Abstract
We report, based on semi-empirical calculations, that Zn2+ binds duplex DNA containing consecutive FdU–dA base pairs in the major groove with distorted trigonal bipyramidal geometry. In this previously uncharacterized binding motif, O4 and F5 on consecutive FdU are axial ligands while three water molecules complete the coordination sphere. NMR spectroscopy confirmed Zn2+ complexation occurred with maintenance of base pairing while a slight hypsochromic shift in circular dichroism (CD) spectra indicated moderate structural distortion relative to B-form DNA. Zn2+ complexation inhibited ethidium bromide (EtBr) intercalation and stabilized FdU-substituted duplex DNA (ΔTm > 15°C). Mg2+ neither inhibited EtBr complexation nor had as strong of a stabilizing effect. DNA sequences that did not contain consecutive FdU were not stabilized by Zn2+. A lipofectamine preparation of the Zn2+–DNA complex displayed enhanced cytotoxicity toward prostate cancer cells relative to the individual components prepared as lipofectamine complexes indicating the potential utility of Zn2+–DNA complexes for cancer treatment.
INTRODUCTION
Zinc has a unique role among metals in regulating the normal function of the prostate gland. Further, the sensitivity of prostatic adenocarcinoma (PCa) cells to Zn2+, particularly in combination with chemotherapy, represents a potentially highly selective approach to prostate cancer treatment (1,2). Zn2+ forms high-affinity complexes with biopolymers, most notably the Zn2+-finger motif that is widely used by transcription factors for sequence-specific DNA recognition. These peptidic motifs frequently have subnanomolar affinity for Zn2+ and presumably persist in cellular environments in which the free Zn2+ concentration is extremely low (3). Zn2+ has also been reported to form specific complexes with DNA with some reports indicating that Zn2+ can mediate base pair formation, particularly for nucleobases with acidic imino hydrogens such as 5-fluorouracil (5FU) (4,5). In principle, Zn2+ complexed to 5-fluoro-2′-deoxyuridine (FdU)-substituted DNA could be useful for cancer treatment in light of the cytotoxic properties of FdUMP[10] (6–8).
The structural motif responsible for Zn2+ complexation by FdU-substituted DNA is not presently known. Previous reports indicated that Zn2+ interacted with N3 of deprotonated FdU forming an ionic base pair with dA similar in geometry to Watson–Crick (4,5). These studies were performed using DNA sequences with alternating FdU and 2′-deoxyadenosine (dA) nucleotides. Subsequent studies using FRET revealed DNA aggregation occurred under similar conditions (9). In the present report, we demonstrate that Zn2+, but not Mg2+, selectively stabilizes DNA containing consecutive FdU–dA base pairs. The Zn2+ complexes were characterized by enhanced thermal stability and exclusion of ethidium bromide (EtBr). Zn2+-mediated stabilization occurs only for DNA sequences containing consecutive FdU–dA base pairs and is not observed for sequences that do not contain FdU or that contain alternating FdU–dA nucleotides.
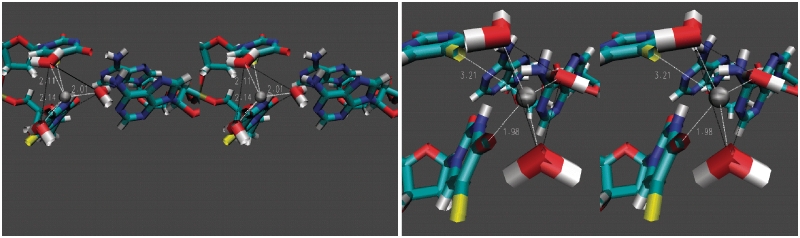
In the present studies, semi-empirical and ab initio calculations were used to determine the energetically preferred geometry for Zn2+ binding to DNA sequences containing consecutive FdU nucleotides (Figure 1). Although crystallographic and NMR studies have indicated a propensity for monovalent cations to occupy the deep, narrow minor groove of A-tract DNA sequences (10,11), the calculations revealed a distinct minimum energy conformation for the FdU–dA dimer complexed with Zn2+ under conditions when the 5′-terminal FdU was deprotonated. In this low energy structure, Zn2+ is localized in the major groove and interacts with F5 from the 5′-terminal FdU and with O4 from the 3′-terminal FdU as axial ligands in a distorted trigonal bipyramidal geometry with three water molecules occupying the equatorial sites. This previously uncharacterized structural motif is consistent with all of the spectroscopic, as well as the computational data.
Figure 1.
Computationally derived model of Zn2+ complexation to the hemi-deprotonated dimeric duplex consisting of consecutive FdU–dA base pairs. Zn2+ is located in the major groove with a distorted trigonal bipyramidal geometry in which the fluorine from the deprotonated FdU is one axial ligand and O4 from the second FdU is the second axial ligand. Water molecules are the equatorial ligands. White dashed lines are used to illustrate the coordinating atoms. (A) View emphasizing the Zn2+–water interactions; (B) View emphasizing Zn2+ interactions with DNA nucleobases. The triangle connecting the three water oxygens provides a visual frame of reference.
