Abstract
The annealing of nucleic acids to (partly) complementary RNA or DNA strands is involved in important cellular processes. A variety of proteins have been shown to accelerate RNA/RNA annealing but their mode of action is still mainly uncertain. In order to study the mechanism of protein-facilitated acceleration of annealing we selected a short peptide, HIV-1 Tat(44–61), which accelerates the reaction efficiently. The activity of the peptide is strongly regulated by mono- and divalent cations which hints at the importance of electrostatic interactions between RNA and peptide. Mutagenesis of the peptide illustrated the dominant role of positively charged amino acids in RNA annealing—both the overall charge of the molecule and a precise distribution of basic amino acids within the peptide are important. Additionally, we found that Tat(44–61) drives the RNA annealing reaction via entropic rather than enthalpic terms. One-dimensional-NMR data suggest that the peptide changes the population distribution of possible RNA structures to favor an annealing-prone RNA conformation, thereby increasing the fraction of colliding RNA molecules that successfully anneal.
INTRODUCTION
As with proteins, the folding of RNA molecules into their native and functional structure is a non-trivial problem (1,2). These folding problems are not only in vitro artefacts but do also exist in vivo (3,4). It is assumed that certain proteins assist refolding events in vivo in a non-specific and ATP-independent manner and thereby support RNA molecules in the process of obtaining their native structure. These proteins have been defined as ‘RNA chaperones’; proteins ‘that resolve misfolded structures’ (1,5). Although most proteins that were shown experimentally to assist nucleic acid folding in a non-specific way were named nucleic acid chaperones (1,6), we recently demonstrated the necessity of discriminating between the mere acceleration of annealing and actual unfolding of RNA structures (7). As proteins can have either only one or both of these two activities, we suggested the following nomenclature: (i) ‘nucleic acid annealers’ are proteins that, with no sequence-specificity, accelerate annealing of complementary nucleic acids but are not able to dissolve RNA structures. (ii) ‘RNA chaperones’ destabilize RNA double-stranded nucleic acid regions (and might in addition accelerate annealing). The necessity of this discrimination is supported by the finding that some DEAD-box RNA helicases exert strand displacement activity in the presence of ATP but are not able to dissolve RNA structures in the absence of ATP, while still retaining their ability to accelerate annealing. For the human RNA helicase II/Gu protein even the physical separation of RNA unwinding and annealing activities was shown (8,9). RNA annealers and chaperones are extremely diverse and do not share a common amino acid motif (5). It is thus not clear which sequence or structural features determine whether a protein exhibits only one or both of the mentioned activities.
Proteins with nucleic acid annealing activity have been found in viruses (10–12), bacteria (13–15) and eukaryotes (6,16–28). The biological role of proteins with annealing activity ranges from bacterial translational regulation to the maturation of eukaryotic transcripts and RNA editing in kinetoplastid organisms. Due to the complexity and difficulty of in situ annealing assays, the potential role of nucleic acid annealers has so far only been assayed in in vitro experiments. Thus, the question whether they also exert the characterized activity on the described substrates in vivo remains unanswered.
All groups that studied protein-accelerated annealing in greater detail concordantly found basic amino acids to be crucial for, and high salt concentrations to be detrimental to, the annealing activity (18,20,26,27). The proposed mechanisms for annealing acceleration are: (i) active increase of local RNA concentration, (ii) stabilization of the annealing transition state by shielding the negative RNA backbone charges, (iii) ‘conversion’ of the RNA into an annealing-prone conformation (17,19,29). However, the following questions remain unanswered: do amino acids other than positively charged ones play a role for the annealing activity? Do nucleic acid annealers share a common mechanism or distinct features? Is there a more precise definition of amino acid composition and arrangement in nucleic acid annealers? Here, we used a model RNA annealer protein to address these questions.
The HIV-1 transactivator of transcription (Tat) is one of the early regulators of the viral life cycle. By binding to the TAR RNA, a structure formed by the nascent viral transcript, Tat recruits cellular factors to the HIV-1 promoter and indirectly stimulates viral transcription. (30,31) Tat was also suggested to play a role in other cellular and viral processes including translation (32), interaction with the RNAi machinery (33–35), PKR regulation (36,37), viral mRNA capping (38) and reverse transcription (39–41).
