Abstract
The Hedgehog (Hh) morphogen directs distinct cell responses according to its distinct signalling levels. Hh signalling stabilizes transcription factor cubitus interruptus (Ci) by prohibiting SCFSlimb-dependent ubiquitylation and proteolysis of Ci. How graded Hh signalling confers differential SCFSlimb-mediated Ci proteolysis in responding cells remains unclear. Here, we show that in COP9 signalosome (CSN) mutants, in which deneddylation of SCFSlimb is inactivated, Ci is destabilized in low-to-intermediate Hh signalling cells. As a consequence, expression of the low-threshold Hh target gene dpp is disrupted, highlighting the critical role of CSN deneddylation on low-to-intermediate Hh signalling response. The status of Ci phosphorylation and the level of E1 ubiquitin-activating enzyme are tightly coupled to this CSN regulation. We propose that the affinity of substrate–E3 interaction, ligase activity and E1 activity are three major determinants for substrate ubiquitylation and thereby substrate degradation in vivo.
 Hedgehog signalling gradients are required for proper wing formation in Drosophila, and Hedgehog is known to regulate the cubitus interruptus transcription factor. Here, the authors show that the COP9 signalosome has a critical role in translating a Hedgehog gradient into a cubitus interruptus gradient.
Hedgehog signalling gradients are required for proper wing formation in Drosophila, and Hedgehog is known to regulate the cubitus interruptus transcription factor. Here, the authors show that the COP9 signalosome has a critical role in translating a Hedgehog gradient into a cubitus interruptus gradient.
In developing tissues, morphogens are produced and secreted from different sources to build concentration gradients that serve as positional information, directing cells of different distances away from the source to adapt distinct fates1,2. To pattern complex tissues with precision, it is conceivable that the intracellular signalling level should scale to the morphogen gradient3,4,5,6. The Hedgehog (Hh) family proteins are evolutionarily conserved. As secreted morphogens, they have been implicated in patterning the formation of vertebrate face, spinal cord and digits, and Drosophila eyes, wings and several others4,7,8,9,10. In Drosophila developing wing discs, Hh signalling involves at least three distinct mechanisms in regulating the zinc-finger transcription factor cubitus interruptus (Ci), namely, blocking the proteolysis of the transcriptional activator full-length Ci (Ci155) (refs 11, 12, 13), enhancing Ci155 nuclear partitioning14,15,16,17 and transactivation activity18,19,20. In the absence of Hh signalling, Ci155 is partially proteolysed to generate the transcriptional repressor Ci (Ci75). Ci155 proteolysis requires consecutive phosphorylation of Ci155 by protein kinase A (PKA)21,22, glycogen synthase kinase 3β (GSK3β)23,24 and casein kinase I (CKI)23,25, resulting in a high affinity for the F-box protein Slimb of the SCFSlimb ubiquitin (Ub) ligase complex for ubiquitylation13,26,27. Although it had been demonstrated that Hh signalling stabilizes Ci155 by precluding phosphorylation-dependent ubiquitylation of Ci155 (refs 23, 24), how graded Hh signals are translated into graded Ci155 proteolysis is not clear.
The Ub E3 ligase activities of SCF (skp1-cullin-F-box protein) and other types of cullin–RING ligases (CRLs) are regulated by the conjugation of the Ub-like polypeptide Nedd8 onto the cullin scaffold component of CRLs, a process known as neddylation28,29. Genetic analyses performed in neddylation pathway mutants of various species indicate that Nedd8 modification of cullins is essential for CRL activities27,30,31,32,33,34. For example, neddylation is required for SCFSlimb-mediated Ci155 proteolysis in cells not receiving Hh, thus suppressing Hh-mediated gene activation12. Nedd8 conjugation at a conserved carboxy-terminal lysine in all cullin proteins facilitates the binding of the RING domain-containing protein Rbx1/ROC1 that recruits Ub-loaded E2s29,35. Furthermore, neddylation induces a conformational change of the Cul5 carboxy terminus, which facilitates the transfer of Ub onto substrates36. Consistently, neddylation reduces the Km for E2 binding to CRL and increases the Kcat for substrate ubiquitylation, thus increasing the processivity of ubiquitylation37. It has been estimated that neddylation of SCF allows the processive addition of more than four Ubs in a productive substrate–SCF encounter38. The Nedd8 moiety on cullins can be deconjugated by the COP9 signalosome (CSN) complex, thus inactivating CRL ligase activities39,40. Similar to other reversible protein modifications, neddylation and deneddylation of cullins occur dynamically in vivo30,41. Little is known whether dynamic deneddylation of CRLs is indeed critical to the substrate stability through modulating the extent of substrate ubiquitylation in vivo.
To better understand the role of deneddylation in Ci155 stability, in response to the Hh gradient, in this study, we used the Drosophila wing disc as a model in which cells display distinct responses to the Hh morphogen because of their positions. We analysed the SCFSlimb-mediated Ci155 proteolysis in mutants for CSN, GSK3β (sgg in Drosophila), Cul1, slimb and E1 Ub-activating enzyme uba1. Taken together, our results suggest that deneddylation by regulating SCFSlimb activity protects a portion of partially phosphorylated Ci155 from proteolysis, thus establishing the low-threshold Hh signalling responses.
Results
CSN deneddylation regulates the protein stability of Slimb
Whereas neddylation of cullins promotes CRL activities, deneddylation of cullins also promotes substrate degradation, mainly through protecting cullins and substrate receptors from neddylation-induced degradation42,43,44,45,46. To test whether protection of substrate receptors by deneddylation is a general rule, we first examined the protein stability of Slimb, a substrate receptor for the SCF complex that ubiquitylates several substrates including Ci155 (refs 13, 47). Mutant clones for the CSN catalytic subunit CSN5 (CSN5null) were generated in third-instar wing discs that also carry the myc-slimb transgene under the control of the ubiquitous tubulin promoter. In CSN5null cells, the protein level of Myc-Slimb is diminished (arrow in Fig. 1a), suggesting that the CSN is required to stabilize Slimb in vivo. In contrast, neddylation promotes Myc-Slimb turnover, as Myc-Slimb accumulates to high levels in cells homozygous for Nedd8AN015 (arrow in Fig. 1b). To test whether the SCF-Slimb interaction is essential for neddylation-mediated Slimb turnover, we examined the protein stability of Myc-SlimbΔfbx that cannot be incorporated into the SCF complex because of the truncation of the F-box47. In contrast to the full-length Slimb, the protein levels of SlimbΔfbx in CSN5null mutant and neighbouring wild-type cells are indistinguishable (Fig. 1c). Taken together, we conclude that the substrate receptor Slimb, when incorporated into the SCF complex, is regulated by neddylation-induced degradation. However, neddylation-induced degradation of substrate receptors is unlikely a general rule, as the protein level of the Flag-conjugated F-box protein Archipelago (Flag-Ago) is unaltered in CSN5null cells (Fig. 1d), suggesting that neddylation and deneddylation regulate the protein stability of some but not all of the SCF substrate receptors in Drosophila.
