Summary
B cell-specific coactivator OCA-B, together with Oct-1/2, binds to octamer sites in promoters and enhancers to activate transcription of immunoglobulin (Ig) genes, although the mechanisms underlying their roles in enhancer-promoter communication are unknown. Here, we demonstrate a direct interaction of OCA-B with transcription factor TFII-I, which binds to DICE elements in IgH promoters, that affects transcription at two levels. First, OCA-B relieves HDAC3-mediated IgH promoter repression by competing with HDAC3 for binding to promoter-bound TFII-I. Second, and most importantly, Igh 3′enhancer-bound OCA-B and promoter-bound TFII-I mediate promoter-enhancer interactions, in both cis and trans, that are important for Igh transcription. These and other results reveal an important function for OCA-B in Igh 3′enhancer function in vivo and strongly favor an enhancer mechanism involving looping and facilitated factor recruitment rather than a tracking mechanism.
Introduction
The expression of immunoglobulin (Ig) genes is the essence of humoral immunity. Hence, it is important to understand their transcriptional regulation. Despite years of research, significant questions regarding its transcriptional regulation still remain unanswered. The immunoglobulin heavy chain (Igh) locus is tightly regulated in a tissue- and stage-specific manner by a combination of ubiquitous and cell type-restricted transcription factors that bind to various promoter and enhancer elements and by various DNA rearrangement events. The VH promoters, Eμ intronic enhancer and 3′ enhancers are the major cis elements responsible for VDJ rearrangement, class switch recombination (CSR), and rearranged Igh gene expression (Calame, 2003). The intronic enhancer exhibits greater activity during pre-B to mature B cell stages, which is required for VH gene assembly and μ-chain expression. The 3′-enhancers, on the other hand, are more active in mature B to plasma cell stages and essential both for the germline transcription of switch regions and for subsequent high level expression of switched Ig genes. In mice, the 3′ enhancers span circa 35 kb and consist of four B cell-specific enhancer elements: HS3A, HS1,2, HS3b and HS4 (Khamlichi et al., 2000). Although individually weak, together these four enhancers show great synergies in CSR and Igh gene transcription (Calame, 2003). Several lines of evidence suggest that 3′enhancers play an important role in Igh gene transcription. The combined deletion of HS3B and HS4 in plasmacytoma cell lines severely diminishes Igh gene transcription (Gregor et al., 1986; Michaelson et al., 1995). In knock-out mice, joint deletion of HS3B and HS4 severely impairs germline transcription and CSR to most immunoglobulin gene isotypes (Pinaud et al., 2001). The genomic deletion of the entire Igh 3′ regulatory region greatly affects CSR and Igh gene transcription of all isotypes (Vincent-Fabert et al., 2010). Finally, in transgenic mice, 3′enhancers are required for proper CSR and germline transcription (Dunnick et al., 2005).
A highly conserved octamer sequence (5′-ATGCAAAT-3′) can be found in all VH promoters and in Eμ and 3′ enhancers (Staudt and Lenardo, 1991). Octamer elements in Ig promoters have been implicated in Ig transcription by a variety of in vitro and cell-based reporter assays (Calame, 2003) and by transgene analysis (Jenuwein and Grosschedl, 1991). Two POU domain transcription factors, the ubiquitously expressed Oct-1 and the more B cell-specific Oct-2, bind directly to octamer motifs (Matthias, 1998). A B cell-specific coactivator, OCA-B, binds together with Oct-1 or Oct-2 to the octamer site to form a ternary complex (Luo et al., 1992). OCA-B was originally identified biochemically as a B cell nuclear extract-derived factor that stimulates Igh promoter transcription through Oct-1/2 interactions (Luo et al., 1992) and subsequently cloned by several laboratories (Gstaiger et al., 1995; Luo and Roeder, 1995; Strubin et al., 1995). Targeted gene knockouts in mice revealed roles for OCA-B in germinal centre formation and in antigen-dependent transcription of switched Ig genes (Kim et al., 1996; Nielsen et al., 1996; Schubart et al., 1996). However, the interpretation of the OCA-B knockout phenotype, especially in relation to switched Ig gene expression, is complicated by the subsequent finding that OCA-B is required for B-cell development at multiple stages (Hess et al., 2001; Samardzic et al., 2002). Recent data have revealed that the Oca-b gene is a direct target of XBP-1, a regulator of the unfolded protein response that is essential for plasma cell development (Shen and Hendershot, 2007), in line with previous reports that OCA-B plays a role in plasma cell differentiation and production of Igh isotypes (Corcoran et al., 2005; Shen and Hendershot, 2007). In addition, transfection assays show that OCA-B plays an important role in Igh 3′enhancer function in cell lines (Stevens et al., 2000; Tang and Sharp, 1999). However, the detailed molecular mechanism by which OCA-B mediates 3′enhancer function is still not known. In this regard, and given that enhancers ultimately affect promoter function and that OCA-B also has the potential to act through promoter-associated octamer elements, it is also important to understand factor interactions at the VH promoters. These promoters, in isolation, show activity only in B lymphoid lineage cells, although the molecular mechanisms responsible for this specificity are also largely unknown.
TFII-I is a ubiquitous transcription factor that was originally identified as a protein that binds to the initiator (Inr) core promoter element and was later shown to bind to various upstream elements that include the E-box (Roy et al., 1997; Roy et al., 1991). Structurally, TFII-I consists of multiple I-repeats, each of which contains a putative helix-loop-helix motif that is potentially important for protein-protein interactions (Roy, 2007). Growth factor signaling leads to rapid tyrosine phosphorylation, nuclear translocation of TFII-I and subsequent activation of target genes (Novina et al., 1998; Novina et al., 1999; Yang and Desiderio, 1997). Recent data suggest that TFII-I plays an important role in B cells and in Ig gene transcription in particular. TFII-I and the related family member BEN bind to DICE (Downstream Immunoglobulin Control Element) sequences that are located downstream of many Igh transcription start sites and play roles in Igh gene activation (Tantin et al., 2004). In addition, non-receptor Bruton’s tyrosine kinase (Btk) interacts with TFII-I in splenic B cells and regulates its tyrosine phosphorylation, nuclear translocation and transcriptional activity (Novina et al., 1999; Yang and Desiderio, 1997). It is known that mutations in Btk cause X-linked agammaglobulinemia (XLA) in human and X-linked immunodeficiency (Xid) in mice (Tsukada et al., 1994). Interestingly, Oca-b and Btk double knockout mice lack peripheral B cells and have severely diminished serum Igs, a phenotype resembling that of human XLA (Schubart et al., 2000).
In this study, we demonstrate that OCA-B directly interacts with TFII-I in several B cell lines. Using a terminally differentiated plasmacytoma cell line as a model system, we functionally show that OCA-B plays multiple roles in Igh gene activation that are independent of other B cell differentiation events such as VDJ rearrangement and CSR. First, OCA-B binding to TFII-I partially relieves the Igh promoter repression caused by HDAC3 recruitment to promoter-bound TFII-I through competitive binding. Second, enhancer-bound Oct-1/2–OCA-B mediates Igh promoter and enhancer juxtaposition through direct interaction with promoter-bound TFII-I. Thus, our data provide important insights into the factors (OCA-B and TFII-I) and mechanisms (chromosome looping) involved in Igh 3′enhancer function in transcription.
Results
OCA-B interacts with TFII-I in B cells
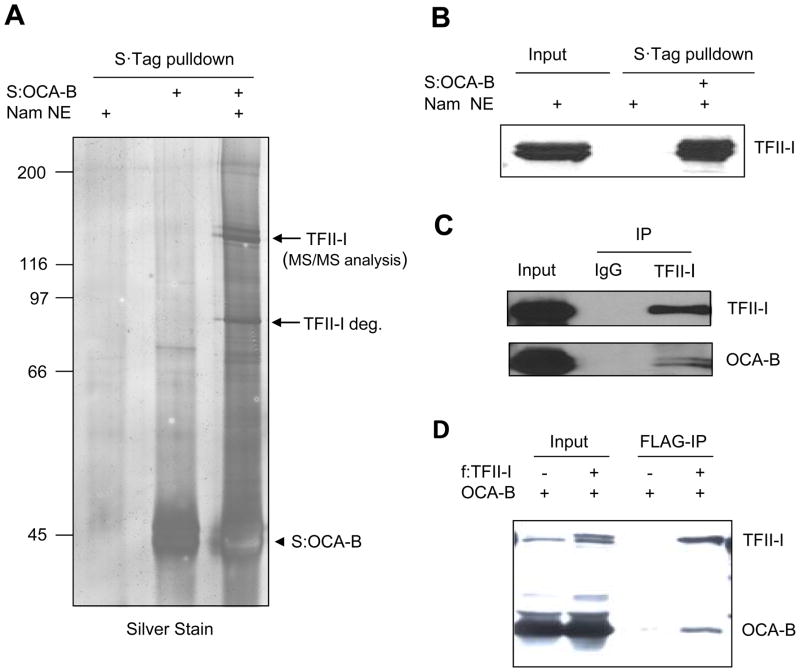
In an attempt to decipher the mechanism of OCA-B–mediated IgH transcription, we employed bacterially expressed OCA-B as a bait to biochemically identify interacting proteins from nuclear extracts. To this end, S-tagged OCA-B was immobilized on S-agarose and incubated with Namalwa B cell nuclear extract. After washing, bead-bound proteins were resolved by SDS-PAGE and identified by mass spectrometry. The proteins that co-purified with OCA-B are shown in Figure 1A. Two dominant 125 kDa polypeptides were identified as isoforms of TFII-I, whereas a circa 80kDa TFII-I degradation product was also identified. The identities of OCA-B interacting peptides were further confirmed by immunoblot analysis with TFII-I specific antibodies (Fig. 1B).
