Abstract
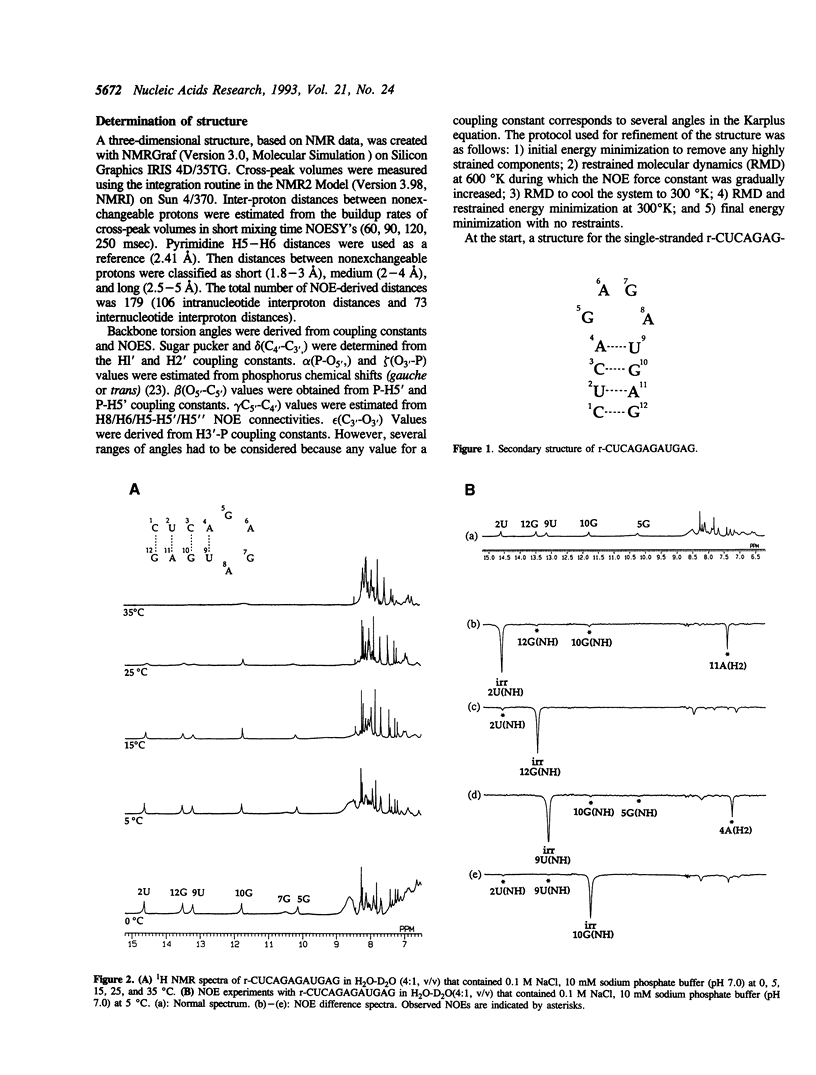
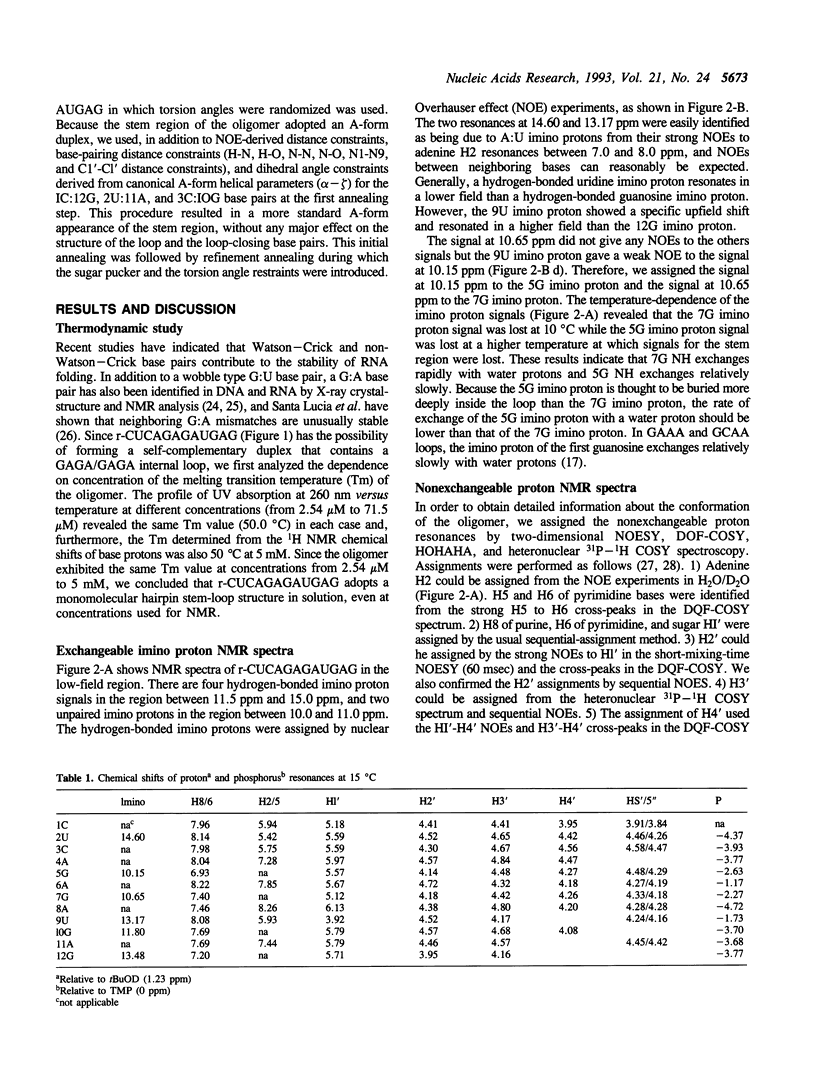
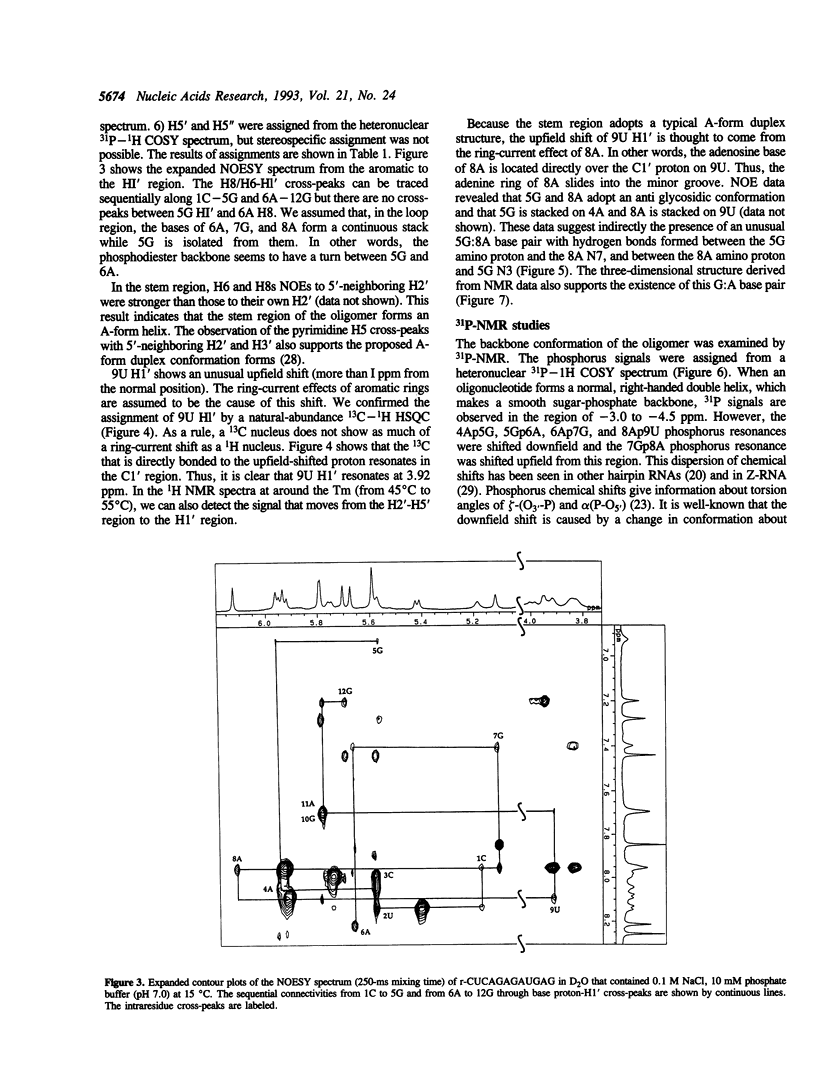
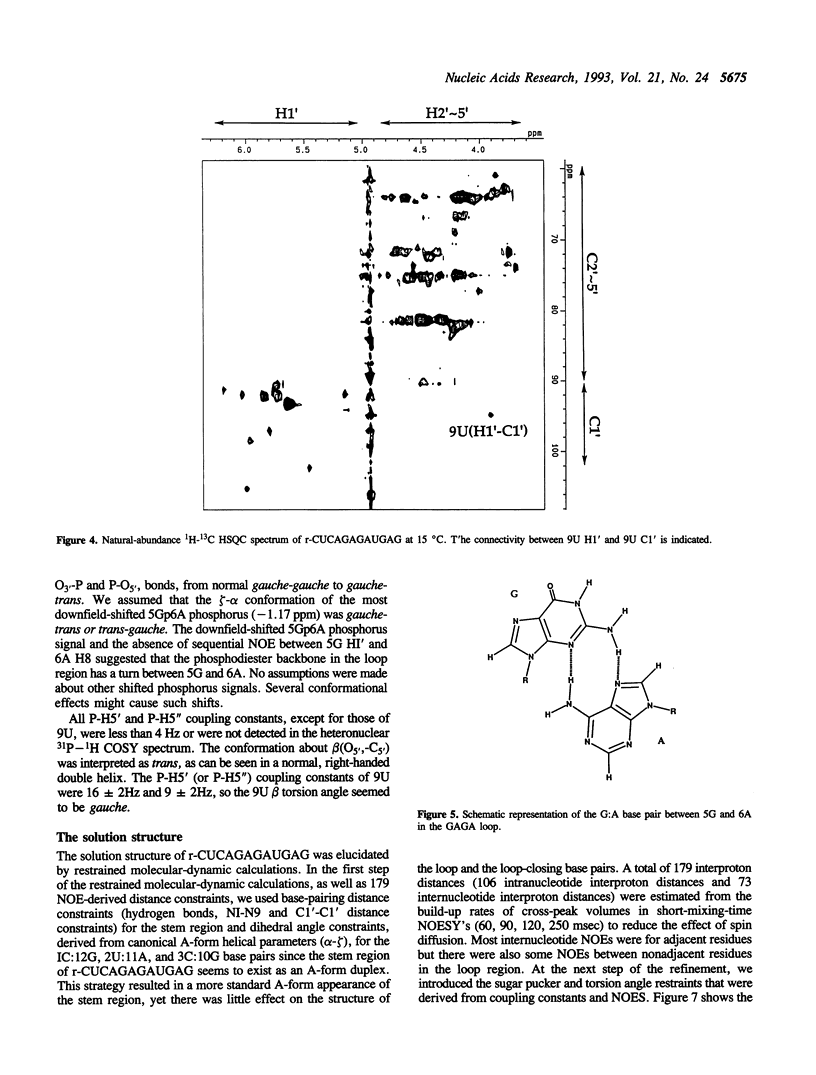
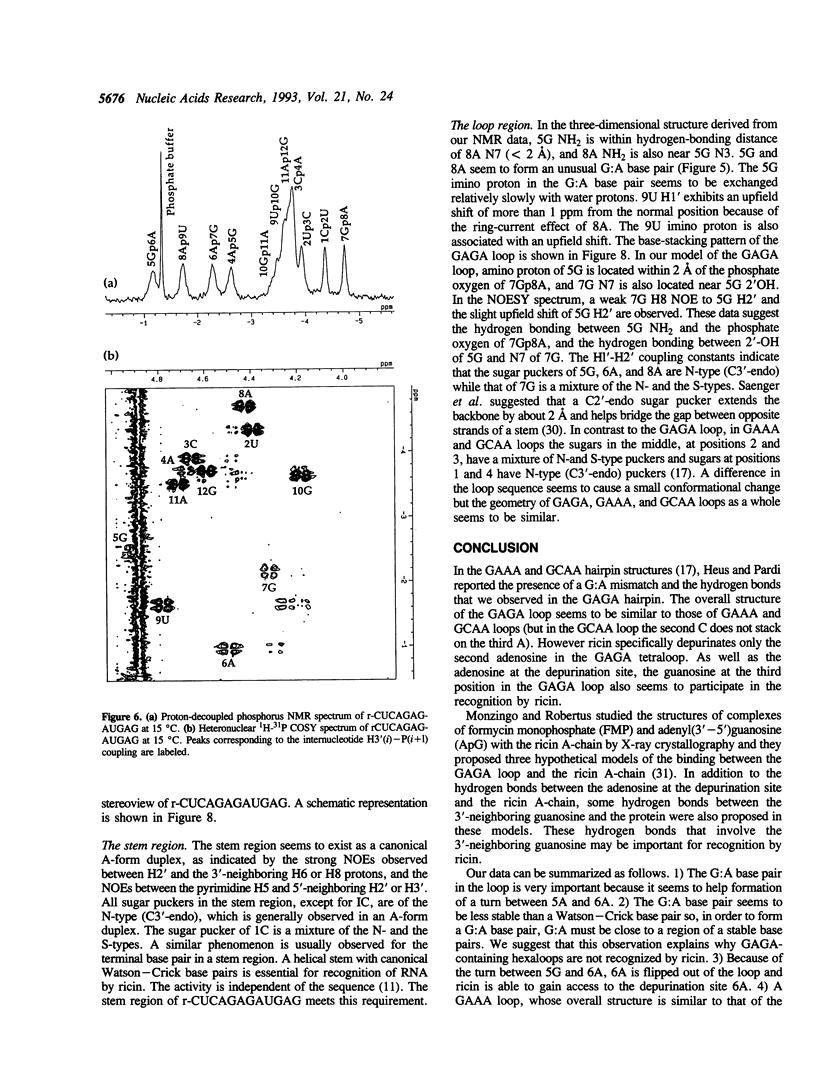
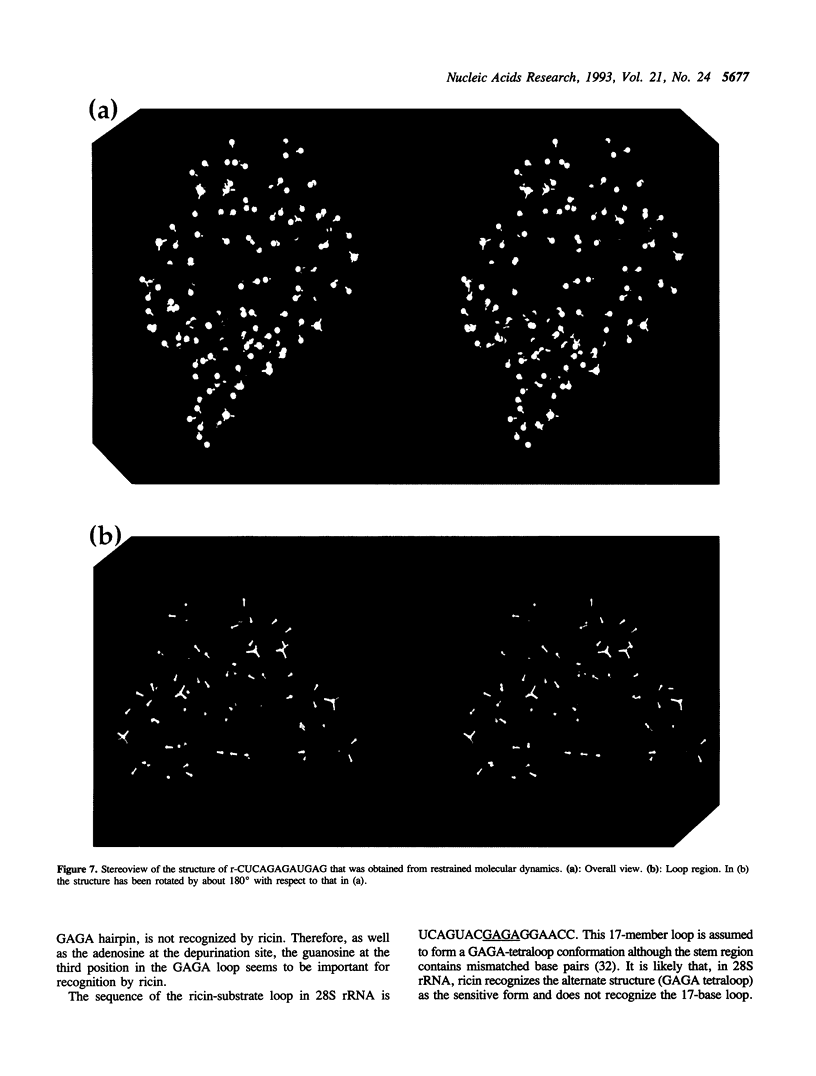
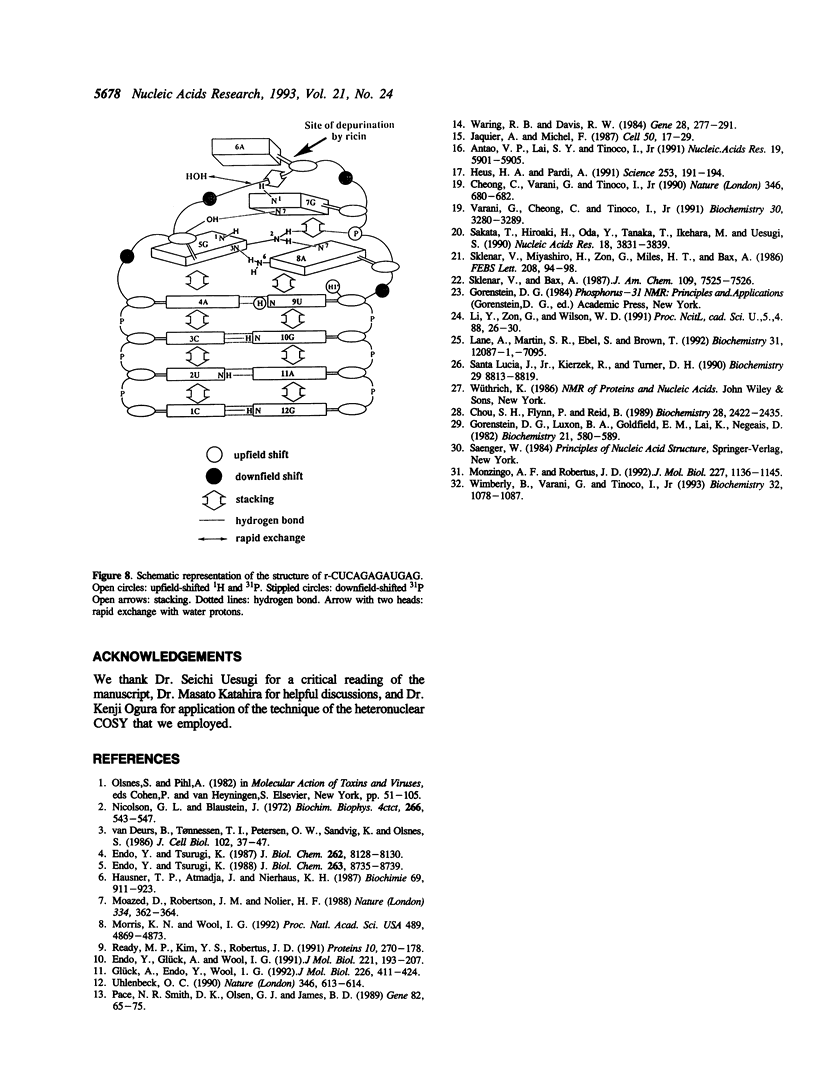
Ricin is a cytotoxic protein that inactivates ribosomes by hydrolyzing the N-glycosidic bond at position A4324 in eukaryotic 28S rRNA. Its substrate domain forms a double helical stem and a 17-base loop that includes the sequence GAGA, the second adenosine of which corresponds to A4324. Recently, studies of mutant RNAs have shown that the four-nucleotide loop, GAGA, can function as a substrate for ricin. To investigate the structure that is recognized by ricin, we studied the properties of a short synthetic substrate, the dodecaribonucleotide r-CUCAGAGAUGAG, which forms a RNA hairpin structure with a GABA loop and a stem of four base pairs. The results of NMR spectroscopy allowed us to construct the solution structure of this oligonucleotide by restrained molecular-dynamic calculations. We found that the stem region exists as an A-form duplex. 