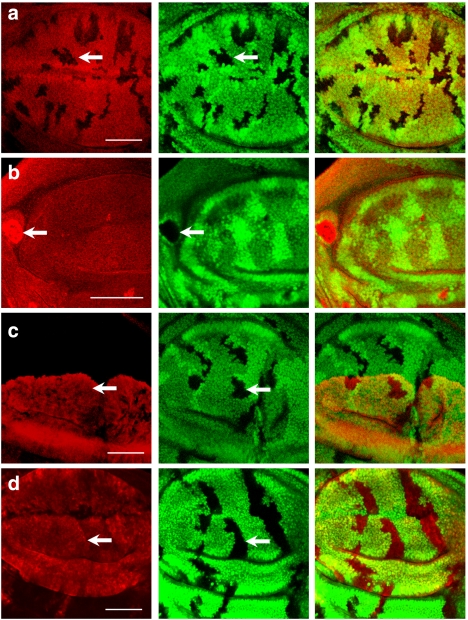
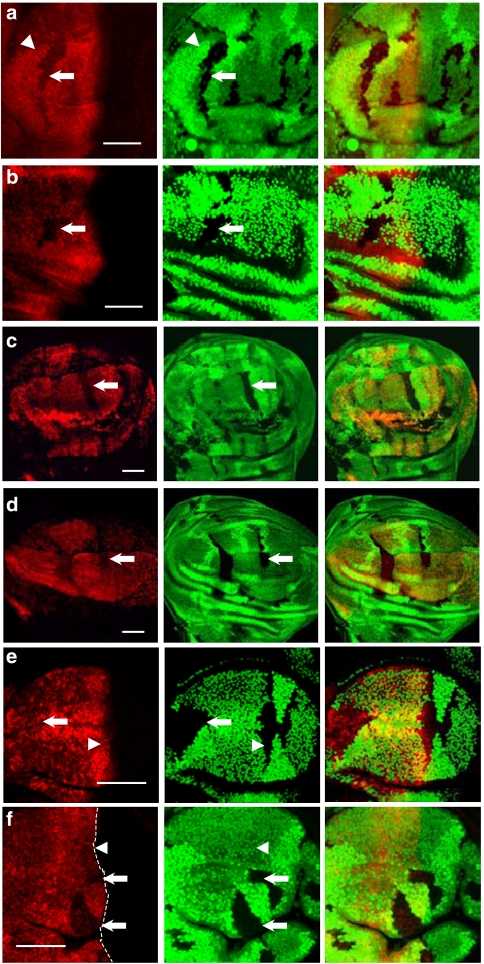
Figure 1. Effects of neddylation and deneddylation on F-box proteins Slimb and Ago.
(a) CSN5null clones generated in Drosophila third-instar larval wing discs are revealed by the absence of GFP (green). The protein level of tubulin promoter-driven myc-slimb (red) decreases in GFP-negative CSN5null clones (arrow). (b) Nedd8AN015 clones marked by the absence of GFP (green) were generated in wing discs expressing Myc-Slimb. The level of Myc-Slimb (red) in Nedd8AN015 clones is higher (arrow) compared with wild type. (c) The level of Myc-SlimbΔfbx (red) expressed by ap-GAL4 is unaltered by CSN5null mutation (arrow) in the dorsal compartment of wing discs. (d) GFP-negative CSN5null clones are generated in wing discs (green), in which Flag-Ago (red) is expressed in wing pouches under the control of ms1096-GAL4. Flag-tag staining of Flag-Ago protein has a comparable signal level in wild-type and CSN5null cells (arrow). Scale bars, 50 μm.
Reduction in the protein levels of Slimb and Cul142 in CSN mutants could promote the accumulation of SCFSlimb substrates. This was tested by examining the protein level of a typical SCFSlimb substrate Armadillo (Arm), the Drosophila ortholog of β-catenin13,27. In line with recent reports48,49, the Arm level in CSN5null cells is slightly but consistently higher than that in wild-type cells (Supplementary Fig. S1).
Deneddylation enhances Ci155 stability
We also tested how deneddylation regulates another SCFSlimb substrate Ci155 in wing discs13,27. Unlike the increased level of Arm, the Ci155 protein level is instead downregulated in both CSN5null and CSN4null mutant cells (Fig. 2a,b), suggesting that neddylated SCFSlimb degrades more Ci155, despite the reduced levels of both Cul142 and the substrate receptor Slimb (Fig. 1a) in CSN mutants. Thus, Ci155 proteolysis is more sensitive to the level of neddylation, but less so to the bulk concentrations of SCFSlimb components. In comparison to mutant clones in the low Hh signalling region (arrowhead in Fig. 2a), the reduction in the Ci155 level is more prominent in low-to-intermediate Hh signalling regions (arrow in Fig. 2a) in which the Ci155 protein level starts to fall but has not yet reached the basal level. Deneddylation upregulates the Ci155 level by a post-transcriptional mechanism, as we found that the protein levels of ci-myc and HA-ci transgenes under the ms1096-GAL4 driver are reduced in CSN mutant cells (arrow in Fig. 2c and Supplementary Fig. S2). Furthermore, the expression of ci-lacZ that recapitulates ci transcription remains constant in CSN5null cells (Fig. 2d).
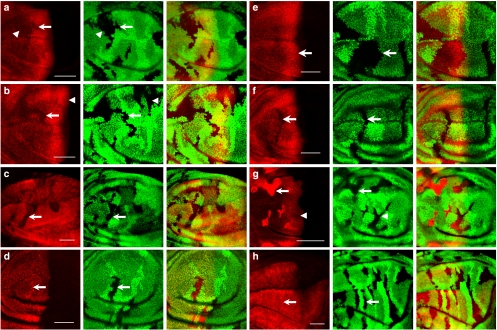
Figure 2. The CSN positively regulates Ci155 protein stability in wing discs.
(a) CSN5null clones are revealed by the absence of GFP (green) in wing discs. Ci155 stained with the 2A1 antibody (red) is reduced in CSN5null clones located in the A compartment, with arrow indicating mutant cells in the low-to-intermediate Hh signalling region and arrowhead indicating cells in the low Hh signalling region. (b) GFP-negative CSN4null clones were generated in wing discs (green). Ci155 staining (red) is lower in the CSN4null cells in the A compartment (arrow) than in the wild-type cells. The Ci155 levels in CSN4null and wild-type cells located in A/P boundary are similar (arrowhead). (c) CSN4null clones revealed by the absence of GFP (green) were generated in wing discs that simultaneously express UAS-ci-myc under the control of ms1096-GAL4. Protein levels of Ci-Myc detected by the anti-Myc antibody (red) are reduced in CSN4null mutant clones (arrow). (d) The ci-lacZ expression (red) is not reduced in CSN4null mutant clones (arrow) generated in wing discs. (e) GFP-negative CSN5null clones (green) were generated in wing discs expressing wild-type CSN5 construct under the control of ms1096-GAL4. Expression of wild-type CSN5 rescues the Ci155 level (red) in CSN5null clones (arrow). (f) Expression of CSN5D148N, which loses deneddylation activity, fails to rescues the Ci155 level (red) in CSN5null clones (arrows). (g) slimbP1493, CSN5null clones revealed by the absence of GFP (green) in wing discs show Ci155 (red) accumulation in the A compartment (arrow) except when located adjacent to the A/P boundary (arrowhead). (h) The Ci-3P mutant protein (red), carrying mutations in three PKA phosphorylation sites, is ectopically expressed in CSN4null mosaic wing pouches under the control of ms1096-GAL4. There is no difference in Ci-3P expression in wild-type and GFP-negative CSN4null cells (arrow). Scale bars in all panels represent 50 μm.