More recently, the Tat protein as well as its fragment Tat(44–61) were found to exhibit nucleic acid chaperone activity (42). In Tat(44–61), which comprises the basic domain and part of the core domain of the full-length protein, the authors claimed to have found the smallest known RNA chaperone to date. Due to its small size we selected Tat(44–61) as a model peptide for our studies on proteins that assist RNA folding.
Although we could not reproduce the protein’s RNA strand displacement activity, the full-length Tat protein and its fragment Tat(44–61) are potent nucleic acid annealers. We subjected Tat(44–61) to a detailed mutational analysis to identify the amino acids that are crucial for the annealing activity of the peptide. We confirmed the essential role of basic amino acids, but also found evidence for the contribution of polar amino acids that can potentially hydrogen bond with the RNA backbone. We found the peptide’s overall charge and the arrangement of basic amino acids within the peptide important for RNA annealing acceleration. Furthermore, we demonstrated that the acceleration of annealing is conferred by an increase in transition state entropy. One-dimensional (1D) 1H NMR data show that the population of possible RNA structures is changed in the presence of Tat(44–61). Thus, either the peptide selectively binds a preexisting RNA conformation and thereby shifts the equilibrium or it changes the structure of the RNA molecule upon binding. In either case the abundance of annealing-prone RNA conformations is increased which raises the fraction of colliding RNA molecules that efficiently anneal with each other.
MATERIALS AND METHODS
RNAs and peptides
All RNAs used were synthesized on solid phase (Eurogentech). Lyophilized RNAs were dissolved in DEPC water and stored at −20 or at −80°C (for long-term storage). The nucleic acid substrates used for the FRET-based assay in this study are listed in Supplementary Table S1.
Synthetic peptides were provided by ThermoFischer Scientific, peptides scr1-3, K7A R13A and K7A R9A R13A were a generous gift of Peter Steinlein, Research Institute for Molecular Pathology Vienna, Austria. The peptides were dissolved in DEPC water, aliquoted, freeze-dried and stored at −20°C. Shortly before use they were dissolved and diluted in 30 mM Tris–HCl pH 7, 30 mM NaCl and 1 mM DTT. Mass spectrometry results confirmed the sequence identity and integrity of Tat(44–61) that had been stored lyophilized at −20°C for several months. Although modifications (such as hydrolysis) of the peptide upon short-time storage in solution are unlikely, we cannot completely exclude this possibility. The fact that the peptide can accelerate annealing of two complementary sequences, however, hints at it being active. The recombinant full-length Tat protein was delivered in a freeze-dried form by Jena Bioscience and dissolved in degassed 100 mM Tris–HCl pH 7.5, 150 mM NaCl and 10 mM DTT. The full-length Tat protein is very sensitive to inactivation through oxidation and aggregation (43) and is thus difficult to handle. We excluded degradation of the protein on SDS–PAGE and confirmed the correct protein sequence by mass spectrometry. Furthermore, we took precautions to avoid oxidation of the protein such as degassing all solutions.
FRET-based annealing and strand displacement assays
Combined nucleic acid annealing and strand displacement assays as well as annealing assays were carried out as described by Rajkowitsch and Schroeder (2007) (44), at 30°C if not indicated otherwise, and using the following concentrations: 10 nM of each RNA strand, 50 mM Tris–HCl in the pH range of 6.5–8, 2 mM DTT and salt concentrations as indicated in the text. Annealing curves were fitted by non-linear regression to the following second-order reaction equation using GraphPadPrism® (with Y being the normalized FRET index and the time X ):
| (1) |
Filter binding assays
Filter binding assays were conducted according to ref. (45) with the following changes: the reaction buffer contained 50 mM Tris–HCl pH 7, 2 mM DTT and 6% glycerol. Reactions were incubated for 5 min at room temperature before filtering. Binding curves were fitted to a Hill equation [Equation (2)] using GraphPadPrism®. (Y, relative amount of filter-bound RNA; Bmax, maximally bound RNA; h, Hill coefficient; KD, binding constant; X, peptide concentration)
 |
(2) |
Due to reproducibility problems of absolute KD values and Hill coefficients which have also been faced by other groups (Michael F. Jantsch, personal communication), we moved on to recording binding curves for the mutant of interest (or the wild-type peptide under the salt condition of interest) in parallel with the wild-type mutant under no salt conditions. We thus obtained relative KD values and Hill coefficients. Other common techniques to determine binding constants, like mobility shift assays, quenching methods and ITC, were not applicable to our system due to their specific limitations.