5G and 8A in the loop region form an unusual G:A base pair, and the phosphodiester backbone has a turn between 5G and 6A. This turn seems to help ricin to gain access to 6A which is the only site of depurination in the entire structure. The overall structure of the GAGA loop is similar to those of the GAAA and GCAA loops that have been described but that are not recognized by ricin. Therefore, in addition to the adenosine at the depurination site, the neighboring guanosine on the 3' side (7G) may also play a role in the recognition mechanism together with 5G and 8A.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antao V. P., Lai S. Y., Tinoco I., Jr A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991 Nov 11;19(21):5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong C., Varani G., Tinoco I., Jr Solution structure of an unusually stable RNA hairpin, 5'GGAC(UUCG)GUCC. Nature. 1990 Aug 16;346(6285):680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- Chou S. H., Flynn P., Reid B. Solid-phase synthesis and high-resolution NMR studies of two synthetic double-helical RNA dodecamers: r(CGCGAAUUCGCG) and r(CGCGUAUACGCG). Biochemistry. 1989 Mar 21;28(6):2422–2435. doi: 10.1021/bi00432a013. [DOI] [PubMed] [Google Scholar]
- Endo Y., Glück A., Wool I. G. Ribosomal RNA identity elements for ricin A-chain recognition and catalysis. J Mol Biol. 1991 Sep 5;221(1):193–207. doi: 10.1016/0022-2836(91)80214-f. [DOI] [PubMed] [Google Scholar]
- Endo Y., Tsurugi K. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J Biol Chem. 1987 Jun 15;262(17):8128–8130. [PubMed] [Google Scholar]
- Endo Y., Tsurugi K. The RNA N-glycosidase activity of ricin A-chain. The characteristics of the enzymatic activity of ricin A-chain with ribosomes and with rRNA. J Biol Chem. 1988 Jun 25;263(18):8735–8739. [PubMed] [Google Scholar]
- Glück A., Endo Y., Wool I. G. Ribosomal RNA identity elements for ricin A-chain recognition and catalysis. Analysis with tetraloop mutants. J Mol Biol. 1992 Jul 20;226(2):411–424. doi: 10.1016/0022-2836(92)90956-k. [DOI] [PubMed] [Google Scholar]