It has been shown that IκBα is less stable in CSN mutants because of the inactivation of the CSN-associated deubiquitinase USP15, rather than a deneddylation-related mechanism50. In the case of Ci155 proteolysis, CSN-mediated deneddylation is indeed required for the regulation of Ci155 levels, as expression of wild-type CSN5, but not deneddylation-defective CSN5D148N, restores the Ci155 level in CSN5null clones (Fig. 2e,f). Downregulation of Ci155 in CSN mutant cells is SCFSlimb dependent, as Ci155 accumulates in slimbP1493CSN5null double-mutant cells (arrow in Fig. 2g). We noted that the Ci155 level remains unchanged in slimbP1493CSN5null double-mutant clones in the anterior/posterior (A/P) boundary region (arrowhead in Fig. 2g), in which Ci155 is subjected to Cul3- but not SCF-mediated proteolysis27,51. The Ci155 level in CSN4null mutant clones that abuts the A/P boundary also remains unaltered (arrowhead in Fig. 2b), suggesting that the CSN regulates only SCFSlimb-dependent Ci155 proteolysis. PKA phosphorylation of Ci155 is required for the interaction between Ci155 and Slimb12,21,22, which should be important for the CSN to regulate SCFSlimb-dependent Ci155 proteolysis. The HA-Ci-3P protein that contains three mutated PKA phosphorylation sites and resistant to SCFSlimb-mediated proteolysis22 appears to have comparable expression levels in CSN4null mutant and wild-type cells (Fig. 2h). This is in contrast to the wild-type Ci-Myc and HA-Ci that are decreased in CSN mutant cells (Fig. 2c and Supplementary Fig. S2). Therefore, the CSN complex regulates the Ci155 levels through the SCFSlimb machinery in the A compartment of wing discs, except at the A/P boundary region.
CSN differentially regulates Ci155 proteolysis
To gain insights into how the CSN regulates SCFSlimb-mediated Ci155 proteolysis, the proportions of proteolysed and stable Ci155 along the A/P axis of wing discs were quantified. We generated random mosaic twin clones from double heterozygotes for the Cul1EX and CSN4null alleles, both located at chromosome 2R (Fig. 3a). One of the twin clones is homozygous for Cul1EX, marked with the double-brightness of green fluorescent protein (GFP) signal, and the other twin clone is homozygous for CSN4null, marked by the complete absence of GFP signal. Cells marked by single brightness of GFP signal are double heterozygous for Cul1EX and CSN4null, which are considered as wild-type control. The Ci155-expressing A compartment of wing pouches was divided into 12 equal regions, with the most anterior cells being in region 1 and cells receiving the highest level of Hh in region 12 (Fig. 3b). We measured the Ci155 protein levels in Cul1EX, CSN4null and wild-type control cells in regions 1–12. The percentage of Ci155 that has undergone SCFSlimb-dependent proteolysis in each region was calculated as the Ci155 level in Cul1EX cells (no proteolysis) subtracted by the Ci155 level in double-heterozygous cells (normal proteolysis in wild-type control), which is then normalized to the Ci155 level in Cul1EX cells: percentage of Ci155 proteolysis=([Ci155]Cul1EX−[Ci155]wild-type)/[Ci155]Cul1EX. We found that the extent of SCFSlimb-dependent Ci155 proteolysis is constantly high across regions 1–8, and gradually declines from region 8 to 12 (Fig. 3c). We also quantified the extents of SCFSlimb-dependent Ci155 proteolysis in CSN4null cells across regions 1–12 using the same methodology: percentage of Ci155 proteolysis=([Ci155]Cul1EX−[Ci155]CSN4null)/[Ci155]Cul1EX. Interestingly, Ci155 proteolysis in CSN4null cells remained similar to wild-type cells in regions 1–8, and graded Ci155 proteolysis in CSN4null cells was also present from regions 8 to 12. Remarkably, Ci155 proteolysis is prominently enhanced in CSN4null cells in regions 9, 10 and 11 compared with wild-type cells in the same regions, with 30, 40 and 24% reductions in the Ci155 levels, respectively (Fig. 3c). In region 12 closest to the A/P boundary in which the protein level of Ci155 is regulated by the Cul3-organized CRL27,51, the proportion of SCFSlimb-dependent Ci155 proteolysis is unaltered in CSN4null mutant cells (arrow in Fig. 3a and arrowhead in Fig. 2b). Mild enhancements of 1–6% of Ci155 proteolysis were noted in CSN4null cells in regions 1–8, although these differences were not statistically significant except for regions 6 and 7 (n≥6, P<0.05 by Student's t-test). On the basis of the proteolytic curves of Ci155 in wild-type and CSN4null cells, we propose that proteolysis-resistant Ci155 in wild-type cells is composed of 'stable' and 'conditionally stable' forms, the latter being stable in wild-type cells but proteolysed in CSN mutant cells by SCFSlimb, whereas the former is insensitive to the absence of CSN activity. The levels of conditionally stable Ci155 are highest in regions 9–11, in which cells are exposed to low-to-intermediate levels of Hh.
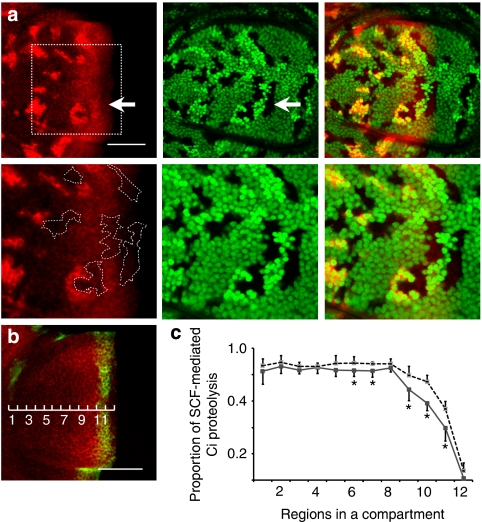
Figure 3. The CSN regulates Ci155 proteolysis predominantly in intermediate Hh signalling regions.
(a) Mutant clones for CSN4null or Cul1EX were generated simultaneously in the same wing discs of the hsflp; FRT42D CSN4null/FRT42D Cul1EX, nlsGFP genotype. Ci155 (red) levels stained by 2A1 antibody was measured in the following cells: CSN4null mutant clones marked by the absence of GFP, Cul1EX mutant clones marked by two copies of GFP and double-heterozygous CSN4null/+, Cul1EX/+ cells marked by one copy of GFP (green). The lower panels in a show close-up images of a region near the A/P boundary (outlined by a dashed square in upper left panel), with some CSN4null clones highlighted by white dashed lines (lower left panel). (b) The A compartment of wing pouches showing expression of Ci155 (red) and dpp-lacZ (green) is divided equally into 12 regions along the A/P axis. Expression of dpp-lacZ is detected in region 11. Scale bars represent 50 μm (a, b). (c) The portions of Ci155 that undergo SCFSlimb-mediated proteolysis in regions 1–12 of wild-type (grey line) and CSN4null mutant cells (dashed line) were calculated. Averaged Ci155 pixel intensities for each region (n >= 6) are shown. Asterisks indicate a significant difference of Ci155 proteolysis between wild-type and CSN4null (P<0.05). Error bars present the standard deviation.
