Abstract
Cytoplasmic initiator tRNAs from plants and fungi possess an unique 2'-phosphoribosyl residue at position 64 of their sequence. In yeast tRNA(iMet), this modified nucleotide located in the T-stem of the tRNA is a 2'-1''-(beta-O-ribofuranosyl-5''-phosphoryl)-adenosine. The phosphoribosyl residue of this modified nucleoside was removed chemically by treatment involving periodate oxidation of tRNA(iMet) and regeneration of the 3'-terminal adenosine with ATP (CTP):tRNA nucleotidyl transferase. The role of phosphoribosylation at position 64 for interaction with elongation factor eEF-1 alpha and initiation factor 2 (eIF-2) was investigated in the homologous yeast system. Whereas the 5'-phosphoribosyl residue prevents the binding of Met-tRNA(iMet) to eEF-1 alpha, it does not influence the interaction with eIF-2. After removal of the ribosyl group, the demodified initiator tRNA showed binding to eEF-1 alpha, but no change was detected with respect to the interaction with the initiation factor eIF-2. This observation is interpreted to mean that a single modification of an eucaryotic initiator tRNA in yeast serves as a negative discriminant for eEF-1 alpha, thus preventing the initiator tRNA(iMet) from entering the elongation cycle of protein biosynthesis.
Full text
PDF
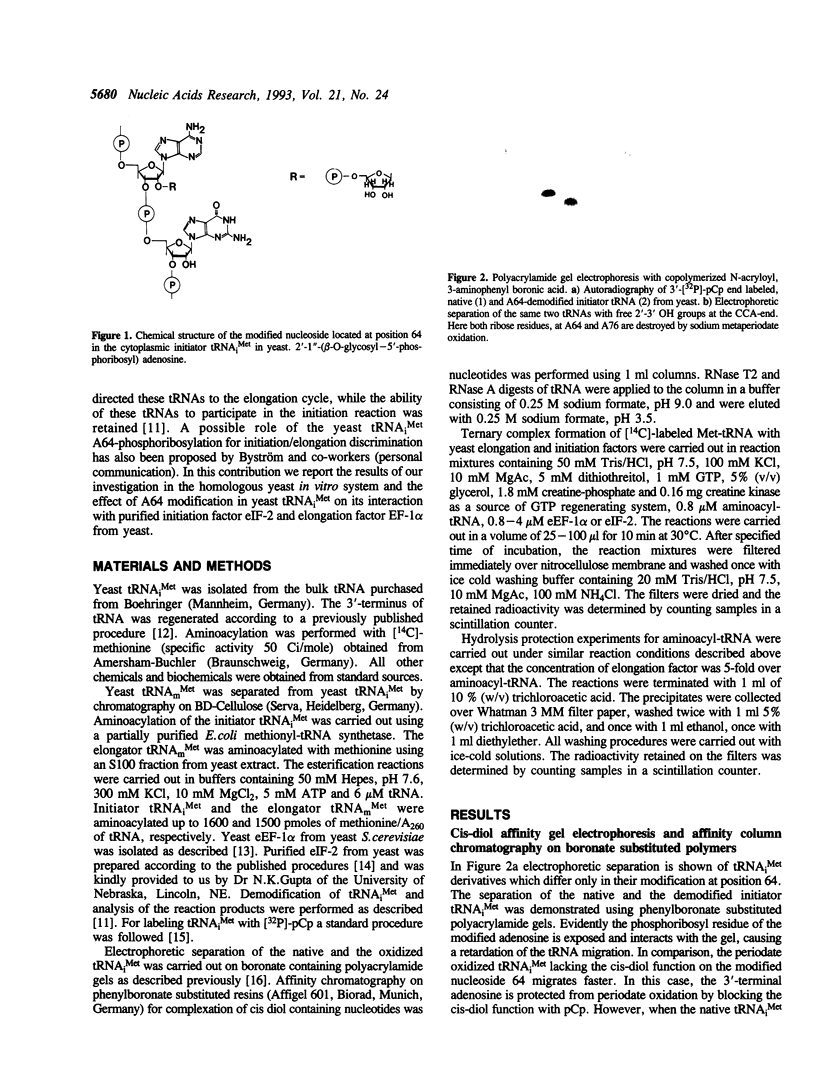



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmad M. F., Nasrin N., Banerjee A. C., Gupta N. K. Purification and properties of eukaryotic initiation factor 2 and its ancillary protein factor (Co-eIF-2A) from yeast Saccharomyces cerevisiae. J Biol Chem. 1985 Jun 10;260(11):6955–6959. [PubMed] [Google Scholar]
- Basavappa R., Sigler P. B. The 3 A crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination. EMBO J. 1991 Oct;10(10):3105–3111. doi: 10.1002/j.1460-2075.1991.tb07864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk G. R., Ericson J. U., Gustafsson C. E., Hagervall T. G., Jönsson Y. H., Wikström P. M. Transfer RNA modification. Annu Rev Biochem. 1987;56:263–287. doi: 10.1146/annurev.bi.56.070187.001403. [DOI] [PubMed] [Google Scholar]
- Dasmahapatra B., Skogerson L., Chakraburtty K. Protein synthesis in yeast. II. Purification and properties of the elongation factor 1 from Saccharomyces cerevisiae. J Biol Chem. 1981 Oct 10;256(19):10005–10011. [PubMed] [Google Scholar]