We also evaluated the cytotoxicity of the Zn2+–DNA complex to determine to what extent the inclusion of Zn2+ in FdU-substituted DNA complexes may be useful for prostate cancer treatment. The Zn2+–DNA complex was prepared using lipofectamine both to stabilize the complex under cell culture conditions and to improve cellular uptake. The results clearly demonstrate that the DNA–Zn2+ complex has greater cytotoxicity toward PCa cells than either DNA alone or Zn2+ alone. Thus, our results demonstrate that Zn2+ interacts with DNA containing consecutive FdU–dA base pairs by binding in the major groove in a previously uncharacterized structural motif. Further, complexation of Zn2+ enhances the cytotoxicity of FdU-substituted DNA toward PCa cells demonstrating the feasibility for using this structural motif for PCa treatment.
MATERIALS AND METHODS
Design and synthesis of DNA hairpins
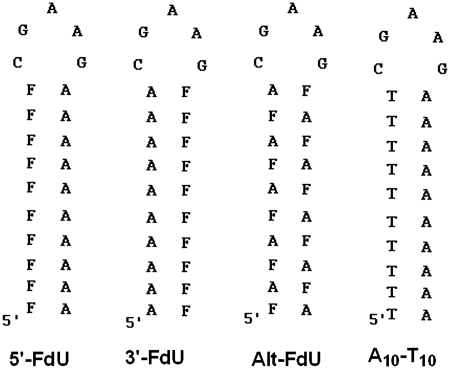
The DNA sequences used for these studies were designed to favor intramolecular hairpin formation and to be stable at physiological temperature. Each of the four hairpin DNA sequences consisted of a 10 bp stem consisting of all A–T (or A–FdU) base pairs. The loop region included the GAA sequence that was previously shown to promote hairpin formation (12). The loop was closed by a single C–G base pair. Gel-electrophoresis studies (15% native PAGE gel) demonstrated a single species was present (Supplementary Figure S8) and subsequent 1H NMR studies demonstrated the chemical shift characteristic for the C–G base pair closing the loop in a similar sequence (13). Four DNA hairpins were designed (Figure 2). The sequence ‘A10–T10’ included 10 consecutive T nucleotides at the 5′-terminus and the corresponding 10 consecutive A nucleotides at the 3′-terminus. The ‘5′-FdU’ sequence was identical to the A10–T10 sequence except all of the T nucleotides were substituted by FdU. The ‘3′-FdU’ hairpin DNA sequence had the location of the 10 consecutive A and FdU nucleotides reversed relative to the 5′-FdU hairpin with the 10 consecutive FdU nucleotides located at the 3′-terminus. The ‘Alt-FdU’ sequence had alternating A and FdU nucleotides throughout the stem and thus lacked consecutive FdU nucleotides. These DNA hairpin sequences were designed to evaluate the structural requirements for FdU substitution on formation of complexes with divalent metal ions, including Zn2+ and Mg2+. All DNA sequences were prepared at the University of Calgary DNA Core laboratory and purified by gel-filtration chromatography.
Figure 2.
Structure of the DNA hairpins investigated in the present studies. Each of the hairpins includes a 10 bp stem capped by the pentanucleotide CGAAG to promote intramolecular hairpin formation. The control sequence A10–T10 consists of all native nucleotides. The three FdU-containing sequences differ in the location of FdU nucleotides as shown.
Formation of Zn2+–DNA complexes
Intramolecular hairpins were formed using 1 mM DNA in dH2O by heating to 80°C for 5 min followed by rapid cooling. Zn2+ (or Mg2+) complexes were formed by incubating 100 µM DNA with 0–5 mM ZnCl2 (or MgCl2) in M-DNA buffer (20 mM sodium cacodylate, pH 8.0, 10 mM NaCl) for 1 h at room temperature. The formation of specific Zn2+ complexes with hairpin DNA sequences was evaluated using an EtBr exclusion fluorescence assay as previously described (5). This assay was performed by combining 20 μl of the above described M-DNA complex with 80 µl of EtBr solution (10 mM NaCl, 20 mM sodium cacodylate, pH 8.0, +25 µM EtBr) followed by incubation for 30 min at room temperature in a 96-well clear bottom, black plate (Costar). The final divalent metal concentration ranged from 0 to 1 mM. EtBr fluorescence was detected using a Typhoon-9210 variable mode imager with Green laser (532 nm) excitation and 610 nm emission filter (with 500 PMT set). Fluorescence intensity was quantified with the software ImageQuant 5.2 and graphed versus Zn2+/Mg2+ concentration.
Analytical characterization of DNA–Zn2+ complexes
DNA–Zn2+ complexes were analyzed by circular dichroism (CD) and NMR spectroscopy to characterize their structural features and any specific changes induced by Zn2+ or Mg2+. A 300 µl aliquot of 20 µM DNA solution with either 0, 0.5 mM, 1 mM Zn2+ (or Mg2+) was prepared in M-DNA buffer as described above and used to obtain the CD spectrum. An Aviv CD spectrometer was used to scan the region from 210 to 310 nm at a rate of 0.5 nm/s. Temperature was controlled at 20°C.