CSN is required for Hh-dependent wing patterning
To examine whether the level of conditionally stable Ci155 is essential for distinct cell fates directed by graded Hh signalling, we assayed the activation of Hh-responsive, position-specific genes, such as ptc and dpp, whose expression represent high and low Hh signalling responses, respectively19. Interestingly, we found that the expression of dpp-lacZ is significantly repressed in CSN4null mutant cells (arrow in Fig. 4a), whereas the Ptc protein level and the expression of ptc-lacZ remain unaltered (arrows in Fig. 4b,c), indicating a critical role of the conditionally stable Ci155 for the Hh-mediated dpp expression. The expression of engrailed (en) is regulated by high Hh signalling activity in the A/P boundary52. We tested whether en expression in the A/P boundary is downregulated in CSN4null mutant because of Ci155 proteolysis. En protein level is instead enhanced not only at the A/P boundary but also in A and P compartments, in which en expression is independent from Hh signalling (Fig. 4d), suggesting that en expression is subjected to another layer of regulation by the CSN.
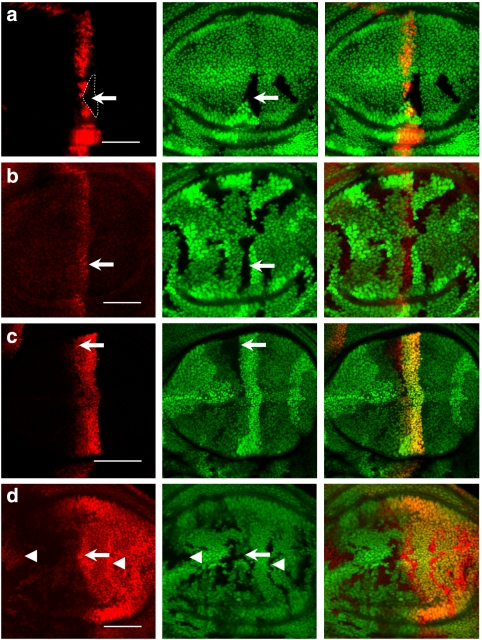
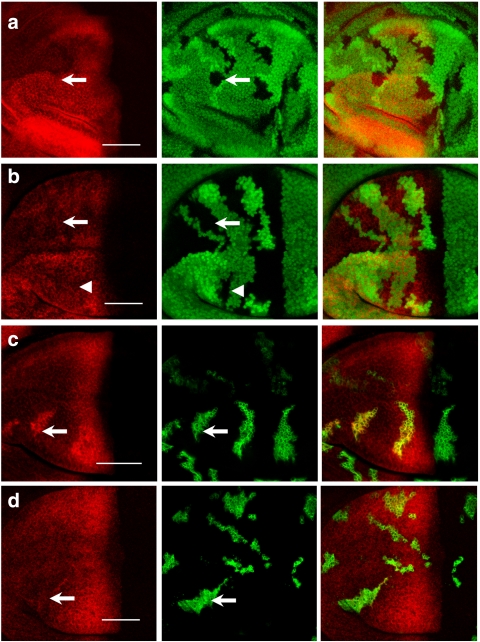
Figure 4. The CSN differentially regulates Hh downstream gene expression.
(a) A CSN4null clone (indicated by the dashed outline) is generated in dpp-lacZ wing discs. dpp-lacZ expression detected by anti-β-gal antibody (red) is markedly repressed in CSN4null cells (arrow). (b) CSN5null clones indicated by the absence of GFP are generated in wing discs. Ptc (red) protein level in CSN5null cells (arrow) detected by the monoclonal antibody Apa-1 is similar to that in wild-type cells. (c) ptc-lacZ staining (red) is comparable in wild-type and CSN5null cells that are marked by the absence of GFP (arrow). (d) En protein levels (red) are increased in the CSN5null cells (arrow) located in the A/P boundary as well as in the A and P compartments (arrowheads). Scale bars in all panels represent 50 μm.
To further substantiate the notion that small perturbations of the Ci155 level can inactivate dpp expression, we tested whether ms1096-GAL4-driven ci-RNAi53 could abolish dpp-lacZ expression. The ms1096-GAL4 driver exhibits differential expression in wing pouches, with higher levels in the dorsal and lower levels in the ventral compartment. As a consequence, dpp-lacZ expression is detectable in the ventral compartment, but diminished in the dorsal compartment, in which the Ci155 level is comparably reduced as in CSN5null cells (Supplementary Fig. S3). In contrast, the expression of ptc-lacZ remains unaltered in the dorsal compartment (Supplementary Fig. S3). Therefore, the dpp-lacZ expression is sensitive to small reductions in Ci155 levels. The expression of dpp-lacZ is downregulated by Ci75, the transcriptional repressor produced by SCFSlimb-mediated proteolysis of Ci155, whereas Ptc expression is not54. Disruption of dpp-lacZ but not Ptc expression in CSN mutant cells suggests an alternative possibility that a higher level of Ci75 accumulates in CSN mutant cells as a result of SCFSlimb-mediated proteolysis.
Proper phosphorylation confers conditionally stable Ci155
Our results suggest that low-to-intermediate Hh signalling (regions 9–11) renders the appearance of conditionally stable Ci155, whereas high (region 12) and low (regions 1–8) Hh signalling activities do not. It has also been suggested that graded Hh signalling activities counteract Ci155 proteolysis by inhibiting different levels of Ci155 phosphorylation55, leading to differential affinities to the F-box protein Slimb for SCFSlimb-mediated proteolysis26. We therefore tested whether the phosphorylation status of Ci155 alters the level of conditionally stable Ci155. Ci155 phosphorylation can be compromised by the ectopic expression of dominant-negative GSK3β (DN-GSK3β), resulting in the inhibition of Ci155 proteolysis in the low Hh signalling region24. Although proteolysed Ci155 in the low Hh region (regions 1–8) is 84±5% of total Ci155 in wild-type cells (Fig. 3c), it is reduced to 38±8% in the DN-GSK3β expression clones (averaged from seven clones in regions 1–8, arrowhead in Fig. 5a). Accumulation of Ci155 in regions 1–8, however, is suppressed by the CSN4null mutation (arrow in Fig. 5a), with 75±11% of Ci155 undergoing proteolysis in CSN4null mutant cells expressing DN-GSK3β (n=8). Thus, the accumulated Ci155 in regions 1–8 on DN-GSK3ββ wings is conditionally stable in nature, undergoing proteolysis in the absence of CSN activity. Likewise, inhibiting CKI kinase activity by expressing dominant-negative Doubletime (DN-DBT) causes Ci155 accumulation25, which is also suppressed by the CSN4null mutation, as shown by reduced Ci155 levels in CSN4null mutant cells expressing DN-DBT (arrow, Fig. 5b) compared with wild-type cells expressing DN-DBT. Thus, lowering the phosphorylation level by inhibiting either GSK3β or CKI produces conditionally stable Ci155 in the low Hh signalling region, mimicking the Ci155 behaviour in the region with low-to-intermediate Hh signalling.
Figure 5. Conditionally stable Ci155 is sensitive to phosphorylation levels.
