed 2B structures. However, there are three residues that differ. Glu 301, which sometimes points into the active site (as in the 4-CPI complex), is directed out of the active site in the tBPA-conjugated structure. This places Glu 301 within hydrogen bonding distance of His 172, a known interaction partner. Additionally, a pair of Phe residues (206 and 297) adopts a conformation where Phe 206 points into the active site, while Phe 297 points out. This is consistent with the 1-CPI and clopidogrel structures, but opposite to the three other closed structures (4-CPI and ticlopidine complexes, and one of the two ligand-free structures), where Phe 297 points in and Phe 206 points out. The flipping of these Phe residues appears to occur to accommodate the ligands in the active site. In the case of tBPA, the ‘in’ orientation of Phe 297 would clash with the tert-butyl and phenyl groups of the inhibitor.
A comparison of this closed form of 2B4-tBPA with the open tBPA-labeled 2B4 crystal structure reveals a distinctly altered protein state. The open form is dimerized as observed in previous structures of 2B4 bound to the large inhibitors 1-PBI and bifonazole. Dimerization of 2B4-tBPA leads to significant changes in the orientation of tBPA and the residues with which it makes contact in the active site. Val 104, Phe 108, Ile 114, Phe 115, Phe 206, Ile 209, Phe 297, and Val 477 are no longer found within van der Waals contact distance. Also, the orientation of the inactivator is such that Arg 98, Trp 121, and Ser 294 are now within contact distance. This dimerization appears to be unique to the CYP2B subfamily and its physiological relevance is currently unknown. However, movement of the F-G region to create more open structures has also been observed in bacterial CYP101A1 (41), CYP101C1 (42), and CYP101D2 (43), suggesting a mechanism used by a variety of P450s to allow for substrate access or perhaps to alter the size and shape of the active site.
To correlate how the two distinct crystal forms of tBPA-modified 2B4 relate to the behavior of the enzyme in solution, we utilized DXMS. Recently, deuterium exchange coupled with mass spectrometry has been used to show that ligand-free 2B4 adopts a predominantly open conformation in solution, while binding of the small molecule 4-CPI slowed the deuteration rates of the protein (32) across previously defined plastic regions (36), which translates to a predominantly closed conformation in solution. Coupled with the DLS results, DXMS experiments show that the enzyme is monomeric under the conditions of the DXMS experiment, and that the labeled enzyme is primarily a closed monomer in solution the majority of the time. However, the two crystal structures suggest an equilibrium exists between the open and closed forms of the labeled enzyme.
With compact conformations of the B/C loop region and F-G helical cassette, and the active site residues packing well against the covalent ligand, the closed form of the labeled enzyme leaves essentially no room for substrate binding and catalysis (Fig. 2A). Therefore, the open form may be more pertinent to explaining the previously reported partial enzymatic activity (13). However, the positioning of the tBPA probe in the active site of this open form leaves questions about substrate access to the heme. In the crystal structure of this conformation, tBPA is seen laying flat against the heme due to the presence of the F′ helix from its dimer pair (Fig. 2B). This would suggest that even when there is sufficient space for a substrate to bind, no catalysis could occur because substrates would lack access to the redox center of the enzyme. However, comparison of the open and closed structures provide physical evidence that tBPA is able to occupy different positions in the 2B4 active site (Fig. 2C). Furthermore, docking experiments using the monomer of the open form, in the absence of its partner’s F′ helix in the active site, show that free rotation of chemical bonds of the Thr 302-tBPA (Fig. 1) adduct allow for the inactivator to move upward to give both testosterone (Figs. 4A, 4B) and 7-EFC (Fig. 4C) access to the heme. In these experiments, testosterone was able to orient itself with carbon 16 or carbon 2 within 5 Å of the heme iron in five and three poses, respectively. These results are consistent with previous reports that 2B4 oxidation of testosterone results primarily in 16α and 2α hydroxylation (44). Additionally, four poses showed 7-EFC oriented with oxygen 13 within 5 Å of the heme iron, which is in agreement with the O-deethylation activity of enzymes in P450 families 1, 2, and 3 (45). These results imply that there could exist other conformations of open forms of 2B4-tBPA that would allow other substrates to enter the active site in the proper orientations, resulting in partial activity.