- Gorenstein D. G., Luxon B. A., Goldfield E. M., Lai K., Vegeais D. Phosphorus-31 nuclear magnetic resonance of double- and triple-helical nucleic acids. Phosphorus-31 chemical shifts as a probe of phosphorus-oxygen ester bond torsional angles. Biochemistry. 1982 Feb 2;21(3):580–589. doi: 10.1021/bi00532a026. [DOI] [PubMed] [Google Scholar]
- Hausner T. P., Atmadja J., Nierhaus K. H. Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie. 1987 Sep;69(9):911–923. doi: 10.1016/0300-9084(87)90225-2. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991 Jul 12;253(5016):191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Michel F. Multiple exon-binding sites in class II self-splicing introns. Cell. 1987 Jul 3;50(1):17–29. doi: 10.1016/0092-8674(87)90658-1. [DOI] [PubMed] [Google Scholar]
- Li Y., Zon G., Wilson W. D. NMR and molecular modeling evidence for a G.A mismatch base pair in a purine-rich DNA duplex. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):26–30. doi: 10.1073/pnas.88.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Robertson J. M., Noller H. F. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988 Jul 28;334(6180):362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- Monzingo A. F., Robertus J. D. X-ray analysis of substrate analogs in the ricin A-chain active site. J Mol Biol. 1992 Oct 20;227(4):1136–1145. doi: 10.1016/0022-2836(92)90526-p. [DOI] [PubMed] [Google Scholar]
- Morris K. N., Wool I. G. Determination by systematic deletion of the amino acids essential for catalysis by ricin A chain. Proc Natl Acad Sci U S A. 1992 Jun 1;89(11):4869–4873. doi: 10.1073/pnas.89.11.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolson G. L., Blaustein J. The interaction of Ricinus communis agglutinin with normal and tumor cell surfaces. Biochim Biophys Acta. 1972 May 9;266(2):543–547. doi: 10.1016/0005-2736(72)90109-5. [DOI] [PubMed] [Google Scholar]
- Pace N. R., Smith D. K., Olsen G. J., James B. D. Phylogenetic comparative analysis and the secondary structure of ribonuclease P RNA--a review. Gene. 1989 Oct 15;82(1):65–75. doi: 10.1016/0378-1119(89)90031-0. [DOI] [PubMed] [Google Scholar]