Aggregation assays
Aggregation through proteins was measured on the basis of the sedimentation assay described by Vo et al. (46). Ten nanomolar 32P-labeled single-stranded 21R+ RNA or double-stranded 21R RNA were incubated at 30°C for 3 min in aggregation buffer (50 mM Tris–HCl pH 7, 2 mM DTT, NaCl and MgCl2 concentrations as indicated in each single experiment) in the presence or absence of 100–300 nM Tat(44–61) or ∼0.1 µg/µl BSA (NEB), total volume 40 µl. After a 10 min centrifugation at 13.4 × g at room temperature, 5 µl of the supernatant were removed carefully and subjected to Scintillation analysis.
Gel annealing and strand displacement assays
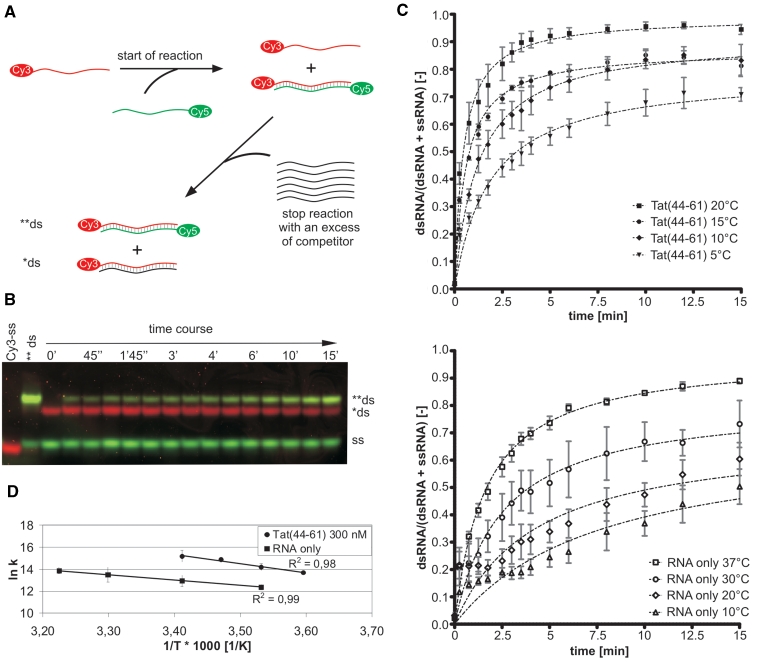
Due to temperature limitations of the multiplate reader, the rate constants’ temperature dependence was measured using gel annealing assays: 10 nM Cy5-labeled 21R+ were pre-incubated with or without 300 nM peptide in 50 mM Tris–HCl pH 7, 0.5 mM MgCl2 and 2 mM DTT final concentrations at the temperature of interest. The annealing reaction was started by adding 10 nM final concentration of Cy3-labeled 21R–. Ten microliter-aliquots were taken after the indicated time points, mixed with 2.5 µl 5× stop buffer (3 µM 21R+, 2% SDS, 150 mM EDTA, 15% sucrose, 12.5% ficoll) and directly loaded onto a running 15% native PAGE (1× TBE). Fluorescent signals of both Cy5 and Cy3 were scanned with a TyphoonTM 9400 (GE Healthcare), quantified with ImageQuantTM and annealing curves were fitted to Equation (1) using GraphPadPrism®.
The full-length Tat protein is susceptible to aggregation (43) which disturbed the fluorescent signal in the FRET-based assay so that an evaluation of the resulting curves was not possible. We thus carried out gel strand displacement assays. Strand displacement gel assays were conducted in a similar way to the annealing gels, apart from the following changes: to start the reaction a 10-fold excess of competitor was added to an already formed double-stranded substrate. The reaction was stopped using a 2.5× stop buffer containing 60 mM EDTA, 0.5 mg/ml yeast tRNA, 0.6% SDS, 30% sucrose and 12.5% ficoll.