(a) CSN4null clones are generated in wing discs that express dominant-negative GSK3β (DN-GSK3β) by ms-1096-GAL4. DN-GSK3β increases Ci155 expression (red) in low Hh signalling regions (arrowhead). The Ci155 level is downregulated in CSN4null cells (arrow) in the presence of DN-GSK3β. (b) Inhibition of CKI activity by C765-GAL4-driven DN-DBT increases the accumulation of Ci155 (red) in wild-type cells. This accumulation of Ci155 is downregulated in CSN4null clones (arrow). (c) CSN4null clones marked by the absence of GFP (green) were generated in wing discs expressing UAS-ci-C1-3E-myc under the control of ms1096-GAL4. The Ci-C1-3E-Myc levels detected by anti-Myc antibody (red) were decreased in CSN4null clones (arrow). (d) CSN4null clones marked by the absence of GFP (green) were generated in wing discs expressing UAS-ci-G2-3E-myc. Ci-G2-3E-Myc (red) staining is decreased in CSN4null clones (arrow). (e) Ectopic expression of PKA-R*, the regulatory subunit of PKA, by C765-GAL4 increases Ci155 levels in the A compartment. The upregulated Ci155 levels (red) either in low or low-to-intermediate Hh regions (arrow and arrowhead, respectively) are not altered in CSN5null clones in wing discs. (f) CSN5null clones were generated in wing discs that express the catalytic subunit of PKA (PKA-mC*) by ms-1096-GAL4. Ci155 levels (red) near the A/P boundary in the high Hh signalling region is downregulated in CSN5null clones (arrows) compared with wild-type cells (arrowhead). Dashed lines indicate the A/P boundary. Scale bars in all panels represent 50 μm.
To examine the critical role of phosphorylation in the appearance of conditionally stable Ci155, we examined whether the CSN regulates the protein levels of Ci mutants in which CKI or GSK3β phosphorylation sites are mutated (Ci-C1-3E and Ci-G2-3E, respectively). Both Ci-C1-3E and Ci-G2-3E cannot be proteolysed efficiently in the absence of Hh signalling26. When ectopically expressed in wing pouches by ms1096-GAL4, the protein levels of Ci-C1-3E or Ci-G2-3E are downregulated by the CSN4null mutation (arrows in Fig. 5c,d), suggesting that Ci-C1-3E and Ci-G2-3E represent the CSN-regulated, conditionally stable forms of Ci.
Expression of the PKA regulatory subunit, PKA-R*, inhibits PKA-dependent Ci155 phosphorylation and precludes PKA-primed sites for further GSK3β and CKI phosphorylation21. We therefore examined the effect of further reducing Ci155 phosphorylation on the CSN-dependent Ci155 stabilization. Under the control of C765-GAL4, the PKA-R* expression enhances Ci155 levels across the A compartment. However, no downregulation of Ci155 levels could be detected in CSN4null mutant cells expressing PKA-R*, including those in intermediate-to-high and low Hh signalling regions (arrowhead and arrow in Fig. 5e, respectively). Therefore, phosphorylation-depleted Ci155 is unlikely to be the CSN-regulated conditionally stable form.
The above analysis also suggests that lack of PKA phosphorylation in the high Hh signalling region 12 may account for the absence of conditionally stable Ci155. To test this, Ci155 phosphorylation by PKA is induced by expressing the catalytic subunit mC* under the control of ms1096-GAL423. Expression of mC* equalizes Ci155 levels across the A compartment, mainly because of the downregulation of Ci155 levels in intermediate-to-high Hh signalling regions and the upregulation in high Hh signalling region 12. However, the Ci155 levels in mC*-expressing CSN5null mutant clones were reduced in large clones covering region 12 (arrows in Fig. 5f), as compared with mC*-expressing wild-type cells (arrowhead in Fig. 5f), an indication of the appearance of conditionally stable Ci155 even in the presence of high Hh signalling. Given that conditionally stable Ci155 appears in the low Hh signalling region on GSK3β or CKI inhibition and in the high Hh signalling region by PKA activation, these results support the idea that partially phosphorylated Ci155 represents the conditionally stable form regulated by the CSN.
Dominant-negative SlimbΔfbx fails to induce CSN regulation
To further corroborate the SCF-dependent mechanism for CSN regulation, we tested whether Ci155 has to be incorporated into the SCF complex for the CSN to confer its conditional stability. To do this, the F-box-truncated SlimbΔfbx protein that can competitively bind Ci155 but cannot be incorporated into SCF was ectopically expressed by ap-GAL4 in wing discs. Higher levels of Ci155 detected in wing discs suggest that SlimbΔfbx-sequestered Ci155 is free from SCF-mediated proteolysis. Unlike the conditionally stable Ci155 upregulated by DN-GSK3β or DN-DBT expression, the upregulated Ci155 levels caused by SlimbΔfbx expression is insensitive to the absence of CSN activity in CSN5null cells (arrow, Fig. 6a). Thus, Ci155 that binds the substrate receptor Slimb, but fails to form SCFSlimb-Ci155 intermediates, is insufficient to produce conditionally stable Ci155.
Figure 6. CSN control of Ci155 levels upregulated by SlimbΔfbx or Uba1 depletion.
(a) CSN5null clones are generated in wing discs expressing Myc-SlimbΔfbx in the dorsal compartment under the control of ap-GAL4. Ectopic expression of SlimbΔfbx increases the levels of Ci155 (red) in the dorsal compartment. Moreover, the upregulated Ci155 is maintained in CSN5null clones (arrow). (b) Under the control of ms1096-GAL4, the moderate level of ectopic Myc-SlimbΔfbx in the dorsal compartment causes similar accumulation of Ci155 (red) in both wild-type and CSN5null cells (arrowhead); the lower level of ectopic Myc-SlimbΔfbx in the ventral compartment also causes similar accumulation of Ci155 in both wild-type and CSN5null cells (arrow) (c) GFP-marked MARCM clones (green) expressing uba1-dsRNA by ms1096-GAL4 generated in wing discs show upregulation of Ci155 (red) levels compared with adjacent wild-type cells that are negative for GFP. (d) GFP-marked uba1-dsRNA MARCM clones that are also CSN5null show Ci155 levels (red) comparable with adjacent GFP-negative wild-type cells. Scale bars in all panels represent 50 μm.
Although SlimbΔfbx-sequestered Ci155 is insensitive to the CSN regulation, the residual Ci155 protein that is free from SlimbΔfbx titration is still insensitive to the CSN regulation, arguing that the CSN regulation does not increase the concentration or availability of the SCFSlimb-Ci155 intermediate. As the strong expression of SlimbΔfbx by ap-GAL4 might sequester Ci155 from SCF regulation, ms1096-GAL4 was used to express moderate levels of SlimbΔfbx in the dorsal compartment and low levels in the ventral compartment. Moderate depletion of the SCFSlimb-Ci155 intermediates in the dorsal compartment by ms1096-GAL4 behaved similarly to the strong depletion by ap-GAL4; no significant difference in Ci155 proteolysis was detected between wild-type and CSN5null cells (arrowhead in Fig. 6b). Interestingly, although the Ci155 levels fluctuate in the ventral compartment because of weak depletion of the SCFSlimb-Ci155 intermediates, they were still insensitive to any CSN modulation (arrow in Fig. 6b). In these experiments, the remaining Ci155 after SlimbΔfbx titration still possessed a high affinity for Slimb in low Hh signalling regions. Thus, in contrast to the perturbation by DN-GSK3β, different extents of perturbation on the concentration of tightly associated SCFSlimb-Ci155 intermediates are invariantly insufficient to generate conditionally stable Ci155.