Figure 1.
Identification of TFII-I as an OCA-B interacting protein. (A) Immobilized OCA-B interacts with transcription factor TFII-I in B cell nuclear extracts. Bacterially purified S-tagged OCA-B was immobilized on S-protein agarose and incubated with Namalwa B cell nuclear extracts. Bound proteins were analyzed by SDS-PAGE and silver staining. Protein bands indicated by arrows were analyzed by MS/MS. (B) Bound proteins in (A) were detected by immunoblot with a specific anti-TFII-I antibody. (C) Immunoblot analysis of anti-TFII-I and control rabbit IgG immunoprecipitates (IP) from Namalwa B cell nuclear extract with antibodies against proteins indicated on the right. Input represents 2.5% of total. (D) Immunoblot analysis of anti-FLAG immunoprecipitates from 293T cells transfected with vectors expressing OCA-B and/or FLAG-TFII-I. Input represents 5% of total.
To substantiate the physiological relevance of these interactions, we next documented similar interactions between endogenous proteins. An affinity-purified antibody directed against the carboxyl terminus of TFII-I was used to pull down endogenous TFII-I and interacting proteins from Namalwa nuclear extract (Fig. 1C). Consistent with the immobilized OCA-B pulldown analysis, endogenous TFII-I was found to specifically interact with endogenous OCA-B. Furthermore, immunoprecipitation analysis revealed copurification of ectopically expressed OCA-B and FLAG-tagged TFII-I in 293T cells (Fig. 1D). Thus, we confirmed TFII-I as a bona fide OCA-B interacting protein in vitro and in vivo.
OCA-B Interacts with Distinct Conserved Regions of the TFII-I I-repeats
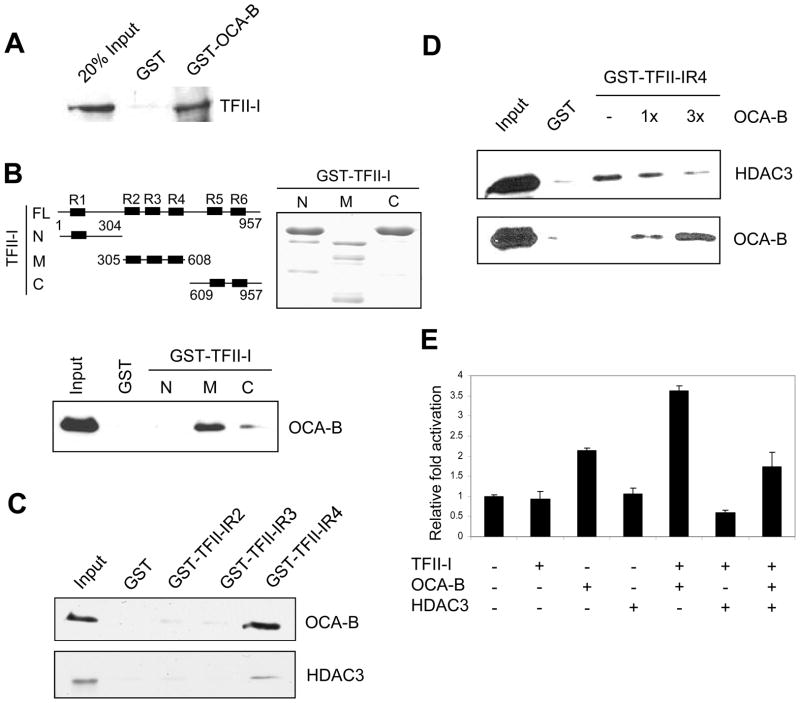
To establish a direct interaction between OCA-B and TFII-I, we performed GST pull-down assays with purified GST-OCA-B fusion and bacterially expressed TFII-I proteins (Supplementary Fig. 1A). TFII-I was found to interact strongly with GST-OCA-B, indicative of a direct interaction (Fig. 2A). This interaction apparently requires a central portion of OCA-B, since neither the N-terminal or nor the C-terminal half of OCA-B showed any interaction with TFII-I (Supplementary Figs. 1B, C). To dissect the TFII-I domain(s) responsible for interaction with OCA-B, the N-terminal third (TFII-IN), middle third (TFII-IM) and C-terminal third (TFII-IC) of TFII-I were expressed as GST fusion proteins and purified (Fig. 2B). When incubated with bacterially expressed OCA-B, only the middle fragment of TFII-I, TFII-IM, showed a significant interaction with OCA-B. An in vitro GST pull-down assay further showed that the I-repeat 4 of TFII-IM (TFII-IR4) strongly interacts with bacterially expressed OCA-B (Fig. 2C). Interestingly, the TFII-IR4 repeat was shown previously to interact with histone deacetylase HDAC3 (Tussie-Luna et al., 2002; Wen et al., 2003). The potential overlapping interaction domain of OCA-B and HDAC3 on TFII-IR4 prompted us to test whether OCA-B and HDAC3 bind to TFII-IR4 simultaneously or in a mutually exclusive fashion. Indeed, the binding of a fixed amount of bacterially expressed HDAC3 to a GST-TFII-IR4 fusion protein was gradually diminished by the addition of increasing amounts of bacterially expressed OCA-B (Fig. 2D), thus indicating that OCA-B and HDAC3 bind to TFII-IR4 in a mutually exclusive (competitive) manner. Furthermore, the TFII-IR4 repeat reduced the OCA-B and TFII-I binding in a dominant negative manner (Supplementary Fig. 1D). Consistent with these observations, the TFII-IR4 repeat partially inhibited the cooperative function of OCA-B and TFII-I on the IgH promoter and, as well, partially relieved the repression of IgH promoter activity by HDAC3 (Supplementary Fig. 1E).
Figure 2.
TFII-I interacts with OCA-B through its conserved I-repeat. (A) Interaction of OCA-B and TFII-I. A purified recombinant TFII-I protein was tested for binding to GST or GST-OCA-B proteins, and bound proteins were detected by anti-TFII-I immunoblot. (B) The middle part of TFII-I (M) interacts with OCA-B. Schematic diagram of full-length (FL) and mutant (N, M, C) TFII-I (top left). Purified GST-tagged TFII-I proteins from bacteria were analyzed by SDS-PAGE and Coomassie blue staining (top right). Recombinant OCA-B proteins were tested for binding to GST or GST-TFII-I mutants, and bound proteins were detected by anti-OCA-B immunoblot (bottom). (C) R4 repeat of TFII-I interacts with both OCA-B and HDAC3. Recombinant OCA-B and HDAC3 were tested for binding to GST and GST-TFII-I mutant proteins, and bound proteins were detected by anti-OCA-B and anti-HDAC3 antibodies, respectively. (D) OCA-B and HDAC3 compete for binding to the R4 repeat of TFII-I. A constant amount of HDAC3 and increasing amounts of OCA-B (0, 1x, 3x relative molar ratio to HDAC3) were tested for binding to a constant amount of GST-TFII-IR4, and bound proteins were detected by anti-OCA-B and anti-HDAC3 antibodies, respectively. (E) TFII-I enhances OCA-B–dependent IgH promoter activation. 293T cells grown in 24-well plates were cotransfected with 20 ng IgH promoter (M17056, −154 to +167bp relative to TSS) luciferase reporter, 1 ng pRL-CMV control vector and 20 ng each of vectors expressing TFII-I, OCA-B and/or HDAC3. Data are presented as the mean ± SD.
Earlier studies have indicated that TFII-I and family member BEN bind to an immunoglobulin heavy chain promoter element (DICE) and are required for optimal promoter activity in transfection experiments (Tantin et al., 2004). Our ChIP and EMSA assays also show that TFII-I binds to the Igh DICE elements both in vitro and in vivo (see below and Supplementary Fig. 2). To assess the physiological role of OCA-B and TFII-I interactions at the Igh promoter, we tested effects of ectopic proteins on expression of a reporter with an Igh promoter that contains multiple DICE elements and is active in the plasmacytoma MPC11 cell line. The results in Figure 2E indicate that while TFII-I alone does not significantly activate Igh transcription in this assay, it acts cooperatively with OCA-B to induce Igh expression. Moreover, in support of the relevance of the above-described competitive OCA-B and HDAC3 binding to TFII-I, coexpression of HDAC3 resulted in a significant repression of the TFII-I and OCA-B enhanced Igh promoter activity (Figure 2E). Related, the phosphorylation of TFII-I by BTK is apparently needed for full Igh promoter activity (data not shown), consistent with earlier studies indicating that phosphorylation of TFII-I is required for its translocation into the nucleus (Novina et al., 1999; Sacristan et al., 2004).
Taken together, these results suggest that the coactivator function of OCA-B on Igh promoters may in part reflect OCA-B–mediated relief, through competitive binding to TFII-I, of the repression caused by TFII-I-associated HDAC activities on the Igh promoter.