Calculations of kinetic parameters
The reaction constants kobs, determined at different temperatures, were divided by the RNA concentration used in the gel annealing assay. From these normalized rate constants, k, an Arrhenius plot was derived (17,47). The slope of the Arrhenius plot yielded the activation energy Ea of the annealing reaction (R—gas constant):
| (3) |
The free activation energy ΔG≠ was derived according to the Eyring–Polyani relation (48) from the normalized rate constant k at a specific temperature T:
| (4) |
(h—Planck’s constant equivalent to 6.626 × 10−34 Js, kB—Boltzmann’s constant equivalent to 1.381 × 10−23 JK−1).
Based on Ea and ΔG≠, the thermodynamic parameter enthalpy ΔH≠ and entropy ΔS≠ were calculated using Equations (5) and (6).
| (5) |
| (6) |
Interaction between RNA and peptide: NMR spectroscopy
To probe the interaction between RNA and peptide, a series of titration NMR experiments were recorded. Spectra were measured at temperatures between 273 and 320 K on either a Varian 600 MHz spectrometer equipped with a z-grad HCN probe or a Bruker 700 MHz spectrometer equipped with a cryogenic z-grad HCN probe. In all samples H2O was exchanged with D2O by multiple cycles of freeze-drying and resolvation in 99.999% D2O (Sigma Aldrich). 1H pulses were applied at the resonance of the water frequency with field strength of 15.6 kHz. One hundred and twenty-eight transients were averaged, the relaxation delay was set to 1.5 s and the spectral width was 10 ppm. Hard power pulses were applied at the water frequency, 16 kpts were recorded for t1. Spectra were processed and analyzed using NMRpipe and NMRdraw (49). For processing, an exponential window function with a LB factor of 1.5 Hz was applied.
Starting from an idealized A-form helical 21R+ RNA (the structure was calculated with CNS using a restrained TAD simulated annealing protocol starting from an extended structure and applying canonical A-form restraints) the chemical shifts were calculated using the program NUCHEMICS (50,51) and were subsequently translated into NMR spectra with the NMRSIM module of TOPSPIN. 1H chemical shifts are referenced directly to TSP as an external reference. Notably, upon addition of peptide to the RNA sample a white precipitate appeared which we assumed to be a fraction of the RNA–peptide complex. Precipitation was probably caused by high working concentrations (400 µM RNA and 400 or 800 µM peptide). Thus, the concentration of peptide and RNA in solution was somewhat smaller than intended.
RESULTS AND DISCUSSIONS
Tat and its fragment Tat(44–61) have nucleic acid annealing activity, but no strand displacement activity
The assays conducted and described by Kuciak et al. (42) implied that the HIV-1 Tat protein possesses two distinct activities: namely acceleration of annealing and destabilization of nucleic acid structures. Although some of these assays made use of potential natural substrates of Tat, both the RNA ribozyme and the RNA trans-splicing assay suggested that the protein has sequence non-specific RNA chaperone activity.
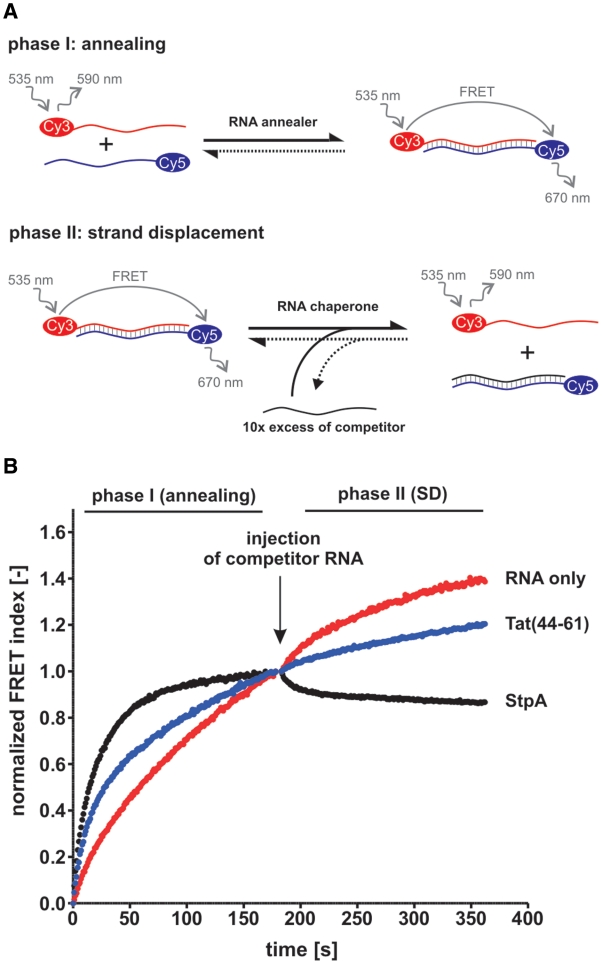