1H, 19F, 31P and 13C NMR spectroscopy were used to investigate specific structural perturbations induced by Zn2+ to the 3′-FdU hairpin. For the NMR analysis, 500 μl of 0.5 mM 3′-FdU in M-DNA buffer modified to contain 40 mM sodium cacodylate were used. NMR samples contained either no metal, 5 mM Zn2+ or 5 mM Mg2+. Also, 10% D2O was included to maintain lock. The final pH of the samples was adjusted to either pH 7 or 8 (±0.1 pH unit). 1H NMR spectra were collected using a Bruker Avance DRX 600 MHz spectrometer with a TXI Cryoprobe at 10°C using the WATERGATE sequence with a 3-9-19 refocusing pulse. Nulls were set at ±18.1 ppm. Spectra were acquired with a spectral width of 33.3 ppm, 8192 total data points and 256 scans. A relaxation delay of 10 s was used to obtain 1H spectra for integration. 13C spectra were collected at 15°C using inverse-gated proton decoupling during acquisition, a 2 s relaxation delay, a spectral width of 200.0 ppm, 8192 total data points and 65,536 scans (30° pulse). 19F and 31P NMR spectra were acquired using a Bruker 300 MHz NMR instrument with 85% H3PO4 (δ = 0 ppm) or 5 mM aqueous solution of KF (δ = 125.3 ppm) as concentric internal standard capillaries (Wilmad).
Temperature-dependant melting analysis
Temperature-dependent melting (Tm) of all four DNA hairpins complexed or not complexed with Zn2+/Mg2+ was determined using UV absorption spectroscopy at 260 nm. Four hundred microliters of a 2.5 µM solution of the annealed hairpins in M-DNA buffer in the presence or absence of 0.5 mM Zn2+ (or Mg2+) were analyzed using a Beckman Coulter DU-800 UV–Vis spectrophotometer in the temperature region 20–85°C. Temperature was increased at a rate of 0.5°C/min. Melting temperatures were determined from the first derivative of the hyperchromicity profiles except for the 3′-FdU and 5′-FdU samples in the presence of Zn2+ for which an inflection point was not observed, consistent with strand slippage during melting, and the Tm calculated from the percent increase in hyperchromicity (Supplementary Figure S4).
Cytotoxicity of FdU hairpins and Zn2+ complexes
The cytotoxicity of the FdU hairpins and Zn2+ complexes was determined using a modified clonogenic assay with an MTT read-out. PC3 prostate cancer cells were seeded in 24-well plates at a cell density of 250 cells/well with 500 μl complete media (Roswell Park Memorial Institute (RPMI) + 10% fetal bovine serum (FBS) + 1% Penicillin & Streptomycin). After 24 h, media was removed and cells were washed twice with 500 µl phosphate-buffered saline (PBS) followed by addition of 500 μl serum-free RPMI. The 3′-FdU–Zn2+ complex was formed by incubating 100 μM 3′-FdU hairpin with 5 mM Zn2+ in M-DNA buffer at room temperature for 1 h. Lipofectamine 2000 (Invitrogen) was diluted in 100 μl Opti-MEM media (pH = 8.0) to 7.5 ng/μl and incubated at room temperature for 5 min. The 3′-FdU–Zn2+ complex was then combined with diluted lipofectamine and incubated at room temperature for 20 min followed by the addition to each well. After 12 h, media was replaced with fresh complete media and incubated at 37°C for a total of 14 days with media changes every 2 days. Cell viability was then assessed using an MTS assay. All experiments were done in triplicate. Each set of data (net absorbance) were then expressed as a percentage, considering the no treatment (i.e., lipofectamine only) group as 100%.
Computational modeling
The initial models for the Zn2+ complexes were derived from coordinates for two consecutive dT–dA base pairs extracted from the NMR solution structure (pdb 1JVE) (13). Conversion to FdU was performed by deleting the thymidine methyl group and replacement with fluorine. Initially, eight complexes were geometry optimized in the absence of Zn2+ and water molecules: (i) fully protonated: both thymidine (or FdU) base nitrogens protonated; (ii) hemi-deprotonated: one protonated thymidine (or FdU) base nitrogen and one unprotonated thymidine (or FdU) base nitrogen; (iii) fully deprotonated: two unprotonated thymidine (or FdU) base nitrogens. There are two non-identical hemi-deprotonated complexes: (i) 5′-dT-(or 5′-FdU-) deprotonated or (ii) 3′-dT-(or 3′-FdU-) deprotonated). Geometry optimizations were performed with the addition of Zn2+ and water molecules to each complex. For all calculations, the NMR structure was used as the starting configuration and two Mg2+ counterions were added in the vicinity of the backbone phosphorus. Zn2+ was added (except in the control computation) between the fluorine atoms of the two FdU. Additional calculations confirmed the structures obtained were independent of initial Zn2+ placement. All modifications were performed using Molden. All structures were geometry optimized with Gaussian 03 using PM3 in Cartesian coordinates. Post-optimization, single-point calculations were performed with a minimal basis set (STO-3g*) to obtain energies and for a Natural Bond Orbital analysis (14) from which the Wieburg bond orders were extracted. Stabilization energies were calculated by subtracting the energy of dA–dT (or FdU) in the appropriate protonation state (doubly, singly or unprotonated) and the energy of Zn2+ with four water molecules from the energy of the full complex.