OCA-B plays an important role in Igh 3′enhancer function
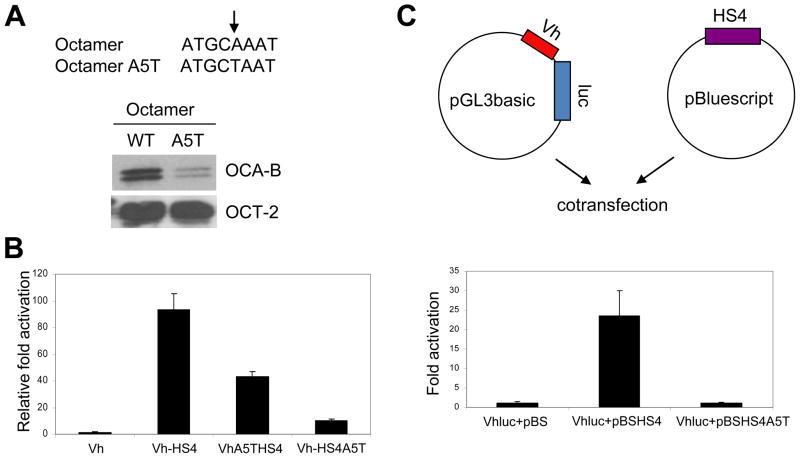
To further elucidate the role of OCA-B in Igh 3′enchancer function, we utilized a previously identified single nucleotide mutation (A5T) in the octamer motif (Cepek et al., 1996; Gstaiger et al., 1996). Consistent with the previous reports, the mutated biotinated template retains Oct-2 binding but dramatically loses the binding of coactivator OCA-B following incubation with B cell nuclear extracts (Fig. 3A). An EMSA assay also indicated that OCA-B cannot bind with Oct-1 to a mutated octamer DNA probe, relative to a wild type octamer DNA probe, to form a ternary complex (data not shown).
Figure 3.
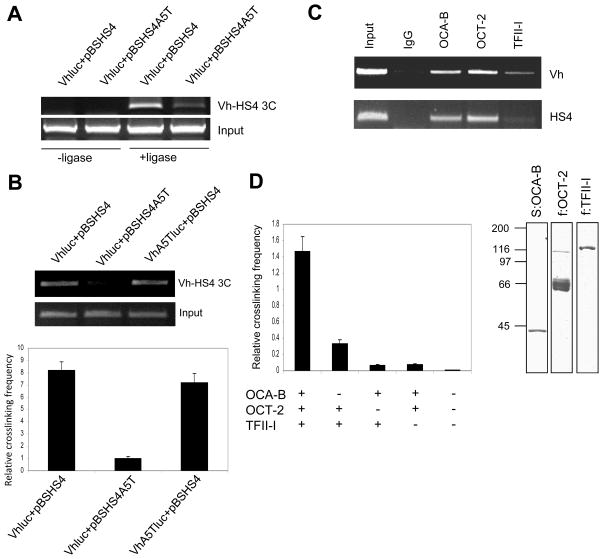
Enhancer-bound OCA-B plays an important role in Igh 3′enhancer function. (A) Diagram of the octamer A5T mutation with the arrow indicating the 5th position of the octamer (top), SDS-PAGE and Coomassie blue staining analyses of OCA-B and Oct-2 proteins expressed in and purified from bacteria and sf9 cells, respectively (lower left), and DNA template recruitment assays (lower right). Assays contained biotinylated DNA fragments with intact or mutant octamer elements and purified Oct-2 and OCA-B. Assays were carried out as described in Methods and bound proteins were detected by immunoblot. (B) An OCA-B–enhancer interaction is required for Igh promoter and HS4 enhancer functions in cis. MPC11 cells were transfected with Igh promoter-HS4 enhancer constructs (cloned in pGL3-basic) that contained either an intact promoter and enhancer octamers (VH-HS4), an A5T mutant octamer in the promoter (VHA5T-HS4) or an A5T mutant octamer in the enhancer (VH-HS4A5T). One reporter (VH) contained only the Igh promoter with an intact octamer cloned in pGL3-basic vector. (C) An OCA-B–enhancer interaction is required for Igh promoter and HS4 enhancer function in trans. MPC11 plasmacytoma cells were transfected with a pGL3-basic vector containing the Igh promoter/reporter (VHluc) and a pBluescript vector that either lacks (pBS) or contains an HS4 enhancer with an intact (pBSHS4) or an A5T mutant (pBSHS4A5T) octamer (Top panel, diagram). The dual luciferase assay result is shown in the bottom panel. Data are presented as the mean ± SD (B,C).
The murine HS4 element of 3′enhancer, which is active throughout B cell development and shows significant independent activity (Madisen and Groudine, 1994; Michaelson et al., 1995), was chosen for mechanistic studies. The full-length Igh promoter from MPC11 cells and the HS4 enhancer were cloned in pGL3-basic vector promoter and enhancer cloning sites, respectively, and assayed in MPC11 cells. The significant activity that was observed with the wild type octamer reporter was reduced about 10-fold by the A5T mutation in HS4, whereas the A5T mutation at the promoter octamer only reduced the total activity by 2-fold (Fig. 3B). This result suggests that OCA-B could have additional roles other than its previously identified coactivator function at the enhancer.
The endogenous Igh promoter and 3′enhancers in B cells are separated by over 100kb. To further mimic the 3′enhancer functions in vivo, we cloned the Igh promoter and HS4 enhancer into pGL3 and pBluescript vectors, respectively (diagramed in Fig. 3C, top). Remarkably, as shown in the luciferase assay (Fig. 3C, bottom), the wild type HS4 enhancer boosted the activity of the Igh promoter in trans by more than 20-fold. In contrast, the A5T mutation in the HS4 enhancer octamer site totally abolished the HS4 enhancer activity. The indicated requirement of an intact octamer site in the HS4 enhancer for full enhancer function in trans suggests that the HS4 enhancer octamer site and its cognate binding proteins play a role in normal promoter-enhancer communication and associated enhancer function.
OCA-B and TFII-I are required for Igh promoter and enhancer interaction in vivo
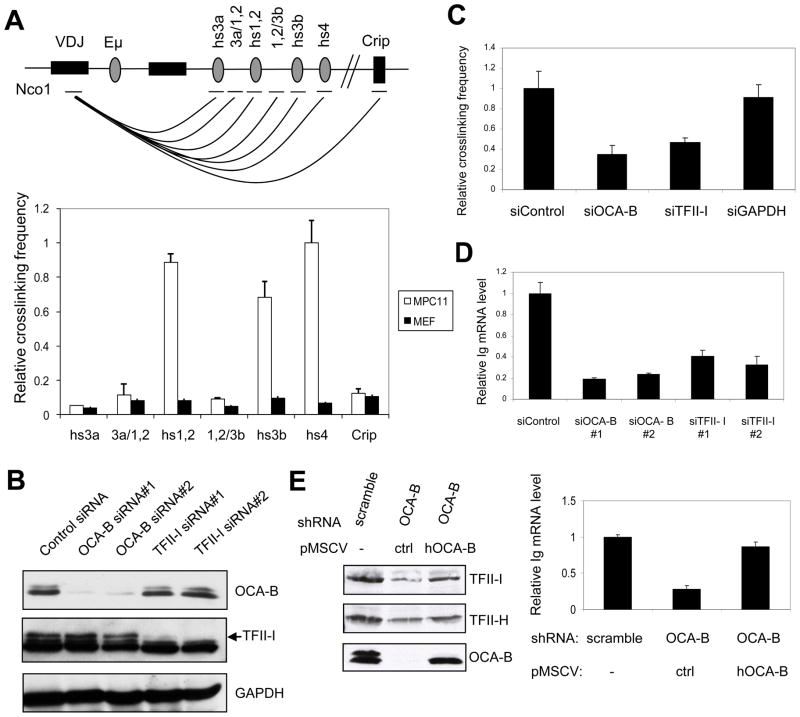
Because our results suggested that OCA-B might play a role in promoter and enhancer communication, we tested this possibility directly by chromatin conformation capture analysis of the endogenous Igh locus in MPC11 cells. These cells were chosen since they show active Igh transcription through known Igh promoter sequences (Gilmore et al., 1987). Relative primer pair efficiencies were measured using an equimolar mixture of 199M11 BAC DNA and Igh promoter sequences as templates, which were linearized by the indicated restriction enzyme and subsequently ligated. 3C assays were performed using real-time PCR Taqman probe methods. A fixed primer and probe located at the Igh promoter and primers located at respective 3′ Igh enhancer elements near the Nco1 sites were chosen to determine their relative interaction strength, as indicated in the diagram of Fig. 4A. The relative interaction strengths between promoter and enhancer elements were normalized to that of the interaction between two non-adjacent fragments of the Ercc3 gene as an internal 3C control. As shown in Fig. 4A, HS1,2, HS3B and HS4 enhancer elements show stronger interactions with the Igh promoter than do other enhancer elements in MPC11 cells, but not in MEF control cells. As a negative control, the Crip gene that lies about 30 kb downstream of the 3′ enhancers showed little interaction with the Igh promoter. Our results confirm previously reported Igh promoter-enhancer interaction in MPC11 cells (Ju et al., 2007). However, our use of smaller genomic fragments containing individual enhancers (reflecting cleavage with a different restriction enzyme) and realtime PCR methodology (rather than standard PCR) likely accounts for the apparent differences in certain individual promoter-enhancer interactions observed in these studies.
Figure 4.