In conclusion, the crystal structures of tBPA-modified 2B4 provide definitive evidence for the covalent modification of the highly conserved threonine residue Thr 302 by tBPA during mechanism-based inactivation. As shown in Figure 2, the OG1 atom of Thr 302 is covalently linked to the terminal carbon of the acetylenic group via an ester bond. Although a number of previous studies pointed to Thr 302 as the possible site for chemical modification by acetylenic compounds, no direct evidence was provided until recently (13). In particular, these crystal structures reveal the structural changes that occur upon covalent adduction of tBPA to Thr 302 and provide insight for the partial activity of the tBPA-modified 2B4. It seems that the remarkable conformational plasticity of 2B4 rescues the partial enzyme activity by allowing substrate entrance in an open conformation. Furthermore, these crystal structures are consistent with the previous in silico studies for predicting the potency of tBPA; the close proximity of the terminal carbon of tBPA to both the heme iron and the hydroxyl oxygen of Thr 302 renders the vulnerability of Thr 302 for chemical modification. Conversely, this unique binding mode can also be explored for rational design of potent mechanism-based inhibitors by targeting this highly conserved Thr 302.
Supplementary Material
Acknowledgments
We thank the staff at the Stanford Synchrotron Radiation Lightsource for assistance with data collection. The Stanford Synchrotron Radiation Lightsource is operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences. The Stanford Synchrotron Radiation Lightsource is supported by the National Institute of Health, the National Center for Research Resources, the Biomedical Technology Program and the U.S. Department of Energy of Biological and Environmental Research.
The abbreviations used are
- tBPA
tert-butylphenylacetylene
- 4-CPI
4-(4-chlorophenyl)imidazole
- 1-CPI
1-(4-chlorophenyl)imidazole
- 1-PBI
1-phenylbenzyl imidazole
- Cymal-5
5-cyclo-hexylpentyl-β-D-maltoside
- 7-EFC
7-ethyoxy-4-trifluoromethylcoumarin
- TFA
trifluoroacetic acid
- EDTA
ethylenediaminetetraacetic acid
- DTT
dithiothreitol
- DLPC
dilauroylphosphatidylcholine
- 232-chol
3α,7α,12α-tris[(β-D-maltopyranosyl)ethyloxy]cholane
- Ni-NTA
nickel-nitrilotriacetic acid
- MBI
mechanism-based inactivation
- DXMS
deuterium exchange mass spectrometry
- DLS
dynamic light scattering
- SSRL
Stanford Synchrotron Radiation Lightsource
- PDB
Protein Data Bank
- RMSD
root mean square deviation
Footnotes
This research was supported by NIH grants ES003619 to J.R.H, CA16954 to P.F.H., GM073197 to Q.Z., and CA099835, CA118595, AI076961, AI081982, AI2008031, GM020501, GM066170, and RR029388 to V.L.W. P.R.W. is supported by T32-DK07233.
Atomic coordinates have been deposited at the Protein Data Bank (accession codes 3R1A, 3R1B).