RESULTS
Formation and stability of DNA–Zn2+ complexes
The propensity of Zn2+ or Mg2+ to increase the thermal stability of four DNA hairpin sequences was evaluated by UV hyperchromicity measurements while the propensity of these metal ions to form specific complexes with these DNA hairpin sequences was evaluated using an EtBr exclusion assay. The results of the thermal stability studies are summarized in Table 1 and representative thermal melting graphs are displayed in supplementary data (Supplementary Figure S4). The control (no-FdU) sequence consisting only of native nucleotides (Figure 2) had a melting temperature of 54.3°C in the absence of divalent metal ions. The native sequence was markedly stabilized by Mg2+ (Tm ≥ 62.2°C); however, Zn2+ addition resulted in non-equilibrium melting conditions for the native sequence with an unexpectedly low apparent Tm (37.6°C). Mg2+ increased the thermal stability of all sequences except Alt–FdU. The extent of stabilization for 3′-FdU or 5′-FdU by Mg2+ was, however, not as great as for the native sequence (Table 1). In contrast to the apparent destabilizing effects of Zn2+ on the native sequence, Zn2+ markedly stabilized both hairpin sequences that contained consecutive FdU nucleotides (3′-FdU and 5′-FdU) (Figure 2). The stabilization of the 3′-FdU and 5′-FdU sequences by Zn2+ was slightly greater than the extent of stabilization of the native sequence by Mg2+. Interestingly, a melting temperature for the Alt–FdU sequence could not be obtained in the presence of Zn2+ as addition of Zn2+-induced precipitation of the Alt–FdU sample during the thermal melting experiments, likely as a consequence of interstrand complex formation as reported in previous FRET studies (9). Zn2+ also qualitatively altered the shape for the thermal melting curves for all four samples, and for 5′-FdU and 3′-FdU the Tm was estimated from the percent change in hyperchromicity rather than from the first derivative of the melting profile (Supplementary Figure S4). The thermal melting results are consistent with selective stabilization of DNA sequences containing consecutive FdU nucleotides by Zn2+.
Table 1.
Melting temperatures for DNA hairpins as a function of metal ion
| Zn2+ | Mg2+ | No metal | |
|---|---|---|---|
| 3′FdU | 66.1 | 47.4 | 44.5 |
| 5′FdU | 60.5 | 45.0 | 39.5 |
| Alt FdU | – | 44.8 | 46.4 |
| Control | 37.6a | 62.2 | 54.3 |
aValue reported is an apparent Tm as addition of Zn2+ resulted in non-equilibrium conditions.
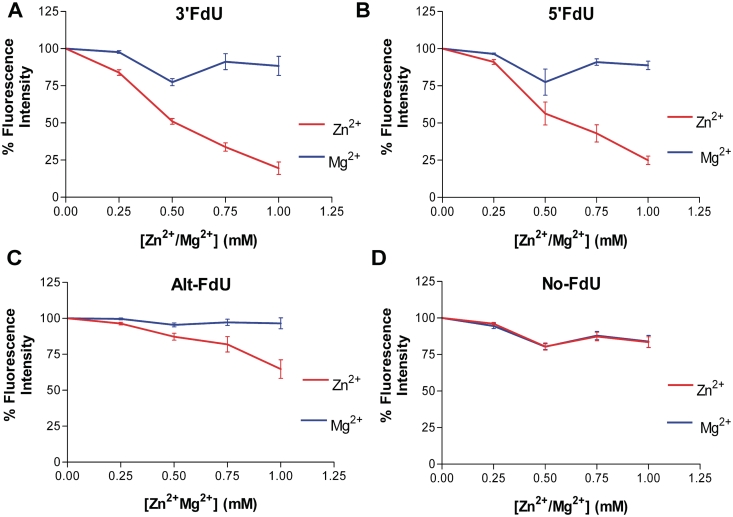
The formation of specific complexes for the four DNA hairpin sequences with Zn2+ and with Mg2+ was evaluated by measuring the quenching of EtBr fluorescence. The results are summarized in Figure 3. Addition of Mg2+ resulted in a slight, concentration-dependent decrease in EtBr fluorescence for all DNA hairpins except Alt–FdU (∼85% of control). In contrast, the effects of Zn2+ were highly sequence dependent. The effects of Zn2+ on the control sequence were similar to the effects of Mg2+ with a moderate concentration-dependent decrease in EtBr fluorescence. The effect of Zn2+ on the ‘Alt–FdU’ sequence was also moderate. In contrast, Zn2+ had a dramatic effect on EtBr fluorescence for the 3′-FdU and 5′-FdU sequences. Addition of 1 mM Zn2+ resulted in attenuation of EtBr fluorescence to <25% of control for these two sequences (Figure 3). The results are consistent with Zn2+ forming specific complexes with DNA sequences containing consecutive FdU nucleotides. Similar complexes do not form with Mg2+ or with sequences containing non-consecutive FdU nucleotides.
Figure 3.
Zn2+, but not Mg2+, inhibits EtBr intercalation into duplex DNA containing consecutive FdU–dA base pairs. EtBr exclusion data are shown for each of the four DNA hairpins in the presence of the indicated concentrations of Zn2+ (red) or Mg2+ (blue). (A) 3′FdU hairpin; (B) 5′ FdU hairpin; (C) Alt-FdU hairpin; (D) No-FdU sequence.