In order to confirm the described activities we used Tat(44–61) in a two-phase FRET-based assay (Figure 1A) that has previously been described and used for several potential RNA chaperone proteins (14,44). As anticipated, Tat(44–61) considerably accelerated annealing of the 21R substrate (Supplementary Table S1), a 21 base-pair long, blunt-ended, artificial RNA duplex [Tm = 72.4°C, calculated using the DINAMelt server (52)] (Figure 1B, phase I). The peptide was however not active in the second (strand displacement) phase of the assay while the positive control StpA efficiently catalyzed strand displacement. Remarkably, Tat(44–61) also accelerated annealing of the DNA substrate 21D while not catalyzing strand displacement by a competitor DNA (Supplementary Figure S1). To exclude the possibility of the 21R double-stranded substrate being too stable for peptide mediated unfolding we used the jm1 substrate which has a lower GC content (Tm = 49.3°C), with the same result (Supplementary Table S1 and Figure S2C). Some proteins are known to need a single-stranded or helical binding platform to perform double-strand unwinding (8). To test this possibility for Tat(44–61) we used a 46 nt RNA, which forms a short hairpin at the 3′-end, together with a partly complementary 21-mer in the combined FRET-based assay (jm1heli substrate). Annealing of this substrate was accelerated by the peptide, whereas no strand displacement was detected (Supplementary Figure S2D). Other variations of the assay included using different temperatures (30 and 37°C), testing several peptide to nucleotide ratios (10 nM to 1 mM peptide per 10 nM RNA), changing the concentrations of MgCl2 and NaCl, using exactly the same buffer composition as described by ref. (42) (buffer A) as well as testing peptide buffers with different ZnCl2 concentrations. While we could always detect an acceleration of annealing, the peptide did not catalyze strand displacement under any of the applied conditions (Supplementary Figure S2A, B, E and F).
Figure 1.
Tat(44–61) accelerates annealing of two complementary RNAs but does not promote strand displacement. (A) Scheme of the FRET-based combined annealing and strand displacement (SD) assay according to Rajkowitsch and Schroeder (44). Two short complementary RNAs, labeled with a Cy3- (donor) and a Cy5-dye (acceptor), respectively, are annealed in a microplate (phase I). The close proximity of the two dyes results in a fluorescence resonance energy transfer (FRET) when the donor dye is excited. Both fluorescent emission signals are measured at 1 s intervals using a microplate reader, and the FRET index is calculated as the ratio of the FRET to the Cy3-signal. Strand displacement is detected in phase II of the assay which is started through the addition of an unlabeled competitor RNA and results in a decrease of the FRET signal. (B) The assay was conducted with 21R RNA in the absence or presence of 1 µM Tat peptide and at 30°C. E. coli StpA (1 µM) was used as a positive control. For clarity, the FRET index was normalized to 1 (phase I and II) and 0 (only phase I).
To test the full-length Tat protein’s ability to promote strand displacement we conducted polyacrylamide gel assays using the 21R substrate, with CsdA as a positive control (Supplementary Figure S3). Again, we could not detect an RNA chaperone activity.
To ensure that the contradiction in results were not caused by a limitation in our assay we carried out the DNA strand displacement assay as described by Kuciak et al. (42) (Supplementary Figure S4) and the trans-splicing assay (53) (Supplementary Figure S5). Despite all positive controls showing activity, we could not reproduce the reported activities of the full-length Tat protein and Tat(44–61) in these RNA chaperone assays.
We therefore conclude that Tat and its fragment are facilitators of annealing but not RNA chaperones. Due to its strong annealing activity, the small size, good availability and easy handling of Tat(44–61) we used it as a model RNA annealer and studied its annealing activity in greater detail.