OCA-B and TFII-I are required for promoter-enhancer interaction and Igh gene transcription. (A) Schematic diagram of the murine Igh locus in MPC11 cells (top) and relative interaction frequencies between Igh promoter and 3′enhancer elements as measured by 3C analysis (bottom). A very low interaction frequency was detected between VH and the Crip gene 30kb downstream of the 3′enhancers. (B) siRNA-mediated knockdown of OCA-B or TFII-I reduces the endogenous Igh transcription level in MPC11 cells. MPC11 cells were treated either with OCA-B or TFII-I siRNAs or a non-specific (control) siRNA and whole cell extracts were analyzed by immunoblot. (C) Effects of OCA-B and TFII-I knockdowns on endogenous Igh promoter-HS4 enhancer interactions in MPC11 cells. Interactions were measured by 3C analysis as described in Methods. (D) Effects of OCA-B and TFII-I knockdowns on endogenous Igh transcription in MPC11 cells. Steady state Igh mRNA levels were analyzed by real-time RT-PCR and normalized to 18S rRNA levels. (E) Rescue of the effects of OCA-B knockdown by overexpression of human OCA-B. MPC11 cells were treated either with control or OCA-B shRNAs and, in the latter case, infected with control or human OCA-B–expressing retroviruses. Whole cell extracts were analyzed by immunoblot (left panel) and Igh mRNA levels were analyzed by qRT-PCR and normalized to 18S rRNA levels (right panel). Data are presented as the mean ± SD (A,C,D,E).
To determine whether OCA-B and TFII-I play a role in Igh promoter and enhancer interactions, we performed similar analyses following siRNA-mediated knockdown in MPC11 cells. Both OCA-B and TFII-I protein levels were greatly reduced by corresponding siRNAs but not by a control siRNA (Fig. 4B). As an internal control, the GAPDH protein level was not perturbed by siRNA treatment. To determine the effects of OCA-B and TFII-I knockdown on Igh promoter and enhancer interactions, 3C assays were carried out in MPC11 cells following siRNA-mediated knockdown of OCA-B or TFII-I. The promoter-HS4 enhancer interaction was significantly reduced by OCA-B knockdown relative to a control siRNA knockdown (Fig. 4C), indicating that OCA-B plays an important role in mediating Igh promoter and enhancer interaction in vivo. Similarly, the knockdown of TFII-I also resulted a more than 2-fold reduction in the promoter-HS4 enhancer interaction (Fig. 4C). Importantly, the control GAPDH siRNA did not show any significant effect on the Igh promoter-HS4 enhancer interaction. As observed for the HS4 enhancer, HS1,2 and HS3B interactions with the Igh promoter were significantly reduced by OCA-B siRNA knockdown (Supplementary Fig. 3A). Notably, all these 3′enhancer elements share conserved octamer sequences capable of OCA-B binding in conjunction with Oct-1/2. In this regard, and while the HS4 enhancer was found to selectively interact with other 3′ enhancers relative to the control Crip gene, these interactions between 3′ enhancers were largely unaffected by OCA-B knockdown (Supplementary Fig. 3B) and must reflect interactions of other enhancer-associated proteins.
The effect of OCA-B and TFII-I knockdown on Igh gene expression in vivo was tested by RT-qPCR. As indicated in Fig. 4D, OCA-B siRNA knockdown significantly reduced the endogenous Igh mRNA level in MPC11 cells. Similar to its effect in reducing the promoter-enhancer interactions, the siRNA-mediated TFII-I knockdown caused a modest (but significant) reduction in the Igh mRNA level compared with OCA-B knockdown. The existence of the closely related TFII-I family member BEN in these cells and their potentially redundant functions at Igh promoters (Tantin et al., 2004) likely account for the more modest effects of TFII-I siRNA knockdown on Igh gene transcription and promoter-enhancer interactions. To rule out indirect effects in the OCA-B knockdown experiments, a rescue experiment was performed. In this analysis MPC11 cells treated with control or OCA-B shRNAs were infected with control retrovirus or retrovirus expressing human OCA-B (Figure 4E, left panel), the coding region of which differs from that of the mouse counterpart in shRNA target sites. Importantly, the IgH promoter activity (measured by qRT-PCR) that was selectively inhibited by the OCA-B shRNA was nearly completely rescued by the overexpressed human OCA-B (right panel), indicating a direct role of OCA-B in Igh gene transcription.
These observations establish functionally relevant interactions between Igh promoter and enhancer elements, and also suggest that OCA-B and TFII-I play an important role in mediating the Igh promoter-enhancer interactions and associated functions. Furthermore, in vivo Igh gene expression is regulated by OCA-B and TFII-I protein levels, and generally correlated with Igh promoter-enhancer interactions.
OCA-B and TFII-I are necessary and sufficient to mediate Igh promoter and HS4 enhancer interaction in vitro
To investigate the role of OCA-B and TFII-I in mediating Igh promoter and enhancer interactions and to rule out potential indirect effects caused by siRNA-mediated knockdowns, we repeated the 3C experiments in MPC11 cells following cotransfection of plasmids containing Igh promoter and enhancer sequences, respectively. Again, we chose the HS4 enhancer as a representative Igh 3′enhancer because it showed strong interactions with the Igh promoter elements in our previous 3C assays and because of the previous demonstration of the independent function of HS4 in activating an Igh promoter in transfected cells (Michaelson et al., 1995). Igh promoter and HS4 enhancer sequences were cloned in pGL3-basic and pBluescript vectors, respectively, and 3C assays were carried out in cells transfected with these vectors. 3C primers were designed so that only signals from transfected vectors, and not genomic sequences, could be amplified. As indicated in Fig. 5A, the Igh promoter interacted strongly with the wild type HS4 enhancer, and this interaction was greatly reduced by the A5T mutation in the HS4 octamer site. The observed 3C signal was dependent upon ligation of the restriction enzyme-linearized DNA template, confirming that it is the PCR product of ligated DNA sequences.
Figure 5.
OCA-B and TFII-I mediate Igh promoter-HS4 enhancer interactions in trans both in MPC11 cells and in vitro. (A,B) Trans interaction between Igh promoter and HS4 enhancer elements in MPC11 cells dependent upon an HS4–OCA-B interaction, but not upon a promoter–OCA-B interaction. MPC11 cells were cotransfected with VHluc reporter and HS4 enhancer (pBSHS4) plasmids with or without A5T mutations in corresponding octamer elements as indicated, and 3C assays were performed. 3C assays in (A) were performed with or without (control) ligase. In (B) the relative interaction frequency (bottom) was measured by real-time PCR. (C) Binding of factors to Igh promoter and HS4 enhancer elements in MPC11 cells. ChIP analysis of MPC11 cells with indicated antibodies using primers that cover the Igh promoter (VH) and HS4 enhancer regions. (D) Trans interactions of Igh promoter and HS4 enhancer regions in vitro. 1 μg of VHluc and pBSHS4 plasmids were incubated with 0.5 μg proteins as indicated, fixed in formaldehyde, digested with HindIII, and analyzed by 3C and real-time PCR assay as described in Methods (left panel). The panel on the right shows an SDS-PAGE (Coomassie blue stain) of affinity purified FLAG-tagged TFII-I expressed in 293T cells. Purified Oct-2 and OCA-B proteins are shown in Fig. 3A. Results are presented as the mean ± SD (B,D).
Since the Igh promoter also has a canonical octamer site, it was of interest to determine whether promoter-bound OCA-B is involved in the Igh promoter-enhancer interactions observed in the 3C assays. As indicated in Fig. 5B, the A5T mutation in the Igh promoter octamer site, which prevents OCA-B binding at the promoter site of the transfected construct (Cepek et al., 1996; Gstaiger et al., 1996), had no effect on promoter and enhancer interactions as judged by 3C assay using conventional PCR and real-time PCR.
To confirm that the OCA-B–TFII-I interaction is sufficient to mediate Igh promoter-enhancer interactions, we set up an in vitro binding and “3C” assay using purified factors and separate plasmids containing Igh promoter and HS4 enhancer sequences. Recruitment of OCA-B, Oct-2 and TFII-I to Igh promoter (Vh) and recruitment of OCA-B and Oct-2 to enhancer (HS4) sites were confirmed by ChIP assay (Fig. 5C). Importantly, OCA-B, Oct-2 and TFII-I together effected a strong promoter-HS4 enhancer interaction in the 3C assay, and omission of any single factor resulted in a significant reduction in this interaction (Fig. 5D). These results indicate that enhancer-bound Oct-2/OCA-B and promoter-bound TFII-I are necessary and sufficient to mediate an interaction between distal Igh promoter and enhancer sequences in vitro.