CD of DNA–Zn2+ complexes
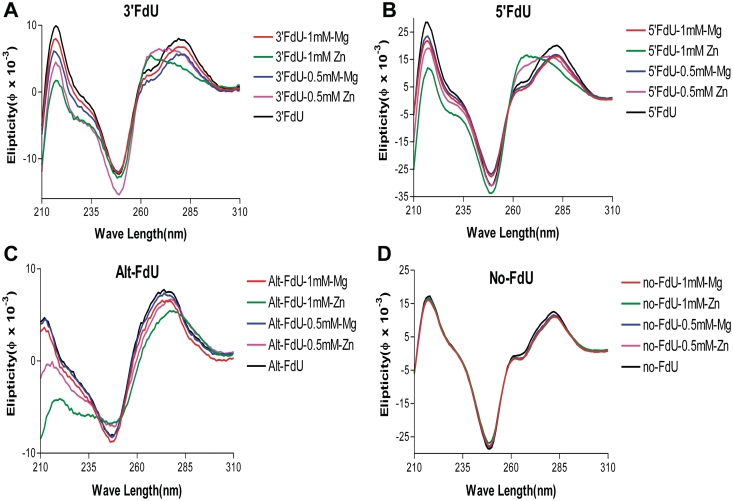
CD was used to evaluate alterations to DNA structure caused by Mg2+ or Zn2+ binding. All four DNA hairpins displayed CD spectral features characteristic of B-form DNA (Figure 4). The addition of Zn2+, but not Mg2+, resulted in a concentration-dependent blue-shift of ∼20 nm for the maximal positive ellipticity near 280 nm selectively for the DNA sequences containing consecutive FdU nucleotides. The CD spectra for the control sequence consisting of only native nucleotides displayed no significant variability upon addition of either Mg2+ or Zn2+ (Figure 4). The maximal ellipticity at 280 nm depends on base pairing, base stacking and base tilting, and shifts in this maximum for the FdU-containing sequences are consistent with alterations in local base pair geometry upon Zn2+ complex formation.
Figure 4.
CD spectra demonstrate Zn2+ selectively induces a hypsochromic shift upon addition to DNA hairpins containing consecutive FdU–dA base pairs. Spectra for each hairpin were acquired in the presence and absence of Zn2+ and Mg2+ at the indicated concentrations. The CD spectra indicate that complexation with Zn2+ alters the structure of DNA hairpins that contain consecutive FdU–dA base pairs. Computational studies indicate base slide and tilt accompany Zn2+ complexation (Figure 1B). (A) 3′FdU hairpin; (B) 5′ FdU hairpin; (C) Alt-FdU hairpin; (D) No-FdU sequence.
Molecular modeling Zn2+–DNA complexes
The preferred geometry for Zn2+ binding DNA sequences containing consecutive FdU nucleotides was calculated using Gaussian 03. Specifically, the coordinates for two consecutive dT–dA base pairs were extracted from the NMR solution structure for a DNA hairpin similar to those used for the present studies (pdb 1JVE) (13). Thymidine methyl groups were replaced with fluorine. The geometry and charge distribution for the 2 bp duplex was optimized under three conditions: (i) ‘high pH state’ in which both FdU were ionized (−1 net charge on each nucleobase); (ii) ‘biological pH state’ in which one FdU was ionized (pKA of FdU ∼7.6) and (iii) a ‘low pH state’ in which neither FdU was ionized. Zn2+ was added to the system and the overall geometry and charge distribution were optimized for minimal energy as described in the methods. The structure of the dinucleotide step under biological pH conditions (i.e. hemi-deprotonated) with the addition of water molecules to coordinate Zn2+ is shown in Figure 1. Surprisingly, Zn2+ is located in the major groove with simultaneous interactions with the O4 carbonyl oxygen of the 3′-terminal FdU and F5 of the 5′-terminal FdU, which is deprotonated in the low energy structure. The Zn2+–F distance is 3.2 Å while the Zn2+–O4 distance is 1.9 Å. The Zn2+–O4 interaction has a bond order of 0.277 and thus is a partial covalent bond (Supplementary Table S3). Zn2+ significantly stabilized the biological pH structure with stabilization energy of 3.84 kcal/mol. Zn2+ coordinates three water molecules with distances of 2.10, 2.12, and 2.14 Å. The overall coordination geometry for Zn2+ is distorted trigonal bipyramidal. A similar geometry is observed in the high pH [fully deprotonated state (Supplementary Figure S1)]. In the ‘low pH state’, Zn2+, unsurprisingly, does not coordinate with FdU stably, but remains coordinated with the three water molecules. The computational model predicts longer than expected hydrogen bonds for the Watson–Crick base pair maintained in the hemi-deprotonated complex (Supplementary Table S1), likely as a result of an absence of base stacking in the dinucleotide step modeled relative to the complete hairpin for which experimental NMR data indicate maintenance of normal Watson–Crick base pairing.
NMR spectroscopy
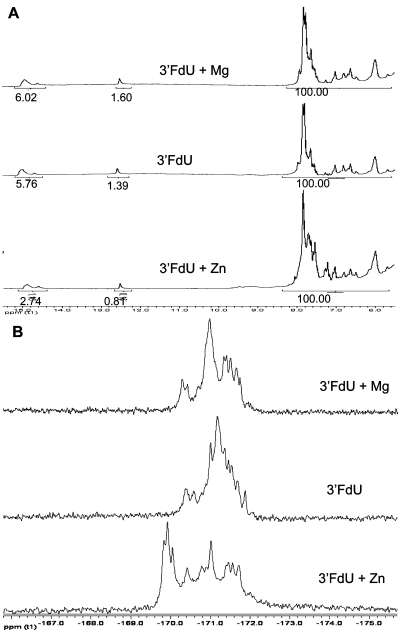
The effects of Zn2+ complex formation on the structure of the 3′-FdU sequence were analyzed using NMR spectroscopy. 1H NMR spectra were obtained in 90% H2O solution. Imino 1H signals were readily apparent for the 3′-FdU hairpin in the presence of Zn2+, Mg2+ and no divalent metal at both pH 7 (Supplementary Figure S5) and pH 8 (Figure 5). Interestingly, although the pKA for monomeric FdU is ∼7.6, there was little change in relative intensity for the FdU imino 1H between pH 7 and pH 8 either in the no metal sample or in the sample that contained Mg2+. Integration of the FdU imino region was performed relative to a region of carbon bound 1H (5.8–8.5 ppm) that was indistinguishable in all samples and arbitrarily set to 100. The well-resolved G-imino at 12.5 ppm varied from 0.81 to 1.60 in all samples relative to this standard while integral values ranged from 5.7 to 6.0 for the 10 FdU imino 1H in this region. End-fraying causes the almost complete loss of the terminal FdU imino 1H and chemical exchange with water is likely responsible, in part, for the integral values being significantly less than the theoretical value of nine. Thus, for the no metal and Mg2+ samples, the FdU imino are mostly protonated and do not titrate significantly between pH 7 and 8.