Supporting Information Available
Confirmation of the chemical modification of 2B4 by tBPA. Structure of the closed monomeric form of tBPA-modified 2B4. Structure of the open dimeric form of tBPA-modified 2B4. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hollenberg PF, Kent UM, Bumpus NN. Mechanism-based inactivation of human cytochromes P450s: Experimental characterization, reactive intermediates, and clinical implications. Chem Res Toxicol. 2008;21:189–205. doi: 10.1021/tx7002504. [DOI] [PubMed] [Google Scholar]
- 2.Shebley M, Kent UM, Ballou DP, Hollenberg PF. Mechanistic analysis of the inactivation of cytochrome P450 2B6 by phencyclidine: Effects on substrate binding, electron transfer, and uncoupling. Drug Metab Disp. 2009;37:745–752. doi: 10.1124/dmd.108.024661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kent UM, Mills DE, Rajnarayanan RV, Alworth WL, Hollenberg PF. Effect of 17-alpha-ethynylestradiol on activities of cytochrome P450 2B (P450 2B) enzymes: Characterization of inactivation of P450s 2B1 and 2B6 and identification of metabolites. J Pharmacol Exp Ther. 2002;300:549–558. doi: 10.1124/jpet.300.2.549. [DOI] [PubMed] [Google Scholar]
- 4.Halpert JR, Miller N, Gorsky L. On the mechanism of the inactivation of the major phenobarbital-inducible isozyme of rat liver cytochrome P450 by chloramphenicol. J Biol Chem. 1985;260:8397–8403. [PubMed] [Google Scholar]
- 5.Kedzie KM, Balfour CA, Escobar GY, Grimm SW, He Y-A, Pepperl DJ, Regan JW, Stevens JC, Halpert JR. Molecular basis for a functionally unique cytochrome P450IIB1 variant. J Biol Chem. 1991;266:22515–22521. [PubMed] [Google Scholar]
- 6.Roberts ES, Alworth WL, Hollenberg PF. Mechanism-based inactivation of cytochromes P450 2E1 and 2B1 by 5-phenyl-1-pentyne. Arch Biochem Biophys. 1998;354:295–302. doi: 10.1006/abbi.1998.0679. [DOI] [PubMed] [Google Scholar]
- 7.Roberts ESNEH, Alworth WL, Hollenberg PF. Mechanism-based inactivation of cytochrome P450 2B1 by 2-ethynylnaphthalene: Identification of an active-site peptide. Chem Res Toxicol. 1993;6:470–479. doi: 10.1021/tx00034a013. [DOI] [PubMed] [Google Scholar]
- 8.Roberts ES, Hopkins NE, Zaluzec EJ, Gage DA, Alworth WL, Hollenberg PF. Identification of active-site peptides from 3H-labeled 2-ethynylnaphthalene-inactivated P450 2B1 and 2B4 using amino acid sequencing and mass spectrometry. Biochemistry. 1994;33:3766–3771. doi: 10.1021/bi00178a037. [DOI] [PubMed] [Google Scholar]
- 9.Regal KA, Schrag ML, Kent UM, Wienkers LC, Hollenberg PF. Mechanism-based inactivation of cytochrome P450 2B1 by 7-ethynylcoumarin: Verification of apo-P450 adduction by electrospray ion trap mass spectrometry. Chem Res Toxicol. 2000;13:262–270. doi: 10.1021/tx990195s. [DOI] [PubMed] [Google Scholar]
- 10.Blobaum AL, Harris DL, Hollenberg PF. P450 active site architecture and reversibility: Inactivation of cytochromes P450 2B4 and 2B4 T302A by tert-butyl acetylenes. Biochemistry. 2005;44:3831–3844. doi: 10.1021/bi0478953. [DOI] [PubMed] [Google Scholar]
- 11.Lin HL, Zhang H, Noon KR, Hollenberg PF. Mechanism-based inactivation of CYP2B1 and its F-helix mutant by two tert-butyl acetylenic compounds: Covalent modification of prosthetic heme versus apoprotein. J Pharmacol Exp Ther. 2009;331:392–403. doi: 10.1124/jpet.109.158782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin HL, Zhang H, Jushchyshyn M, Hollenberg PF. Covalent modification of Thr302 in cytochrome P450 2B1 by the mechanism-based inactivator 4-tert-butylphenylacetylene. J Pharmacol Exp Ther. 2010;333:663–669. doi: 10.1124/jpet.109.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Lin H-L, Walker VJ, Hamdane D, Hollenberg PF. tert-Butylphenylacetylene is a potent mechanism-based inactivator of cytochrome P450 2B4: Inhibition of cytochrome P450 catalysis by steric hinderance. Mol Pharmacol. 2009;76:1011–1018. doi: 10.1124/mol.109.059808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kent UM, Juschyshyn MI, Hollenberg PF. Mechanism-based inactivators as probes of cytochrome P450 structure and function. Curr Drug Metab. 2001;2:215–243. doi: 10.2174/1389200013338478. [DOI] [PubMed] [Google Scholar]