Proper enhancer activity correlates with distinct promoter histone modifications and general transcription factor recruitment
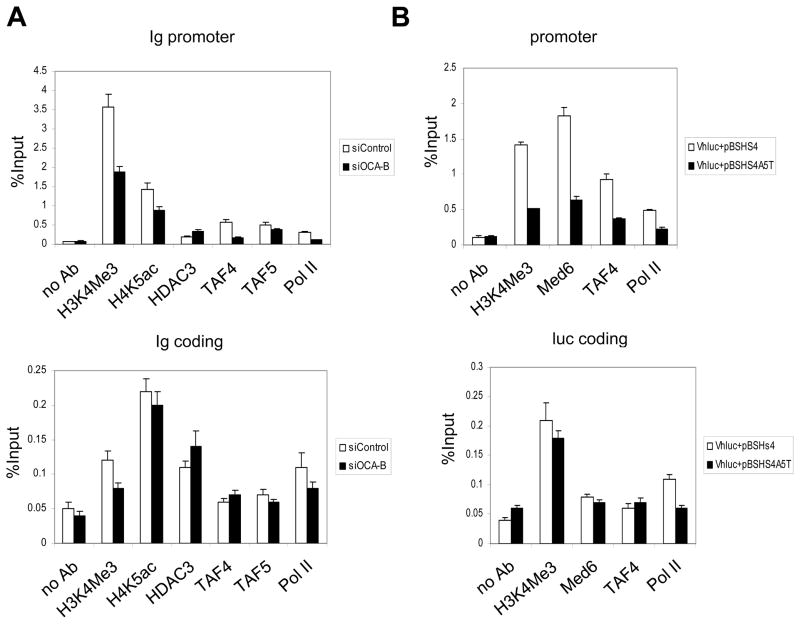
To better understand the molecular mechanism by which reduced OCA-B levels lead to reduced Igh promoter-enhancer interactions and Igh gene transcription, we carried out ChIP assays in MPC11 cells to determine histone modification status and factor recruitment. As indicated in Fig. 6A, OCA-B knockdown by siRNA resulted in a modest but significant reduction of the histone H3K4 trimethylation mark that is associated with transcription activation. Similarly, TAF/TFIID and Pol II recruitments to the Igh promoter were also reduced by an OCA-B siRNA knockdown relative to a control siRNA knockdown. Interestingly, HDAC3 binding at the Igh promoter was inversely correlated with the OCA-B level. Because our previous studies showed that OCA-B and HDAC3 binding to TFII-I are mutually exclusive, the ChIP analyses further indicate a possible physiological role for the mutually exclusive binding of OCA-B and HDAC3 to the Igh promoter. H4K5 acetylation, which is regulated by HDAC3, correlates with the OCA-B level in cells. The histone modification marks and factors such as TAF4 and TAF5 were markedly lower in the coding region relative to the promoter region and largely unchanged by OCA-B knockdown relative to the control siRNA knockdown (Fig. 6A).
Figure 6.
Histone modifications and factor recruitment at the Igh promoter and coding regions mediated by OCA-B. (A) MPC11 cells were treated with OCA-B–specific or control siRNA, and histone modifications and factor recruitment at Igh promoter (top) or coding (bottom) regions were analyzed by quantitative ChIP with the indicated antibodies. (B) MPC11 cells were cotransfected with VHluc and pBSHS4 or pBSHS4A5T plasmids. Histone modifications at the transfected VH promoter (top) and luciferase coding (bottom) regions were measured by ChIP and real-time PCR with indicated antibodies. Data are presented as the mean ± SD.
To rule out potential indirect effects of siRNA-mediated knockdowns, ChIP experiments were also carried out using MPC11 cells cotransfected with separate plasmids containing promoter/reporter (Vhluc) and HS4 enhancer (pBSHS4) sequences (Fig. 6B). In the cotransfection assay with the wild type HS4 enhancer, histone H3K4 trimethylation and Mediator (MED6), TFIID (TAF4) and Pol II recruitments at the Igh promoter were readily detected. However, replacement of the wild type HS4 enhancer with the HS4 A5T mutant enhancer resulted in significant reductions in H3K4 trimethylation and recruitments of Mediator, TFIID and Pol II at the transfected promoter. Again, these histone modifications and factor recruitments are promoter-specific, with the signals in the coding region being much weaker and largely unchanged upon transfection with the mutant enhancer. Overall, these results reveal a strong correlation between OCA-B–dependent enhancer-promoter interactions, OCA-B–dependent Igh transcription and OCA-B–dependent H3K4 methylation and factor recruitment to the Igh promoter.
OCA-B is required for proper Igh gene transcription in primary cells
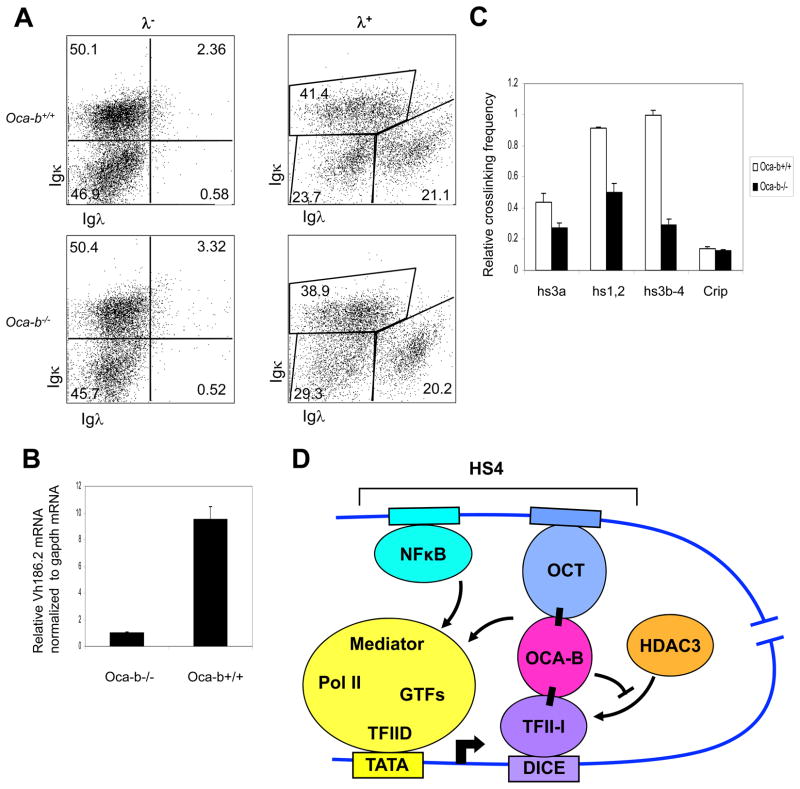
Previously, we and others have established and characterized Oca-b null mice. To determine an OCA-B requirement for Igh gene transcription in primary cells, we utilized an NP immunization protocol to specifically induce expression of NP-reactive VH186.2 family genes (Jacob et al., 1993; McHeyzer-Williams et al., 1993). Single spleen B cells from NP-immunized wild type and Oca-b null mice were analyzed by flow cytometry as shown in Fig. 7A. The λ+ population that contains the NP-positive, VH186.2-expressing B cells was enriched. Importantly, the VH186.2 mRNA level, which was quantified by RT-qPCR and normalized to endogenous GAPDH mRNA, was greatly reduced in OCA-B null cells relative to cells from wild type mice (Fig. 7B).
Figure 7.
OCA-B requirement for endogenous Igh expression in primary B cells. (A) Cellular populations from NP-immunized Oca-b+/+ and Oca-b−/− splenocytes were analyzed by FACS using antibodies against the indicated cell surface markers. (B) VH186.2 mRNA levels in Oca-b+/+ and Oca-b−/− splenocytes were analyzed by quantitative RT-PCR after normalization to Gapdh mRNA. Results are presented as the mean ± SD. (C) Effects of OCA-B on Igh promoter-enhancer interactions in wild type and OCA-B null primary cells were measured by 3C as described in Methods. After HindIII digestion, all Igh genes containing JH2 or JH3 segments after VDJ recombination were analyzed. (D) A proposed model for OCA-B and TFII-I interaction that facilitates Igh promoter and HS4 enhancer communication. First, binding of OCA-B to promoter-bound TFII-I relieves HDAC3 binding and the associated repression. Second, an interaction between enhancer-bound OCA-B and TFII-I facilitates juxtaposition, by looping, between Igh promoter and enhancer elements. Proper enhancer function is achieved by facilitated factor recruitment to the promoter and/or by a direct transfer mechanism.
To extend the earlier analyses of enhancer-promoter interactions in cell lines, the relative Igh promoter and enhancer interaction strengths were compared in primary B cells from wild type and null mice. This analysis used the method described by Ju et al (2007) to monitor Igh genes that contain the JH3 segment after VDJ recombination. The relative Igh promoter and enhancer interactions were decreased in OCA-B null cells compared with wild type cells (Fig. 7C). These results indicate that OCA-B plays an important role in Igh gene expression and Igh promoter-enhancer communications in primary B cells.
DISCUSSION
Direct OCA-B and TFII-I interactions that are important for Igh gene promoter-enhancer communication and Ig gene transcription
In eukaryotic cells, transcriptional activation involves the coordinated recruitment of the basal transcription machinery in concert with changes in chromatin structure, including nucleosome remodeling and local histone posttranslational modifications. In metazoans, enhancers and locus control regions (LCRs) can activate high level transcription of linked genes from tens of kilobases away. Recent technological advances that include 3C (Dekker et al., 2002) and RNA trap (Carter et al., 2002) have provided direct evidence that these distal regulatory elements physically interact with their target genes in vivo (Spilianakis et al., 2005; Tolhuis et al., 2002). Thus, large genomic distances along chromosomes that separate genes and their control elements are overcome by direct spatial clustering, resulting in the formation of chromosome loops. Immunoglobulin gene expression, which is tightly regulated in B cells, is also under the control of multiple cis regulatory elements. Recent data reveal that both Ig heavy and light chain promoters physically interact with corresponding distal enhancer elements in vivo (Ju et al., 2007; Liu and Garrard, 2005). It is believed that such specific interactions between enhancers and their cognate promoters are mediated by defined proteins associated with these cis elements, and long-range chromosome interactions of the entire human genome have been mapped by the Hi-C method (Lieberman-Aiden et al., 2009). However, only in rare cases have the molecular entities that mediate these interactions been established (Dekker et al., 2002; Vakoc et al., 2005).