Sensitivity of Ci155 proteolysis to the Ub supply
A compromised affinity between partially phosphorylated Ci155 and Slimb can result in reduced availability of SCFSlimb-Ci155 intermediates. However, as shown in the SlimbΔfbx titration experiment, reduced availability of SCFSlimb-Ci155 intermediates was insufficient to confer CSN regulation. We hypothesized that partial Ci155 phosphorylation may compromise the duration of SCFSlimb-Ci155 association, disrupting substrate polyubiquitylation. In line with this idea, recent findings suggest that most of the encounters between SCF and its substrate are unproductive, with the dissociation rate koff being much larger than the reaction rate of adding the first Ub (kub1)38. Although most of the encounters between SCF and its substrate are unproductive, neddylation of SCF allows polyubiquitylation (kub2–4) to proceed at higher rates once the first Ub is added38. Therefore, CSN deneddylation could be critical at the Ub chain elongation step when the affinity between SCF and substrate is reduced, such as in the case of partially phosphorylated Ci155. Processive Ci155 ubiquitylation would demand a constant supply of activated Ub, which could be sensitive to the level of the Ub-activating enzyme. In mosaic analysis with a repressible cell marker (MARCM) clones, in which uba1-dsRNA was expressed to knockdown the expression of the Ub-activating enzyme, accumulation of Ci155 (arrow in Fig. 6c) suggests that the insufficiency in supplying activated Ub leads to the disruption of Ci155 polyubiquitylation and proteolysis. The defect of Ci155 accumulation in uba1-dsRNA cells is likely caused by reduced ubiquitylation, which could be compensated by constitutively neddylated SCF that promotes processivity of ubiquitylation. Indeed, the elevated Ci155 level in uba1-dsRNA knockdown cells was almost completely suppressed by introducing the CSN5null mutation in the uba1-dsRNA MARCM clones (arrow in Fig. 6d). This result is consistent with the idea that inactivating the deneddylation machinery could compensate for inefficient polyubiquitylation in uba1-dsRNA knockdown cells, thereby restoring the assembly of proteasome-targeting polyubiquitin chains on Ci155.
Discussion
In this study, we show that CSN-mediated deneddylation of SCFSlimb is critical for the Ci155 stability in low-to-intermediate Hh regions in which partially phosphorylated Ci155 has a reduced affinity for the ubiquitylation machinery SCFSlimb. The effect of CSN deneddylation on SCFSlimb likely reduces ubiquitylation processivity, thus preserving 'conditionally stable Ci155' in these low-to-intermediate Hh regions. We propose a broader view for CRL substrate degradation in vivo, determined by three interdependent factors: substrate-enzyme affinity, deneddylation-regulated CRL activity and the supply of Uba1-activated Ub. We propose that steady-state Ci155 levels are low in the low Hh signalling regions because processive polyubiquitylation of the tightly associated SCFSlimb-Ci155 complex proceeds even with intermittent SCFSlimb inactivation by CSN-mediated deneddylation (Fig. 7a). In low-to-intermediate Hh regions, the steady-state Ci155 levels are sensitive to CSN-mediated deneddylation as a result of weakened SCFSlimb-Ci155 association, as in DN-GSK3β cells (Fig. 7b). In uba1-dsRNA cells, the Ci155 levels are sensitive to CSN-mediated deneddylation because of insufficient activated Ub (Fig. 7c). In these cases, we envision that processive polyubiquitylation becomes more difficult because of constant neddylation–deneddylation cycling by the CSN, and can be aborted by the dissociation of Ci155 from weakly associated SCFSlimb-Ci155 or the lack of activated Ub. However, an insufficiency in Ci155 downregulation can be offset by constitutive neddylated SCFSlimb in CSN mutants, which ubiquitylates substrates processively on a single SCFSlimb-Ci155 encounter (Fig. 7d,e). This model explains the expression of CSN-dependent conditionally stable Ci155 in the low-to-intermediate Hh signalling regions, in which Ci155 may be released from the labile SCFSlimb-Ci155 complex before SCF re-neddylation.
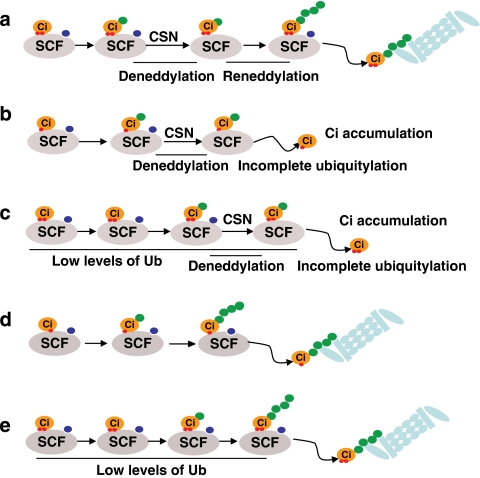
Figure 7. Models for SCFSlimb-mediated Ci155 proteolysis.
Successful substrate polyubiquitylation requires collaboration of three components: adequate substrate–enzyme interaction time, persistent neddylated E3 ligase activity and high levels of activated ubiquitin supply. Green dot, ubiquitin; blue dot, Nedd8; and red dot, phosphate group. (a) For fully phosphorylated Ci155 that binds strongly to SCF in no-to-low Hh signalling regions, long substrate–enzyme binding duration allows complete polyubiquitylation, despite temporary inactivation of SCF ligase by CSN-mediated deneddylation. (b) In low-to-intermediate Hh signalling regions as well as in the presence of DN-GSK3β, partially phosphorylated Ci155 associates with SCF weakly. Dissociation of Ci155 from the SCFSlimb complex following CSN-mediated deneddylation of SCF disrupts processive ubiquitylation of Ci155. (c) Limited supply of activated ubiquitin could disrupt processive ubiquitylation of fully phosphorylated Ci155 in no-to-low Hh regions in the presence of CSN deneddylation. (d) CSN mutations allow the processivity of polyubiquitylation mediated by constantly neddylated SCF, which compensates for the short duration of interaction between SCFSlimb and Ci155 in low-to-intermediate Hh signalling regions or in the presence of DN-GSK3β. (e) In no-to-low Hh regions, limited supply of activated ubiquitin in uba1-dsRNA cells interrupts Ci155 polyubiquitylation, which is suppressed by constitutively neddylated SCF in CSN mutants.
Substrate–SCF affinity is controlled by substrate phosphorylation and, in the case of Ci155, the number of phospho groups on substrates. It was predicted that switch-like protein degradation could be achieved for substrates bearing multiple phosphorylation sites. For example, SCF-mediated Sic1 polyubiquitylation and degradation ensues at the G1–S transition when the number of phosphorylated sites on Sic1 reaches six, as Sic1 binding affinity to the F-box protein CDC4 increases dramatically from Sic1-5p to Sic1-6p56. Ci155 also bears multiple phosphorylation sites. Unlike Sic1, however, partial phosphorylation of Ci155 is meaningful in establishing low-to-intermediate responses to Hh signalling. The amount of Ci155 binding to Slimb increases incrementally with the number of phosphorylated sites26, which precludes decisive proteolysis and allows the meta-stable SCFSlimb-Ci155 intermediates for CSN regulation. In CSN mutant wing discs, responses to Hh signalling become more switch-like, as only low and high Hh signalling responses are detected. Thus, this study and previous studies23,24,25,26,57 indicate that partial phosphorylation of Ci155 is essential, but not sufficient, for non-switch-like substrate proteolysis. CSN-mediated deneddylation has to be incorporated into the machinery for specific enhancement of Ci155 levels over a low threshold. Whether it is the enhanced levels of Ci155 or the reduced levels of the proteolysed Ci75 repressor that affect dpp-lacZ expression is unclear. This specific requirement of deneddylation in developing wings underscores the developmental role of CSN deneddylation at a tissue level. We also envision that CSN-mediated deneddylation of cullin-based ligases may modulate cellular levels of many other proteins in a context-dependent manner, thereby adjusting their biological readouts to meet distinct requirements of different cell contexts in a multicellular organism.