- Desgrès J., Keith G., Kuo K. C., Gehrke C. W. Presence of phosphorylated O-ribosyl-adenosine in T-psi-stem of yeast methionine initiator tRNA. Nucleic Acids Res. 1989 Feb 11;17(3):865–882. doi: 10.1093/nar/17.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser A. L., Desgres J., Heitzler J., Gehrke C. W., Keith G. O-ribosyl-phosphate purine as a constant modified nucleotide located at position 64 in cytoplasmic initiator tRNAs(Met) of yeasts. Nucleic Acids Res. 1991 Oct 11;19(19):5199–5203. doi: 10.1093/nar/19.19.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi C. O., Severini M., Spurio R., La Teana A., Pon C. L. Molecular dissection of translation initiation factor IF2. Evidence for two structural and functional domains. J Biol Chem. 1991 Sep 5;266(25):16356–16362. [PubMed] [Google Scholar]
- Igloi G. L., Kössel H. Use of boronate-containing gels for electrophoretic analysis of both ends of RNA molecules. Methods Enzymol. 1987;155:433–448. doi: 10.1016/0076-6879(87)55029-7. [DOI] [PubMed] [Google Scholar]
- Keith G., Glasser A. L., Desgrès J., Kuo K. C., Gehrke C. W. Identification and structural characterization of O-beta-ribosyl-(1"----2')-adenosine-5"-phosphate in yeast methionine initiator tRNA. Nucleic Acids Res. 1990 Oct 25;18(20):5989–5993. doi: 10.1093/nar/18.20.5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesewetter S., Ott G., Sprinzl M. The role of modified purine 64 in initiator/elongator discrimination of tRNA(iMet) from yeast and wheat germ. Nucleic Acids Res. 1990 Aug 25;18(16):4677–4682. doi: 10.1093/nar/18.16.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J., Abelson J. tRNA identity. Annu Rev Biochem. 1989;58:1029–1049. doi: 10.1146/annurev.bi.58.070189.005121. [DOI] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Sternbach H., von der Haar F., Cramer F. Enzymatic incorporation of ATP and CTP analogues into the 3' end of tRNA. Eur J Biochem. 1977 Dec;81(3):579–589. doi: 10.1111/j.1432-1033.1977.tb11985.x. [DOI] [PubMed] [Google Scholar]
- Steinberg S., Misch A., Sprinzl M. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1993 Jul 1;21(13):3011–3015. doi: 10.1093/nar/21.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney U., Lee C. P., Seong B. L., RajBhandary U. L. Mutants of initiator tRNA that function both as initiators and elongators. J Biol Chem. 1991 Sep 25;266(27):18018–18024. [PubMed] [Google Scholar]
- von Pawel-Rammingen U., Aström S., Byström A. S. Mutational analysis of conserved positions potentially important for initiator tRNA function in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Apr;12(4):1432–1442. doi: 10.1128/mcb.12.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]