The results reported here identify the Igh promoter-binding factor TFII-I as an OCA-B–interacting protein. A direct interaction between OCA-B and TFII-I has been confirmed by coimmunoprecipitation of both endogenous and ectopically expressed proteins and by in vitro binding assays with purified factors. In addition, coupled transfection and chromatin conformation capture assays have demonstrated that promoter-bound TFII-I and 3′enhancer-bound OCA-B mediate Igh promoter and 3′enhancer interactions. Related, we have shown that purified OCA-B, Oct-2 and TFII-I are necessary and sufficient to mediate an Igh promoter-enhancer interaction in an in vitro reconstituted system. In further studies with MPC11 mouse plasmacytoma cells, which express the endogenous γ2b heavy chain, OCA-B knockdown and site-directed mutagenesis analyses indicate an important role of OCA-B in mediating Igh promoter-enhancer interactions and Igh expression in vivo. In view of these results, and the demonstrated physical association of OCA-B and TFII-I with Igh regulatory elements shown by ChIP assay, the OCA-B and TFII-I effect on Igh expression in MPC11 cells appears to be direct.
Octamer elements with demonstrated functions in various in vitro and cell-based assays are found in both promoter proximal and distal enhancers region in Ig genes (Luo et al., 1992; Tang and Sharp, 1999; Wirth et al., 1987). Using various complementary analyses, our results have not only confirmed the previously reported Igh promoter and 3′enhancer loop formation in MPC11 cells, but most importantly, have demonstrated an important role for OCA-B and TFII-I in mediating these promoter-enhancer interactions in vivo. Our results also suggest that several of the individual Igh 3′enhancer elements might use similar mechanisms to interact with promoter elements and to form loop structures dependent upon enhancer-bound OCA-B and promoter-bound TFII-I. This is in line with mouse genetic analyses indicating little disruption to overall Igh gene expression upon deletion of individual 3′enhancer sites, since intronic enhancer and other 3′enhancer elements with bound OCA-B should still able to interact with the Igh promoter via promoter-bound TFII-I and provide at least some enhancer functions (Bebin et al., 2010; Vincent-Fabert et al., 2009). On the other hand, since the DICE elements to which TFII-I binds are not found in all Igh promoter regions (Johnston et al., 2006), it will be important to determine whether TFII-I is able to bind to these DICE-less Igh gene promoters through cis elements such as the initiator elements that are more commonly found in Igh promoters (Johnston et al., 2006). Moreover, our current studies do not exclude the possibility that other factors might also contribute to facilitation of Igh promoter-enhancer interactions in vivo. In this regard, a functional interaction of the Igκ 3′ enhancer with a cognate Vκ promoter was shown to be facilitated by tethering of a POU-domain to a promoter proximal octamer site (Bertolino and Singh, 2002).
In addition to our demonstration of a more direct role for enhancer-bound OCA-B in Igh transcription through facilitation of enhancer-promoter interactions, we also have shown that OCA-B binds to the same TFII-I domain that previously was reported to interact with histone deacetylase HDAC3. Consistent with this observation, OCA-B was found to compete with HDAC3 for binding to the TFII-I IR4 domain in an in vitro binding assay and to relieve a possible repression of TFII-I by HDAC3 on an Igh promoter in a transfection assay. These results are indicative of a second and more indirect OCA-B coactivator function at the promoter involving neutralization of a negative regulatory mechanism that operates through a histone deacetylase. In more physiological tests of OCA-B function, we showed that OCA-B is required for proper Igh gene expression in vivo by comparing Oca-b null B cells with wild type cells from mice immunized with NP antigens. It is noteworthy that this immunization protocol specifically induces the expression of Igh genes containing the VH186.2 promoter. This promoter contains three potential DICE elements for TFII-I binding (Johnston et al., 2006). Thus, the severely decreased expression of Igh genes in Oca-b null primary B cells likely reflects the combined effects of losing both the anti-HDAC function of OCA-B at promoter regions and OCA-B–dependent promoter-enhancer interactions.
Igh 3′enhancer function through looping and facilitated recruitment mechanism
Proposed models of enhancer action include tracking, linking and looping (Bulger and Groudine, 1999). Our results support a model in which the Igh 3′enhancer exerts its function through a combined looping and facilitated recruitment mechanism. First, physical interactions between Igh promoter and 3′enhancer elements have been documented with 3C assays, suggesting that looping occurs between Igh promoter and 3′enhancers. Second, in transfection assays, the enhancer provided in trans still strongly activates the Igh promoter, which indicates not only an important role for enhancer-bound OCA-B in mediating promoter-enhancer interactions, but also that the function of the 3′enhancer is not likely through a tracking mechanism. Third, in line with the observation that knockdown of OCA-B or TFII-I results in reduced promoter-enhancer interactions, as well as impaired enhancer function, knockdown of OCA-B also results in reduction in histone modifications associated with activation and reduced factor recruitment. Thus, the histone H4K5 acetylation and H3K4 trimethyl marks, as well as Mediator, TFIID and RNA polymerase II recruitments, are all significantly reduced at the promoter region following OCA-B depletion. From a mechanistic view point, these results suggest that proper enhancer function provided by chromatin looping is required for Igh gene promoter histone modifications, general transcription factor/cofactor recruitment and transcription initiation.
It is noteworthy that the M17056 and VH186.2 Igh genes analyzed in this study both belong to the VHJ558 family, which is the most DH-distal group, and multiple TFII-I binding DICE elements are found in these VH promoters (Johnston et al., 2006; Tantin et al., 2004). Although there is at present no direct evidence that TFII-I occupies all these DICE elements in vivo, our EMSA results indicate that TFII-I indeed binds to all three M17036 DICE elements in vitro (Supplementary Fig. 2). Thus, the concomitant binding of several TFII-I molecules to a single VH promoter could in principle stabilize their binding to target sites through TFII-I protein-protein interactions and/or enhance the physical interactions with multiple 3′enhancers that share conserved octamer binding sites. Further analyses are needed to address these possibilities.
Redundancy of the Igh 3′enhancer elements
Several studies have indicated the importance of the entire group of 3′enhancer elements in achieving proper Igh gene transcription and CSR (Stevens et al., 2000; Vincent-Fabert et al., 2010). In knock-out mice, combined deletions of the whole group of 3′enhancer elements greatly impair CSR and Igh gene transcription (Vincent-Fabert et al., 2010), whereas single deletions of HS3A, HS1,2, HS3B or HS4 show little or no effect on Igh synthesis and CSR (Bebin et al., 2010; Manis et al., 1998; Vincent-Fabert et al., 2009). In a BAC transgenic mouse, deletion of the entire group of 3′enhancer sequences affects germline transcription and CSR (Dunnick et al., 2005). Also in plasmacytoma cells, deletion of more than two 3′enhancer elements correlates with reduction of Igh expression (Shi and Eckhardt, 2001). Finally in transfection assays, and despite the weak activity when assessed individually, the grouped 3′enhancers show significant synergism (Stevens et al., 2000).
As discussed earlier, our current 3C analysis and that of Ju et al (Ju et al., 2007) have revealed direct interactions of an Igh promoter with several Igh 3′enhancer regions that all contain octamer elements. Furthermore, also as discussed earlier, we show that promoter-enhancer interactions are dependent on OCA-B binding at 3′enhancer sites. In addition, interactions between individual 3′enhancers have been previously reported (Ju et al., 2007). Taken together, these results suggest that single deletions in the 3′enhancer may not greatly affect overall 3′enhancer activity because of the possibility that promoter-bound TFII-I can still mediate promoter-enhancer interactions through interactions with OCA-B bound to other (functionally redundant) 3′enhancers. Therefore, in this model, synergism of the 3′enhancer elements and overall enhancer function would be greatly impaired only by deletion of multiple 3′enhancer elements,
In summary, our results show OCA-B functions at multiple levels in Igh transcription. At the Igh promoter, OCA-B competes with HDAC3 for binding to promoter-bound TFII-I, thus exerting a coactivator function by relieving the repression function of HDAC3. Importantly, enhancer-bound OCA-B also serves as a structural protein that juxtaposes (bridges) the Igh promoter and 3′enhancer through chromatin looping and interaction with Igh promoter-bound TFII-I. Subsequently, these promoter-constrained enhancer elements can exert their functions by facilitating recruitment of general transcription (co)factors to the promoter and consequent PIC formation.
Experimental Procedures
Plasmids, Mutagenesis, Recombinant Protein Preparation and Antibodies
cDNAs for OCA-B, HDAC3 and TFII-I have been previously described. For bacterially expressed OCA-B, a full-length cDNA with an S-tag sequence was subcloned into the pET28 vector. For transfection, full-length cDNAs with or without the FLAG-epitope sequence were subcloned into pCIneo vector. Octamer mutants were generated by site-directed mutagenesis (Stragagene). All PCR products and subcloning products were verified by DNA sequencing. Recombinant S-tagged OCA-B was expressed in bacteria and purified on S-protein agarose. Recombinant FLAG-Oct-2 was expressed in Sf9 cells and purified on M2 agarose. Recombinant FLAG-TFII-I was transiently expressed in 293T cells and purified on M2 agarose. Affinity-purified rabbit anti-OCA-B and anti-HDAC3 were purchased from Santa Cruz, and mouse anti-TFII-I was from BD transduction laboratory.