Methods
Fly genetics and fly stocks
Cul1EX12, Nedd8AN01512, UAS-CSN542, UAS-CSN5D148N42 and UAS-flag-ago58; CSN4null and CSN5null59; UAS-HA-ci-3P22, Tub-myc-slimb, UAS-myc-slimb-Δfbx47, UAS-DN-GSK3β24, UAS-DN-DBT25, UAS-ci-myc26, UAS-ci-C1-3E-myc26, UAS-ci-G2-3E-myc26 and slimbP149313; UAS-PKA-R* and UAS-PKA-mC* 60 have been described previously. BS3.0 dpp-LacZ and ci-LacZ were obtained from Bloomington stock center. UAS-uba1-dsRNA (stock ID: 1782R-2) was obtained from Fly Stocks of National Institute of Genetics. hsflp; FRT42D ubi-nlsGFP; C765-GAL4/TM6B, ms1096-GAL4, hsflp; FRT42D ubi-nlsGFP, hsflp; ap-GAL4/CyO; FRT82B ubi-nlsGFP and ms1096-GAL4, hsflp; FRT82B ubi-nlsGFP were used to generate CSN4null and CSN5null mutant clones that simultaneously express transgenes of interest. ms1096-GAL4, hsflp; FRT82B tub-GAL80 was used to generate CSN5null MARCM clones in wing pouches. hsflp/+, FRT42D Cul1EX, ubi-nlsGFP/FRT42D CSN4null was used to generate twin mutant clones for Cul1EX and CSN4null. ms1096-GAL4, hsflp/+; FRT42D Cul1EX, ubi-nlsGFP/FRT42D CSN4null; UAS-DN-GSK3β/+ was used to generate twin mutant clones for Cul1EX and CSN4null in DN-GSK3β-expressing wing discs.
Flies were kept at 25 °C. At 48–72 h after egg laying, larvae were heat shocked at 37 °C for 1 h to generate CSN4null and CSN5null mutant clones. To generate uba1-shRNA-expressing CSN5null MARCM clones, 72 h after egg laying, ms1096-GAL4, hsflp/+; UAS-uba1-dsRNA/+; FRT82B CSN5null/FRT82B tub-GAL80 larvae were heat shocked at 37 °C for 1 h.
Immunostaining and image processing
For immunostaining, dissected imaginal wing discs from wandering third-instar larvae were fixed in 4% formaldehyde in phosphate-buffered saline (PBS) solution for 17 min, washed with 0.3% triton X-100 in PBS (PBST) for 10 min (three times), blocked in PBST supplemented by 5% normal donkey serum for 30 min and incubated with primary antibodies of various dilutions in PBST containing 5% normal donkey serum for 16 h at 4 °C. These imaginal discs were washed with PBST for 10 min (three times), followed by a 2-h secondary antibody incubation before washes (three times in PBST). The stained wing discs were mounted on slides in PBS + 50% glycerol. All procedures were performed at 25 °C unless specifically described. The primary antibodies used were mouse anti-Flag (M2, 1:3,000; Sigma), mouse anti-Myc (9E10, 1:200; Santa Cruz Biotechnology), rat anti-full-length Ci (2A1, 1:10; Developmental Studies Hybridoma Bank), mouse anti-β-gal (1:1,000; Sigma) and mouse anti-Arm and mouse anti-En (1:100, N2-7A1 and 1:30, 4D9, respectively, Developmental Studies Hybridoma Bank). The secondary antibodies used were Cy3-conjugated goat anti-mouse IgG, Cy3-conjugated goat anti-rabbit IgG and Cy3-conjugated goat anti-rat IgG (1:1,000, Jackson ImmunoResearch Laboratories).
Images were acquired by Zeiss LSM510 META Confocal Imaging System (Zeiss). To quantify Ci155 expression levels in wild-type, CSN4null and Cul1EX cells, the maximal pixel intensities of Ci155 immunoreactivities in the A compartment were below 255 and set to be zero in the P compartment as background control by using Photoshop (Adobe).
Author contributions
J.-T.W. designed and performed experiments, analysed data and wrote the paper. W.-H.L. helped design experiments for Figures 3 and 6 and wrote the paper. W.-Y.C. performed experiments for Supplementary Figure S3, Y.-C.H. performed experiments for Figure 4d, C.-Y.T. for microinjection, M.S.H. provided UAS-flag-ago transgene, H.P. designed and supervised experiments and analysed data for Figure 4d. C.-T.C. designed experiments, analysed data and wrote the paper.
Additional information
How to cite this article: Wu, J.-T. et al. CSN-mediated deneddylation differentially modulates Ci155 proteolysis to promote Hedgehog signalling responses. Nat. Commun. 2:182 doi: 10.1038/ncomms1185 (2011).
Supplementary Material
Supplementary Figures S1–S3
Acknowledgments
We thank H.H. Lee for detail discussion and editing the manuscript. We thank D. Kalderon, J. Jiang, Y.H. Liao, W.J. Wang and T.P. Yao for reagents and critical suggestions. We thank the staff of the Sixth Core Lab and DNA Sequencing Facility, Department of Medical Research, National Taiwan University Hospital for technical support. We also thank the Taiwan Fly Core Facility and Fly Core Facility in National Taiwan University for technical support. This work is supported by grant NSC 97-2320-B-002-054-MY3 from National Science Council, Taiwan and grant NHRI-EX99-9926SC from National Health Research Institutes, Taiwan to J.-T.W.