Figure 5.
(A) 1H NMR spectra showing no significant intensity change for the imino region for the 3′-FdU hairpin in the presence of Zn2+ or Mg2+; (B) 19F NMR spectra showing that Zn2+, but not Mg2+, results in significant downfield shifts.
In contrast to the no metal and Mg2+ samples, the integral values for the FdU imino 1H region for the Zn2+ complex display a marked pH dependence. The integral value of 5.87 at pH 7 for the Zn2+ sample is similar to that observed for the no metal and Mg2+ samples and likely indicates a mostly protonated state for the FdU imino 1H. EtBr is not excluded from the 3′-FdU in the presence of Zn2+ at pH 7. At pH 8, the integral value for the FdU imino 1H region decreases to 2.74, about one-half of the value observed at pH 7. Under these conditions, the 3′-FdU forms a specific complex with Zn2+ that excludes EtBr. Thus, Zn2+ perturbs the equilibrium to favor a partly deprotonated state (e.g. hemi-deprotonated) and forms a specific complex with the 3′-FdU hairpin. This complex formation occurs with no discernible changes in chemical shift, only in intensity, for the FdU imino 1H, indicating the overall structure of the DNA does not undergo major conformational change consistent with the CD data.
In contrast to the relatively minor changes in chemical shift observed for imino 1H resonances in the presence of Zn2+, 19F NMR spectra displayed significant downfield shifts in the presence of Zn2+ at pH 8 (Supplementary Figure 5B) but not pH 7 (Supplementary Figure S5). The entire 19F region was downfield shifted by 0.2–0.5 ppm and increased spectral dispersion was also evident in the presence of Zn2+. These results are consistent with the computationally derived structure in which one fluorine is an axial ligand for Zn2+. The 19F resonances were not shifted by Mg2+ (Supplementary Figure 5B). Since the calculations also indicated Zn2+ interacted with an O4 carbonyl oxygen, the one dimensional 13C spectrum was also acquired. Detectable downfield chemical shift changes were observed for resonances having chemical shifts consistent with the C4 carbonyl position (∼160 ppm) (Supplementary Figure S6). No other significant changes were evident in the 1D 13C NMR spectrum. The 19F and 13C NMR results are consistent with the structural calculations indicating Zn2+ interacts with O4 and F5 of consecutive FdU nucleotides. In contrast to the 19F and 13C spectra in which Mg2+ had no significant effect, Mg2+, but not Zn2+, resulted in marked sharpening of the 31P resonances and some increase in spectral dispersion (Supplementary Figure S7).
Cytotoxicity of DNA–Zn2+ to PCa cells
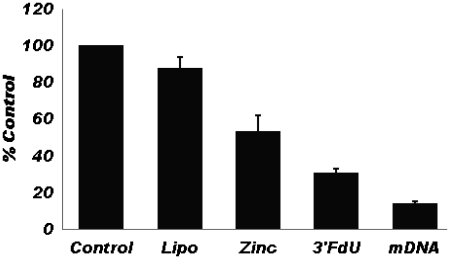
Although normal prostate tissue has the highest Zn2+ concentration in the human body, prostate cancer (PCa) cells have extremely low intracellular Zn2+ and Zn2+ is highly cytotoxic to PCa cells (15). Thus, Zn2+ complexes of FdU-containing DNA sequences may be cytotoxic to PCa cells both from the intracellular release of Zn2+ and from the release of the cytotoxic nucleotide analog FdUMP (6,7). The 3′-FdU DNA hairpin was prepared with and without Zn2+ using lipofectamine to promote cellular internalization. Cytotoxicity was evaluated using a modified clonogenic assay with an MTS readout. The results are shown in Figure 6. As is evident from the data, the 3′-FdU hairpin sequence is cytotoxic toward PCa cells. The lipofectamine complex containing Zn2+, but did not include the FdU-substituted DNA, was also cytotoxic toward PCa cells. The cytotoxicity of the Zn2+–DNA complex prepared with lipofectamine displayed the greatest cytotoxicity toward PC3 cells (Figure 6). All samples were prepared under identical conditions. We conclude that inclusion of Zn2+ in lipofectamine complexes of FdU-containing DNA sequences enhances the cytotoxicity of FdU-substituted DNA toward prostate cancer cells.
Figure 6.