S-tagged OCA-B Pulldown Assay and Mass Spectrometric Protein Identification
50μg of S-tagged OCA-B were immobilized on S-protein agarose (Novagen) and incubated with Namalwa B cell nuclear extract (1 mg protein) in BC250/0.1% NP-40 (20 mN HEPES [pH 7.9], 250 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 20% glycerol, and 0.1% NP-40). After extensive washing with BC250/0.1% NP-40, the bound proteins were eluted with SDS-PAGE loading buffer and resolved by SDS-PAGE. After staining with Gelcode Blue (Thermo Scientific), protein bands were excised and identified by MS/MS analysis (Rockefeller University Proteomics Resource Center).
Protein Interaction Assays
For co-immunoprecipation of ectopically expressed OCA-B and FLAG-tagged TFII-I, 293T cells were transfected with corresponding vectors using FuGENE 6 reagent (Roche Molecular Biochemical) according to the manufacturer’s instruction. Whole-cell extracts in BC300 (300 mM KCl, 1mM EDTA, 20 mM HEPES [pH7.9], 0.5% NP-40, and 20% glycerol) containing protease inhibitor cocktail (Roche) were incubated with M2-agarose beads (Sigma) for 4 hr at 4°C. Bound proteins were eluted with BC100/0.2% NP-40 containing 0.5 mg/ml FLAG peptide and subjected to immunoblot analysis. For co-immunoprecipation of endogenous proteins, Namalwa nuclear extract (500 μg protein) in BC300 was incubated with 5 μg anti-TFII-I overnight at 4°C. After incubation with protein A/G beads for 2 hr, the bound proteins were boiled in SDS-PAGE loading buffer and analyzed by immunoblot. For GST pull-down assays, 2 μg of GST-fused proteins and 200 ng of purified factors were mixed with glutathione-Sepharose 4B beads in 20 mM Tris-Cl (pH 7.5), 150 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.05% NP-40, 0.5 mg/ml BSA, and 0.5 mM phenylmethylsulfornyl fluroride (PMSF). After 4 hr at 4°C, the beads were washed and bound proteins were eluted and analyzed by immunoblot. For the assay in Fig. 2D, 200 ng of GST-fused IR4 were mixed with 200 ng of HDAC3 and increasing amounts of OCA-B.
ChIP and 3C Assays
ChIP assays were carried out as described previously (Kim et al., 2003). 3C assays were performed as described (Hagege et al., 2007). Briefly, 107 cells were subjected to formaldehyde cross-linking and nuclei were prepared. After overnight digestion with Nco I at 37°C, the temperature was raised to 65°C to denature the restriction enzyme. The digested nuclei were diluted in 1x T4 DNA ligase buffer to a concentration of about 3 ng DNA/ul and ligated with T4 DNA ligase for 4 hr at 16°C. After proteinase K digestion and 65°C incubation overnight to reverse the cross-linking, DNA was purified by phenol:chloroform extraction. 100 ng of ligated DNA samples were analyzed by realtime PCR TaqMan probe methods. For in vitro binding and “3C” assay, 1 μg each of proteins and DNA were incubated in 20 mM Hepes (pH 7.9), 0.1 mM EDTA, 100 mM KCl, 10% glycerol, 5 mM MgCl2, 0.05% NP-40 and 1 mM DTT for 30 min at room temperature, followed by formaldehyde cross-linking and 3C assay.
Supplementary Material
1. Supplementary figure S1: TFII-I requires a central OCA-B domain for interaction, and TFII-IR4 fragment interferes with the interaction between OCA-B and TFII-I. This figure is associated with figure 2 and provide information regarding OCA-B domain requirement for TFII-I interaction and function of TFII-IR4 fragment.
2. Supplementary figure S2: In vitro binding of TFII-I to DICE elements in the VH promoter. This figure provides data regarding in vitro binding of TFII-I to DICE elements in IgH promoter and is related to figure 2.
3. Supplementary figure S3: SiRNA-mediated knockdown of OCA-B reduces endogenous IgH promoter-enhancer interaction, but not interactions between hs4 enhancer with other 3′enhancers. This figure provides complementary data to figure 4.
Acknowledgments
We thank Rockefeller proteomics and flow cytometry resource centers for their valuable technical assistance. This work was supported by National Institutes of Health grant CA113872 (to R.G.R.), a Lymphoma and Leukemia Society Special Fellowship (to U.K.) and a Cancer Research Institute Postdoctoral Fellowship (to X.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bebin AG, Carrion C, Marquet M, Cogne N, Lecardeur S, Cogne M, Pinaud E. In vivo redundant function of the 3′ IgH regulatory element HS3b in the mouse. J Immunol. 2010;184:3710–3717. doi: 10.4049/jimmunol.0901978. [DOI] [PubMed] [Google Scholar]
- Bertolino E, Singh H. POU/TBP cooperativity: a mechanism for enhancer action from a distance. Mol Cell. 2002;10:397–407. doi: 10.1016/s1097-2765(02)00597-x. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- Calame K, Sen R. Transcription of immunoglobulin genes. In: Honjo FAT, Neuberger M, editors. Molecular Biology of B cells. New York: Elsevier Science Publishing Co. Inc; 2003. pp. 79–96. [Google Scholar]
- Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- Cepek KL, Chasman DI, Sharp PA. Sequence-specific DNA binding of the B-cell-specific coactivator OCA-B. Genes Dev. 1996;10:2079–2088. doi: 10.1101/gad.10.16.2079. [DOI] [PubMed] [Google Scholar]
- Corcoran LM, Hasbold J, Dietrich W, Hawkins E, Kallies A, Nutt SL, Tarlinton DM, Matthias P, Hodgkin PD. Differential requirement for OBF-1 during antibody-secreting cell differentiation. J Exp Med. 2005;201:1385–1396. doi: 10.1084/jem.20042325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dunnick WA, Shi J, Graves KA, Collins JT. The 3′ end of the heavy chain constant region locus enhances germline transcription and switch recombination of the four gamma genes. J Exp Med. 2005;201:1459–1466. doi: 10.1084/jem.20041988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore GL, Yang JQ, Marcu KB, Birshtein BK. Absence of somatic mutation in the variable region of MPC 11 variants expressing a different heavy chain isotype. J Immunol. 1987;139:619–624. [PubMed] [Google Scholar]
- Gregor PD, Kobrin BJ, Milcarek C, Morrison SL. Sequences 3′ of immunoglobulin heavy chain genes influence their expression. Immunol Rev. 1986;89:31–48. doi: 10.1111/j.1600-065x.1986.tb01471.x. [DOI] [PubMed] [Google Scholar]
- Gstaiger M, Georgiev O, van Leeuwen H, van der Vliet P, Schaffner W. The B cell coactivator Bob1 shows DNA sequence-dependent complex formation with Oct-1/Oct-2 factors, leading to differential promoter activation. EMBO J. 1996;15:2781–2790. [PMC free article] [PubMed] [Google Scholar]
- Gstaiger M, Knoepfel L, Georgiev O, Schaffner W, Hovens CM. A B-cell coactivator of octamer-binding transcription factors. Nature. 1995;373:360–362. doi: 10.1038/373360a0. [DOI] [PubMed] [Google Scholar]
- Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- Hess J, Nielsen PJ, Fischer KD, Bujard H, Wirth T. The B lymphocyte-specific coactivator BOB.1/OBF.1 is required at multiple stages of B-cell development. Mol Cell Biol. 2001;21:1531–1539. doi: 10.1128/MCB.21.5.1531-1539.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J, Przylepa J, Miller C, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. III. The kinetics of V region mutation and selection in germinal center B cells. J Exp Med. 1993;178:1293–1307. doi: 10.1084/jem.178.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Grosschedl R. Complex pattern of immunoglobulin mu gene expression in normal and transgenic mice: nonoverlapping regulatory sequences govern distinct tissue specificities. Genes Dev. 1991;5:932–943. doi: 10.1101/gad.5.6.932. [DOI] [PubMed] [Google Scholar]
- Johnston CM, Wood AL, Bolland DJ, Corcoran AE. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J Immunol. 2006;176:4221–4234. doi: 10.4049/jimmunol.176.7.4221. [DOI] [PubMed] [Google Scholar]
- Ju Z, Volpi SA, Hassan R, Martinez N, Giannini SL, Gold T, Birshtein BK. Evidence for physical interaction between the immunoglobulin heavy chain variable region and the 3′ regulatory region. J Biol Chem. 2007;282:35169–35178. doi: 10.1074/jbc.M705719200. [DOI] [PubMed] [Google Scholar]
- Khamlichi AA, Pinaud E, Decourt C, Chauveau C, Cogne M. The 3′ IgH regulatory region: a complex structure in a search for a function. Adv Immunol. 2000;75:317–345. doi: 10.1016/s0065-2776(00)75008-5. [DOI] [PubMed] [Google Scholar]
- Kim U, Qin XF, Gong S, Stevens S, Luo Y, Nussenzweig M, Roeder RG. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature. 1996;383:542–547. doi: 10.1038/383542a0. [DOI] [PubMed] [Google Scholar]