References
- Wolpert L. Positional information and the spatial pattern of cellular differentiation. J. Theor. Biol. 25, 1–47 (1969). [DOI] [PubMed] [Google Scholar]
- Osterfield M., Kirschner M. W. & Flanagan J. G. Graded positional information: interpretation for both fate and guidance. Cell 113, 425–428 (2003). [DOI] [PubMed] [Google Scholar]
- Ibanes M. & Belmonte J. C. Theoretical and experimental approaches to understand morphogen gradients. Mol. Syst. Biol. 4, 176 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob L. & Lum L. Hedgehog signaling pathway in Drosophila. Sci. STKE 2007, cm7 (2007). [DOI] [PubMed] [Google Scholar]
- Ashe H. L. & Briscoe J. The interpretation of morphogen gradients. Development 133, 385–394 (2006). [DOI] [PubMed] [Google Scholar]
- Wang Y., McMahon A. P. & Allen B. L. Shifting paradigms in Hedgehog signaling. Curr. Opin. Cell Biol. 19, 159–165 (2007). [DOI] [PubMed] [Google Scholar]
- Motoyama J. Essential roles of Gli3 and sonic hedgehog in pattern formation and developmental anomalies caused by their dysfunction. Congenit. Anom. (Kyoto) 46, 123–128 (2006). [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A., Mas C. & Stecca B. The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol. 17, 438–447 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N. E. Patterning signals and proliferation in Drosophila imaginal discs. Curr. Opin. Genet. Dev. 17, 287–293 (2007). [DOI] [PubMed] [Google Scholar]
- Jiang J. & Hui C. C. Hedgehog signaling in development and cancer. Dev. Cell 15, 801–812 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aza-Blanc P., Ramirez-Weber F. A., Laget M. P., Schwartz C. & Kornberg T. B. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89, 1043–1053 (1997). [DOI] [PubMed] [Google Scholar]
- Ou C. Y., Lin Y. F., Chen Y. J. & Chien C. T. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 16, 2403–2414 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J. & Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature 391, 493–496 (1998). [DOI] [PubMed] [Google Scholar]
- Methot N. & Basler K. Suppressor of fused opposes hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development 127, 4001–4010 (2000). [DOI] [PubMed] [Google Scholar]
- Wang Q. T. & Holmgren R. A. Nuclear import of cubitus interruptus is regulated by hedgehog via a mechanism distinct from Ci stabilization and Ci activation. Development 127, 3131–3139 (2000). [DOI] [PubMed] [Google Scholar]
- Wang G., Amanai K., Wang B. & Jiang J. Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of cubitus interruptus. Genes Dev. 14, 2893–2905 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisson B. E., Ziegenhorn S. L. & Holmgren R. A. Regulation of Ci and Su(fu) nuclear import in Drosophila. Dev. Biol. 294, 258–270 (2006). [DOI] [PubMed] [Google Scholar]
- Ingham P. W. & McMahon A. P. Hedgehog signaling in animal development: paradigms and principles. Genes. Dev. 15, 3059–3087 (2001). [DOI] [PubMed] [Google Scholar]
- Hooper J. E. & Scott M. P. Communicating with Hedgehogs. Nat. Rev. Mol. Cell Biol. 6, 306–317 (2005). [DOI] [PubMed] [Google Scholar]
- Jiang J. Regulation of Hh/Gli signaling by dual ubiquitin pathways. Cell Cycle 5, 2457–2463 (2006). [DOI] [PubMed] [Google Scholar]
- Price M. A. & Kalderon D. Proteolysis of cubitus interruptus in Drosophila requires phosphorylation by protein kinase A. Development 126, 4331–4339 (1999). [DOI] [PubMed] [Google Scholar]
- Wang G., Wang B. & Jiang J. Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes Dev. 13, 2828–2837 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. A. & Kalderon D. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell 108, 823–835 (2002). [DOI] [PubMed] [Google Scholar]
- Jia J. et al. Shaggy/GSK3 antagonizes Hedgehog signalling by regulating Cubitus interruptus. Nature 416, 548–552 (2002). [DOI] [PubMed] [Google Scholar]
- Jia J. et al. Phosphorylation by double-time/CKIepsilon and CKIalpha targets cubitus interruptus for Slimb/beta-TRCP-mediated proteolytic processing. Dev. Cell 9, 819–830 (2005). [DOI] [PubMed] [Google Scholar]
- Smelkinson M. G. & Kalderon D. Processing of the Drosophila hedgehog signaling effector Ci-155 to the repressor Ci-75 is mediated by direct binding to the SCF component Slimb. Curr. Biol. 16, 110–116 (2006). [DOI] [PubMed] [Google Scholar]
- Chou Y. H. & Chien C. T. Scabrous controls ommatidial rotation in the Drosophila compound eye. Dev. Cell 3, 839–850 (2002). [DOI] [PubMed] [Google Scholar]
- Lammer D. et al. Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 12, 914–926 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T. et al. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene 18, 6829–6834 (1999). [DOI] [PubMed] [Google Scholar]
- Wu J. T., Chan Y. R. & Chien C. T. Protection of cullin-RING E3 ligases by CSN-UBP12. Trends Cell Biol. 16, 362–369 (2006). [DOI] [PubMed] [Google Scholar]
- Osaka F. et al. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 19, 3475–3484 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C., Britton J. H. & Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2, 1071–1080 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo J. C. et al. AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis Cullin AtCUL1 is required for auxin response. Plant Cell 14, 421–433 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri S., Dharmasiri N., Hellmann H. & Estelle M. The RUB/Nedd8 conjugation pathway is required for early development in Arabidopsis. EMBO J. 22, 1762–1770 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T. et al. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20, 4003–4012 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda D. M. et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134, 995–1006 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A. & Deshaies R. J. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32, 21–31 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce N. W., Kleiger G., Shan S. O. & Deshaies R. J. Detection of sequential polyubiquitylation on a millisecond timescale. Nature 462, 615–619 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S. et al. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292, 1382–1385 (2001). [DOI] [PubMed] [Google Scholar]
- Schwechheimer C. & Deng X. W. COP9 signalosome revisited: a novel mediator of protein degradation. Trends Cell Biol. 11, 420–426 (2001). [DOI] [PubMed] [Google Scholar]
- Bosu D. R. & Kipreos E. T. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 3, 7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. T., Lin H. C., Hu Y. C. & Chien C. T. Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat. Cell Biol. 7, 1014–1020 (2005). [DOI] [PubMed] [Google Scholar]
- Gusmaroli G., Figueroa P., Serino G. & Deng X. W. Role of the MPN subunits in COP9 signalosome assembly and activity, and their regulatory interaction with Arabidopsis Cullin3-based E3 ligases. Plant Cell 19, 564–581 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Cheng P., He Q. & Liu Y. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev. 19, 1518–1531 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee S., Geyer R. K., Toda T. & Wolf D. A. CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat. Cell Biol. 7, 387–391 (2005). [DOI] [PubMed] [Google Scholar]
- Cope G. A. & Deshaies R. J. Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 7, 1 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko H. W., Jiang J. & Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature 420, 673–678 (2002). [DOI] [PubMed] [Google Scholar]
- Huang X., Langelotz C., Hetfeld-Pechoc B. K., Schwenk W. & Dubiel W. The COP9 signalosome mediates beta-catenin degradation by deneddylation and blocks adenomatous polyposis coli destruction via USP15. J. Mol. Biol. 391, 691–702 (2009). [DOI] [PubMed] [Google Scholar]
- Panattoni M. et al. Targeted inactivation of the COP9 signalosome impairs multiple stages of T cell development. J. Exp. Med. 205, 465–477 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer K., Bozko P. M., Dubiel W. & Naumann M. CSN controls NF-kappaB by deubiquitinylation of IkappaBalpha. EMBO J. 26, 1532–1541 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. et al. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev. Cell 10, 719–729 (2006). [DOI] [PubMed] [Google Scholar]
- Blair S. S. Engrailed expression in the anterior lineage compartment of the developing wing blade of Drosophila. Development 115, 21–33 (1992). [DOI] [PubMed] [Google Scholar]
- Ou C. Y., Wang C. H., Jiang J. & Chien C. T. Suppression of Hedgehog signaling by Cul3 ligases in proliferation control of retinal precursors. Dev. Biol. 308, 106–119 (2007). [DOI] [PubMed] [Google Scholar]
- Methot N. & Basler K. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell 96, 819–831 (1999). [DOI] [PubMed] [Google Scholar]
- Aikin R. A., Ayers K. L. & Therond P. P. The role of kinases in the Hedgehog signalling pathway. EMBO Rep. 9, 330–336 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash P. et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414, 514–521 (2001). [DOI] [PubMed] [Google Scholar]
- Smelkinson M. G., Zhou Q. & Kalderon D. Regulation of Ci-SCFSlimb binding, Ci proteolysis, and hedgehog pathway activity by Ci phosphorylation. Dev. Cell 13, 481–495 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M. S. et al. Gcm protein degradation suppresses proliferation of glial progenitors. Proc. Natl Acad. Sci. USA 106, 6778–6783 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oron E. et al. COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development 129, 4399–4409 (2002). [DOI] [PubMed] [Google Scholar]
- Li W., Ohlmeyer J. T., Lane M. E. & Kalderon D. Function of protein kinase A in hedgehog signal transduction and Drosophila imaginal disc development. Cell 80, 553–562 (1995). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S3