- 15.Nagano S, Poulos TL. Crystallographic study on the dioxygen complex of wild-type and mutant cytochrome P450cam. Implications for the dioxygen activation mechanism. J Biol Chem. 2005;280:31659–31663. doi: 10.1074/jbc.M505261200. [DOI] [PubMed] [Google Scholar]
- 16.Schlichting I, Berendzen J, Chu K, Stock AM, Maves SA, Benson DE, Sweet RM, Ringe D, Petsko GA, Sligar SG. The catalytic pathway of cytochrome P450cam at atomic resolution. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 17.Vaz ADN, Pernecky SJ, Raner GM, Coon MJ. Peroxo-iron and oxenoid-iron species as alternative oxyegenating agents in cytochrome P450-catalyzed reactions: Switching by threonin-302 to alanine mutagenesis of cytochrome P450 2B4. Proc Natl Acad Sci USA. 1996;93:4644–4648. doi: 10.1073/pnas.93.10.4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimata Y, Shimada H, Hirose T, Ishimura Y. Role of Thr-252 in cytochrome P450cam: A study with unnatural amino acid mutagenesis. Biochem Biophys Res Commun. 1995;208:96–102. doi: 10.1006/bbrc.1995.1310. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda T, Imai Y, Komori M, Nakamura M, Kusunose E, Satouchi K, Kusunose M. Replacement of Thr-303 of P450 2E1 with serine modifies the regioselectivity of its fatty acid hydroxylase activity. J Biochem. 1993;113:7–12. doi: 10.1093/oxfordjournals.jbchem.a124006. [DOI] [PubMed] [Google Scholar]
- 20.Mak PJ, Zhang H, Hollenberg PF, Kincaid JR. Defining the structural consequences of mechanism-based inactivation of mammalian cytochrome P450 2B4 using resonance Raman spectroscopy. J Am Chem Soc. 2010;132:1494–1495. doi: 10.1021/ja910276s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q, Ma X, Ward A, Hong W, Jaakola V, Stevens RC, Finn MG, Chang G. Designing facial amphiphiles for the stabilization of integral membrane proteins. Angew Chem Int Ed. 2007;46:7023–7025. doi: 10.1002/anie.200701556. [DOI] [PubMed] [Google Scholar]
- 22.Gay SC, Roberts AG, Maekawa K, Talakad JC, Hong W-X, Zhang Q, Stout CD, Halpert JR. Structures of cytochrome P450 2B4 complexed with the antiplatelet drugs ticlopidine and clopidogrel. Biochemistry. 2010;49:8709–8720. doi: 10.1021/bi100914z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Kenaan C, Hamdane D, Hoa GH, Hollenberg PF. Effect of conformational dynamics on substrate recognition and specificity as probed by the introduction of a de novo disulfide bond into cytochrome P450 2B1. J Biol Chem. 2009;284:25678–25686. doi: 10.1074/jbc.M109.032748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leslie AGW. Integration of macromolecular diffraction data. Acta Crystallogr Sect D Biol Crystallogr. 1999;D55:1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch W. Evaluation of single-crystal X-ray diffraction data from a position-sensitive detector. J Appl Crystallogr. 1988;21:916–924. [Google Scholar]
- 26.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr Sect D Biol Crystallogr. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Lohkamp B, Scott W, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D Biol Crystallogr. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helms V, Wade RC. Thermodynamics of water mediating protein-ligand interactions in cytochrome P450cam: A molecular dynamics study. Biophys J. 1995;69:810–824. doi: 10.1016/S0006-3495(95)79955-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilderman PR, Shah MB, Liu T, Li S, Hsu S, Roberts AG, Goodlett DR, Zhang Q, Woods VLJ, Stout CD, Halpert JR. Plasticity of cytochrome P450 2B4 as investigated by hydrogen-deuterium exchange mass spectrometry and X-ray crystallography. J Biol Chem. 2010;285:38601–38611. doi: 10.1074/jbc.M110.180646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeLano WL MacPyMOL. The PyMOL molecular graphics system. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- 34.Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9 A resolution: Insight into the range of P450 conformations and coordination of redox partner binding. J Biol Chem. 2004;279:27294–27301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y, Sun L, Muralidhara BK, Kumar S, White MA, Stout CD, Halpert JR. Structural and thermodynamic consequences of 1-(4-chlorophenyl)imidazole binding to cytochrome P450 2B4. Biochemistry. 2007;46:11559–11567. doi: 10.1021/bi7011614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y, White MA, Muralidhara BK, Sun L, Halpert JR, Stout CD. Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: Insight into P450 conformational plasticity and membrane interaction. J Biol Chem. 2006;281:5973–5981. doi: 10.1074/jbc.M511464200. [DOI] [PubMed] [Google Scholar]
- 37.Gay SC, Sun L, Maekawa K, Halpert JR, Stout CD. Crystal structures of cytochrome P450 2B4 in complex with the inhibitor 1-biphenyl-4-methyl-1H-imidazole: Ligand induced structural response through α-helical repositioning. Biochemistry. 2009;48:4762–4771. doi: 10.1021/bi9003765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muralidhara BK, Negi S, Chin CC, Braun W, Halpert JR. Conformational flexibility of mammalian cytochrome P450 2B4 in binding imidazole inhibitors with different ring chemistry and side chains. Solution thermodynamics and molecular modeling. J Biol Chem. 2006;281:8051–8061. doi: 10.1074/jbc.M509696200. [DOI] [PubMed] [Google Scholar]
- 39.Halpert J. Covalent modification of lysine during the suicide inactivation of rat liver cytochrome P-450 by chloramphenicol. Biochem Pharmacol. 1981;30:875–881. doi: 10.1016/s0006-2952(81)80010-x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Lin HL, Kenaan C, Hollenberg PF. Targeting of the highly conserved threonine 302 residue of cytochromes P450 2B family during mechanism-based inactivation by aryl acetylenes. Arch Biochem Biophys. 2011;507:135–143. doi: 10.1016/j.abb.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee YT, Wilson RF, Rupniewski I, Goodin DB. P450cam visits an open conformation in the absence of substrate. Biochemistry. 2010;49:3412–3419. doi: 10.1021/bi100183g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma M, Bell SG, Yang W, Hao Y, Rees NH, Bartlam M, Zhou W, Wong LL, Rao Z. Structural analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444. ChemBioChem. 2011;12:88–99. doi: 10.1002/cbic.201000537. [DOI] [PubMed] [Google Scholar]
- 43.Yang W, Bell SG, Wang H, Zhou W, Bartlam M, Wong LL, Rao Z. The structure of CYP101D2 unveils a potential path for substrate entry into the active site. Biochem J. 2010;433:85–93. doi: 10.1042/BJ20101017. [DOI] [PubMed] [Google Scholar]
- 44.Hernandez CE, Kumar S, Liu H, Halpert JR. Investigation of the role of cytochrome P450 2B4 active site residues in substrate metabolism based on crystal structures of the ligand-bound enzyme. Arch Biochem Biophys. 2006;455:61–67. doi: 10.1016/j.abb.2006.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waxman DJ, Lapenson DP, Aoyama T, Gelboin HV, Gonzalez FJ, Korzekwa K. Steroid hormone hydroxylase specificities of eleven cDNA-expressed human cytochrome P450s. Arch Biochem Biophys. 1991;290:160–166. doi: 10.1016/0003-9861(91)90602-f. [DOI] [PubMed] [Google Scholar]
- 46.Cupp-Vickery J, Anderson R, Hatziris Z. Crystal structures of ligand complexes of P450eryF exhibiting homotropic cooperativity. Proc Natl Acad Sci USA. 2000;97:3050–3055. doi: 10.1073/pnas.050406897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.