- Kim U, Siegel R, Ren X, Gunther CS, Gaasterland T, Roeder RG. Identification of transcription coactivator OCA-B-dependent genes involved in antigen-dependent B cell differentiation by cDNA array analyses. Proc Natl Acad Sci U S A. 2003;100:8868–8873. doi: 10.1073/pnas.1033108100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Garrard WT. Long-range interactions between three transcriptional enhancers, active Vkappa gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol Cell Biol. 2005;25:3220–3231. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Fujii H, Gerster T, Roeder RG. A novel B cell-derived coactivator potentiates the activation of immunoglobulin promoters by octamer-binding transcription factors. Cell. 1992;71:231–241. doi: 10.1016/0092-8674(92)90352-d. [DOI] [PubMed] [Google Scholar]
- Luo Y, Roeder RG. Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol Cell Biol. 1995;15:4115–4124. doi: 10.1128/mcb.15.8.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Groudine M. Identification of a locus control region in the immunoglobulin heavy-chain locus that deregulates c-myc expression in plasmacytoma and Burkitt’s lymphoma cells. Genes Dev. 1994;8:2212–2226. doi: 10.1101/gad.8.18.2212. [DOI] [PubMed] [Google Scholar]
- Manis JP, van der Stoep N, Tian M, Ferrini R, Davidson L, Bottaro A, Alt FW. Class switching in B cells lacking 3′ immunoglobulin heavy chain enhancers. J Exp Med. 1998;188:1421–1431. doi: 10.1084/jem.188.8.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthias P. Lymphoid-specific transcription mediated by the conserved octamer site: who is doing what? Semin Immunol. 1998;10:155–163. doi: 10.1006/smim.1998.0117. [DOI] [PubMed] [Google Scholar]
- McHeyzer-Williams MG, McLean MJ, Lalor PA, Nossal GJ. Antigen-driven B cell differentiation in vivo. J Exp Med. 1993;178:295–307. doi: 10.1084/jem.178.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson JS, Giannini SL, Birshtein BK. Identification of 3′ alpha-hs4, a novel Ig heavy chain enhancer element regulated at multiple stages of B cell differentiation. Nucleic Acids Res. 1995;23:975–981. doi: 10.1093/nar/23.6.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PJ, Georgiev O, Lorenz B, Schaffner W. B lymphocytes are impaired in mice lacking the transcriptional co-activator Bob1/OCA-B/OBF1. Eur J Immunol. 1996;26:3214–3218. doi: 10.1002/eji.1830261255. [DOI] [PubMed] [Google Scholar]
- Novina CD, Cheriyath V, Roy AL. Regulation of TFII-I activity by phosphorylation. J Biol Chem. 1998;273:33443–33448. doi: 10.1074/jbc.273.50.33443. [DOI] [PubMed] [Google Scholar]
- Novina CD, Kumar S, Bajpai U, Cheriyath V, Zhang K, Pillai S, Wortis HH, Roy AL. Regulation of nuclear localization and transcriptional activity of TFII-I by Bruton’s tyrosine kinase. Mol Cell Biol. 1999;19:5014–5024. doi: 10.1128/mcb.19.7.5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinaud E, Khamlichi AA, Le Morvan C, Drouet M, Nalesso V, Le Bert M, Cogne M. Localization of the 3′ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity. 2001;15:187–199. doi: 10.1016/s1074-7613(01)00181-9. [DOI] [PubMed] [Google Scholar]
- Rajaiya J, Nixon JC, Ayers N, Desgranges ZP, Roy AL, Webb CF. Induction of immunoglobulin heavy-chain transcription through the transcription factor Bright requires TFII-I. Mol Cell Biol. 2006;26:4758–4768. doi: 10.1128/MCB.02009-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AL. Signal-induced functions of the transcription factor TFII-I. Biochim Biophys Acta. 2007;1769:613–621. doi: 10.1016/j.bbaexp.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AL, Du H, Gregor PD, Novina CD, Martinez E, Roeder RG. Cloning of an inr- and E-box-binding protein, TFII-I, that interacts physically and functionally with USF1. EMBO J. 1997;16:7091–7104. doi: 10.1093/emboj/16.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AL, Meisterernst M, Pognonec P, Roeder RG. Cooperative interaction of an initiator-binding transcription initiation factor and the helix-loop-helix activator USF. Nature. 1991;354:245–248. doi: 10.1038/354245a0. [DOI] [PubMed] [Google Scholar]
- Sacristan C, Tussie-Luna MI, Logan SM, Roy AL. Mechanism of Bruton’s tyrosine kinase-mediated recruitment and regulation of TFII-I. J Biol Chem. 2004;279:7147–7158. doi: 10.1074/jbc.M303724200. [DOI] [PubMed] [Google Scholar]
- Samardzic T, Marinkovic D, Nielsen PJ, Nitschke L, Wirth T. BOB.1/OBF.1 deficiency affects marginal-zone B-cell compartment. Mol Cell Biol. 2002;22:8320–8331. doi: 10.1128/MCB.22.23.8320-8331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubart DB, Rolink A, Kosco-Vilbois MH, Botteri F, Matthias P. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature. 1996;383:538–542. doi: 10.1038/383538a0. [DOI] [PubMed] [Google Scholar]
- Schubart DB, Rolink A, Schubart K, Matthias P. Cutting edge: lack of peripheral B cells and severe agammaglobulinemia in mice simultaneously lacking Bruton’s tyrosine kinase and the B cell-specific transcriptional coactivator OBF-1. J Immunol. 2000;164:18–22. doi: 10.4049/jimmunol.164.1.18. [DOI] [PubMed] [Google Scholar]
- Shen Y, Hendershot LM. Identification of ERdj3 and OBF-1/BOB-1/OCA-B as direct targets of XBP-1 during plasma cell differentiation. J Immunol. 2007;179:2969–2978. doi: 10.4049/jimmunol.179.5.2969. [DOI] [PubMed] [Google Scholar]
- Shi X, Eckhardt LA. Deletional analyses reveal an essential role for the hs3b/hs4 IgH 3′ enhancer pair in an Ig-secreting but not an earlier-stage B cell line. Int Immunol. 2001;13:1003–1012. doi: 10.1093/intimm/13.8.1003. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Staudt LM, Lenardo MJ. Immunoglobulin gene transcription. Annu Rev Immunol. 1991;9:373–398. doi: 10.1146/annurev.iy.09.040191.002105. [DOI] [PubMed] [Google Scholar]
- Stevens S, Ong J, Kim U, Eckhardt LA, Roeder RG. Role of OCA-B in 3′-IgH enhancer function. J Immunol. 2000;164:5306–5312. doi: 10.4049/jimmunol.164.10.5306. [DOI] [PubMed] [Google Scholar]
- Strubin M, Newell JW, Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- Tang H, Sharp PA. Transcriptional regulation of the murine 3′ IgH enhancer by OCT-2. Immunity. 1999;11:517–526. doi: 10.1016/s1074-7613(00)80127-2. [DOI] [PubMed] [Google Scholar]
- Tantin D, Tussie-Luna MI, Roy AL, Sharp PA. Regulation of immunoglobulin promoter activity by TFII-I class transcription factors. J Biol Chem. 2004;279:5460–5469. doi: 10.1074/jbc.M311177200. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Tsukada S, Rawlings DJ, Witte ON. Role of Bruton’s tyrosine kinase in immunodeficiency. Curr Opin Immunol. 1994;6:623–630. doi: 10.1016/0952-7915(94)90151-1. [DOI] [PubMed] [Google Scholar]
- Tussie-Luna MI, Bayarsaihan D, Seto E, Ruddle FH, Roy AL. Physical and functional interactions of histone deacetylase 3 with TFII-I family proteins and PIASxbeta. Proc Natl Acad Sci U S A. 2002;99:12807–12812. doi: 10.1073/pnas.192464499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Vincent-Fabert C, Fiancette R, Pinaud E, Truffinet V, Cogne N, Cogne M, Denizot Y. Genomic deletion of the whole IgH 3′ regulatory region (hs3a, hs1,2, hs3b, hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood. 2010 doi: 10.1182/blood-2010-01-264689. [DOI] [PubMed] [Google Scholar]
- Vincent-Fabert C, Truffinet V, Fiancette R, Cogne N, Cogne M, Denizot Y. Ig synthesis and class switching do not require the presence of the hs4 enhancer in the 3′ IgH regulatory region. J Immunol. 2009;182:6926–6932. doi: 10.4049/jimmunol.0900214. [DOI] [PubMed] [Google Scholar]
- Wen YD, Cress WD, Roy AL, Seto E. Histone deacetylase 3 binds to and regulates the multifunctional transcription factor TFII-I. J Biol Chem. 2003;278:1841–1847. doi: 10.1074/jbc.M206528200. [DOI] [PubMed] [Google Scholar]
- Wirth T, Staudt L, Baltimore D. An octamer oligonucleotide upstream of a TATA motif is sufficient for lymphoid-specific promoter activity. Nature. 1987;329:174–178. doi: 10.1038/329174a0. [DOI] [PubMed] [Google Scholar]
- Yang W, Desiderio S. BAP-135, a target for Bruton’s tyrosine kinase in response to B cell receptor engagement. Proc Natl Acad Sci U S A. 1997;94:604–609. doi: 10.1073/pnas.94.2.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1. Supplementary figure S1: TFII-I requires a central OCA-B domain for interaction, and TFII-IR4 fragment interferes with the interaction between OCA-B and TFII-I. This figure is associated with figure 2 and provide information regarding OCA-B domain requirement for TFII-I interaction and function of TFII-IR4 fragment.
2. Supplementary figure S2: In vitro binding of TFII-I to DICE elements in the VH promoter. This figure provides data regarding in vitro binding of TFII-I to DICE elements in IgH promoter and is related to figure 2.
3. Supplementary figure S3: SiRNA-mediated knockdown of OCA-B reduces endogenous IgH promoter-enhancer interaction, but not interactions between hs4 enhancer with other 3′enhancers. This figure provides complementary data to figure 4.