- Ready M. P., Kim Y., Robertus J. D. Site-directed mutagenesis of ricin A-chain and implications for the mechanism of action. Proteins. 1991;10(3):270–278. doi: 10.1002/prot.340100311. [DOI] [PubMed] [Google Scholar]
- Sakata T., Hiroaki H., Oda Y., Tanaka T., Ikehara M., Uesugi S. Studies on the structure and stabilizing factor of the CUUCGG hairpin RNA using chemically synthesized oligonucleotides. Nucleic Acids Res. 1990 Jul 11;18(13):3831–3839. doi: 10.1093/nar/18.13.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia J., Jr, Kierzek R., Turner D. H. Effects of GA mismatches on the structure and thermodynamics of RNA internal loops. Biochemistry. 1990 Sep 18;29(37):8813–8819. doi: 10.1021/bi00489a044. [DOI] [PubMed] [Google Scholar]
- Sklenár V., Miyashiro H., Zon G., Miles H. T., Bax A. Assignment of the 31P and 1H resonances in oligonucleotides by two-dimensional NMR spectroscopy. FEBS Lett. 1986 Nov 10;208(1):94–98. doi: 10.1016/0014-5793(86)81539-3. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C. Tetraloops and RNA folding. Nature. 1990 Aug 16;346(6285):613–614. doi: 10.1038/346613a0. [DOI] [PubMed] [Google Scholar]
- Varani G., Cheong C., Tinoco I., Jr Structure of an unusually stable RNA hairpin. Biochemistry. 1991 Apr 2;30(13):3280–3289. doi: 10.1021/bi00227a016. [DOI] [PubMed] [Google Scholar]
- Waring R. B., Davies R. W. Assessment of a model for intron RNA secondary structure relevant to RNA self-splicing--a review. Gene. 1984 Jun;28(3):277–291. doi: 10.1016/0378-1119(84)90145-8. [DOI] [PubMed] [Google Scholar]
- Wimberly B., Varani G., Tinoco I., Jr The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry. 1993 Feb 2;32(4):1078–1087. doi: 10.1021/bi00055a013. [DOI] [PubMed] [Google Scholar]
- van Deurs B., Tønnessen T. I., Petersen O. W., Sandvig K., Olsnes S. Routing of internalized ricin and ricin conjugates to the Golgi complex. J Cell Biol. 1986 Jan;102(1):37–47. doi: 10.1083/jcb.102.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]