MTS assay results demonstrating the cytotoxicity of Zn2+, the 3′-FdU DNA hairpin and the Zn2+ complex of the 3′-FdU hairpin, towards PC3 prostate cancer cells. The Zn2+–DNA complex (mDNA) displays the greatest cytotoxicity.
DISCUSSION
The present work demonstrates that Zn2+ forms specific complexes with duplex DNA sequences containing consecutive FdU nucleotides. The evidence for complex formation includes significant increase in thermal stability and exclusion of EtBr intercalation. The effects of Zn2+ are highly specific for DNA duplexes containing consecutive FdU nucleotides and do not occur for analogous sequences that contain similar numbers of FdU in non-sequential positions nor occur for sequences consisting of native nucleotides. While Mg2+ was found to stabilize all of the DNA hairpins included in the present study except Alt-FdU, stabilization by Mg2+ was greatest for the sequence consisting of only native nucleotides. Further, Mg2+ was not as efficient as Zn2+ at stabilizing DNA sequences that included consecutive FdU nucleotides. Mg2+ was also not as effective as Zn2+ at excluding EtBr binding to DNA sequences containing consecutive FdU nucleotides. Thus, Zn2+ interacts with DNA sequences containing consecutive FdU nucleotides in a manner that is distinct from Mg2+.
Although previous reports have indicated that Zn2+ causes intermolecular aggregates from DNA containing alternating FdU–dA at basic pH, the present work is the first to demonstrate that intramolecular complex formation with Zn2+ occurs for DNA hairpins containing consecutive FdU nucleotides. As complex formation was not observed with the corresponding dT hairpin DNA sequences and is pH dependent, it is likely that Zn2+ preferentially interacts, in part, with ionized FdU nucleotides. Specific interaction of Zn2+ with the O2 carbonyl would place Zn2+ in the minor groove, a location that has been demonstrated to bind Na+ ions in other A–T rich DNA sequence (10). The basis for the requirement of Zn2+ binding to DNA sequences that include consecutive FdU nucleotides was elucidated from ab initio and semi-empiricial Gaussian calculations that demonstrated, for the first time, that Zn2+ coordinates with F5 of the 5′-FdU nucleotide and the O4 carbonyl oxygen of the 3′-neighboring FdU. Activation of F5 for Zn2+ complexation requires FdU deprotonation, while the O4–Zn2+ interaction occurs with minimal disruption to Watson–Crick geometry. The overall geometry of Zn2+ in complex with consecutive FdU nucleotides is trigonal bipyramidal and the ligands not donated by FdU (e.g. F5 and O4) are occupied by water molecules. This mode of Zn2+ binding with nucleic acids has not been described previously and is distinct from the binding mode postulated previously in which Zn2+ replaced hydrogen in A–FdU base pairs (4,5).
The unique binding mode of Zn2+ identified in the present studies is supported by all of the spectral data. First, the binding motif is consistent with the EtBr exclusion data as interactions of positively charged Zn2+ with consecutive nucleotides would be expected to exclude positively charged EtBr from intercalating into the duplex. The computationally preferred Zn2+ binding motif is also consistent with all of the 1H, 19F, 31P and 13C NMR data as well as the CD data. In contrast to previous studies that indicated imino 1H were not evident in Zn2+ complexes of FdU-containing DNA sequences (4,5), the present study demonstrated Zn2+ complex formation occurred under conditions when about one-half the FdU imino 1H were present (e.g., hemi-deprotonated). Thus, under the solution conditions employed in these studies, the FdU nucleotides are not uniformly ionized, Zn2+ does not mediate base pair formation and A-FdU base pairing is present although reduced in intensity. The 19F spectra, however, show a downfield shift upon Zn2+ complex formation. No changes in the 19F spectra occurred upon addition of Mg2+. The observed downfield shift is consistent with reduced electron density about 19F in the Zn2+ complex. 19F resonances are very sensitive to electronic perturbations and while the observed downfield shift for these resonances does not prove a direct interaction between Zn2+ and F5 as predicted by the calculations, the observed chemical shifts are consistent with such an interaction. Downfield shifts in the 13C resonances for the C4 carbonyl region of FdU nucleotides were also observed following Zn2+ complex formation consistent with the computationally-derived low energy structure. In contrast, Mg2+ induced significant chemical shift perturbations to the 31P NMR spectra while Zn2+ did not. Thus, all of the NMR spectral data are consistent with the computationally derived binding mode for Zn2+ with DNA sequences including consecutive FdU nucleotides.
The present study has demonstrated a previously uncharacterized binding mode for Zn2+ with DNA sequences that include consecutive FdU nucleotides. Zn2+ binding is energetically favored under conditions where half of the FdU are deprotonated, or slightly above physiological pH (the pKA of FdU is ∼7.6). As both Zn2+ and FdU-containing DNA are cytotoxic toward prostate cancer cells, it is of interest whether Zn2+ might be complexed with FdU-containing DNA for therapeutic purposes (16). In the present study, we have demonstrated that lipofectamine enhances the cytotoxicity of Zn2+–DNA complexes towards PCa cells. The lipofectamine-stabilized Zn2+–DNA complexes demonstrate enhanced cytotoxicity relative to the components under standard tissue culture conditions. As both Zn2+ and FdU-containing DNA are well-tolerated in vivo, the complexes are likely to be well-tolerated in vivo as well, and may provide a new and more efficacious approach to prostate cancer treatment.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Defense Prostate Cancer Research Program (093606 to W.H.G.); National Institutes of Health (CA102532 to W.H.G.); The North Carolina Biotechnology Center (2P30 CA12197-32); National Institutes of Health (CA12937 and P30 CA12197-32 to F.R.S.). Funding for open access charge: National Institutes of Health (NCI CA102532 DOD CDMRP 093606).
Conflict of interest statement. There is a potential conflict of interest in that Dr Gmeiner is the primary owner of Salzburg Therapeutics which holds the licenses to FdUMP [N] technology.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Dr Marcus Wright for help with acquisition of 31P and 19F NMR data and Bree Berry and Dr Evan Gomes for technical assistance. All calculations were performed on the DEAC cluster which is supported by WFU Information systems and the WFU Provost’s office.
REFERENCES
- 1.Liang JY, Liu YY, Zou J, Franklin RB, Costello LC, Feng P. Inhibitory effect of zinc on human prostatic carcinoma cell growth. Prostate. 1999;40:200–207. doi: 10.1002/(sici)1097-0045(19990801)40:3<200::aid-pros8>3.0.co;2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uzzo RG, Leavis P, Hatch W, Gabai VL, Dulin N, Zvartau N, Kolenko VM. Zinc inhibits nuclear factor-kappa B activation and sensitizes prostate cancer cells to cytotoxic agents. Clin. Cancer Res. 2002;8:3579–3583. [PubMed] [Google Scholar]
- 3.Shi Y, Beger RD, Berg JM. Metal binding properties of single amino acid deletion mutants of zinc finger peptides: studies using cobalt(II) as a spectroscopic probe. Biophys. J. 1993;64:749–753. doi: 10.1016/S0006-3495(93)81435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JS, Latimer LJ, Reid RS. A cooperative conformational change in duplex DNA induced by Zn2+ and other divalent metal ions. Biochem. Cell Biol. 1993;71:162–168. doi: 10.1139/o93-026. [DOI] [PubMed] [Google Scholar]
- 5.Wood DO, Dinsmore MJ, Bare GA, Lee JS. M-DNA is stabilised in G*C tracts or by incorporation of 5-fluorouracil. Nucleic Acids Res. 2002;30:2244–2250. doi: 10.1093/nar/30.10.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao ZY, Sordet O, Zhang HL, Kohlhagen G, Antony S, Gmeiner WH, Pommier Y. A novel polypyrimidine antitumor agent FdUMP[10] induces thymineless death with topoisomerase I-DNA complexes. Cancer Res. 2005;65:4844–4851. doi: 10.1158/0008-5472.CAN-04-1302. [DOI] [PubMed] [Google Scholar]
- 7.Bijnsdorp IV, Comijn EM, Padron JM, Gmeiner WH, Peters GJ. Mechanisms of action of FdUMP[10]: metabolite activation and thymidylate synthase inhibition. Oncol. Rep. 2007;18:287–291. doi: 10.3892/or.18.1.287. [DOI] [PubMed] [Google Scholar]
- 8.Gmeiner WH, Reinhold WC, Pommier Y. Genome-Wide mRNA and microRNA profiling of the NCI 60 cell-Line screen and comparison of FdUMP[10] with fluorouracil, floxuridine, and topoisomerase1 poisons. Mol. Cancer Ther. 2010;9:3105–3114. doi: 10.1158/1535-7163.MCT-10-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spring BQ, Clegg RM. Fluorescence measurements of duplex DNA oligomers under conditions conducive for forming M-DNA (a metal-DNA complex) J. Phys. Chem. B. 2007;111:10040–10052. doi: 10.1021/jp0725782. [DOI] [PubMed] [Google Scholar]
- 10.Cesare Marincola F, Denisov VP, Halle B. Competitive Na(+) and Rb(+) binding in the minor groove of DNA. J. Am. Chem. Soc. 2004;126:6739–6750. doi: 10.1021/ja049930z. [DOI] [PubMed] [Google Scholar]
- 11.Valentina Tereshko GM, Martin E. A “Hydrat-Ion” spine in a B-DNA minor groove. J. Am. Chem. Soc. 1999;121:3590–3595. [Google Scholar]
- 12.Hirao I, Kawai G, Yoshizawa S, Nishimura Y, Ishido Y, Watanabe K, Miura K. Most compact hairpin-turn structure exerted by a short DNA fragment, d(GCGAAGC) in solution: an extraordinarily stable structure resistant to nucleases and heat. Nucleic Acids Res. 1994;22:576–582. doi: 10.1093/nar/22.4.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ulyanov NB, Bauer WR, James TL. High-resolution NMR structure of an AT-rich DNA sequence. J. Biomol. NMR. 2002;22:265–280. doi: 10.1023/a:1014987532546. [DOI] [PubMed] [Google Scholar]
- 14.Reed AE, Weinstock RB, Weinhold F. Natural-population analysis. J. Chem. Phys. 1985;83:735–746. [Google Scholar]
- 15.Ghosh SK, Kim P, Zhang XA, Yun SH, Moore A, Lippard SJ, Medarova Z. A novel imaging approach for early detection of prostate cancer based on endogenous zinc sensing. Cancer Res. 2010;70:6119–6127. doi: 10.1158/0008-5472.CAN-10-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gmeiner WH. Genetic determinants for activated fluoropyrimidine chemotherapy. Drug Devel. Res. 2006;66:1–11. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.