The annealing activity of Tat(44–61) is dependent on the concentration of mono- and divalent cations
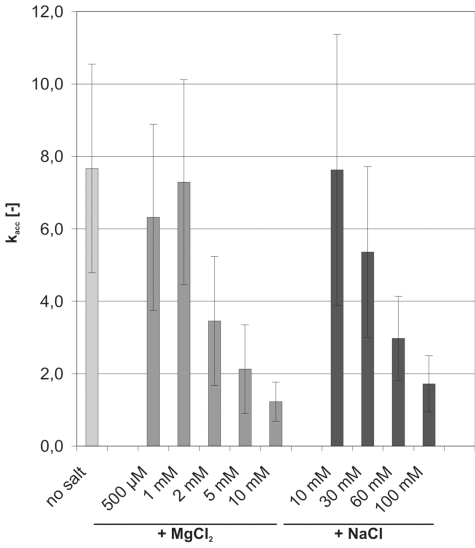
Tat(44–61) accelerated annealing with very similar kobs between pH 6.5 and 7.5 (Supplementary Figure S6). We thus measured acceleration of annealing at pH 7. We also tested the influence of mono- and divalent cations on the peptide’s activity (Figure 2). Consistent with the kinetic salt effect, the reaction constant of the ‘RNA only’ reaction increased with rising NaCl or MgCl2 concentration (47). In comparison to ‘no salt’ conditions 100 mM NaCl and 10 mM MgCl2 accelerated annealing 1.5- and 3-fold, respectively (data not shown). Tat(44–61) does not require any cations for its annealing activity: the peptide accelerated annealing ∼7- to 8-fold when no or only low amounts of ions were present. These kacc values [with kacc = kobs(peptide)/kobs(RNA only)] are in good agreement with the calculated acceleration of annealing measured with gel annealing assays (kacc = 6 and 9.5 at 10 or 20°C, respectively, as derived from the kobs values in Figure 6).
Figure 2.
Mono- and divalent cations are detrimental to the peptide’s annealing activity. FRET-based annealing assays with 21R RNA were carried out in the absence or presence of 300 nM Tat(44–61), and at different MgCl2 and NaCl concentrations. The reaction constants (kobs) of the reaction in the presence of peptide were divided by the kobs of the ‘RNA only’ reaction, yielding kacc. Acceleration of annealing by the peptide was strongly impaired at MgCl2 concentrations above 2 mM and NaCl concentrations >60 mM.
Figure 6.
Tat(44–61) increases the transition state entropy of the annealing reaction. (A) RNA gel annealing assays were conducted in the absence or presence of protein and at different temperatures. To start the reaction a Cy3-labeled RNA strand was added to the Cy5-labeled complementary RNA. Aliquots were drawn after certain time points and the reaction was stopped with a 60× excess of non-labeled competitor strand. (B) The samples were applied directly onto a running native PAGE. Since the Cy5-dyes add ‘size’ to the RNAs the heteroduplex of two labeled strands (**ds) runs higher than the complex of the Cy3- and the unlabeled strands (*ds). The Cy5- and Cy3-fluorescent signals were scanned and the Cy5-signals from single and double strands were quantified. (C) Plotting the ratio of double-stranded RNA to total amount of RNA for each time point of a reaction yielded annealing curves which were fitted with a monophasic second-order reaction equation with equimolar initial reaction concentration. With rising temperature the kobs of the annealing reaction in the presence or absence of 300 nM Tat(44–61) increased. The rate constants k, normalized to the RNA concentration, were as follows: Tat(44–61) 5°C 9.0 × 105 M−1s−1 (filled inverted triangle), 10°C 14.5 × 105 M−1s−1 (filled diamond), 15°C 29.0 × 105 M−1 s−1 (filled circle), 20°C 40.0 × 105 M−1s−1 (filled square); RNA only 10°C 2.4 × 105 M−1s−1 (open triangle), 20°C 4.2 × 105 M−1s−1 (open diamond), 30°C 7.3 × 105 M−1s−1 (open circle), 37°C 10.6 × 105 M−1s−1 (open square). Means of curves and rate constants k were calculated from three to four independent experiments. (D) The natural logarithms of the normalized reaction constants k were plotted against the inverse temperature demonstrating the linear correlation between the two values in the measured temperature range (Arrhenius plot).
Acceleration of annealing by Tat(44–61) was strongly impaired at MgCl2 concentrations >2 mM and NaCl concentrations above 60 mM (Figure 2). Similar dependences on MgCl2 concentrations have been reported for HIV-1 NCp7 although the nucleocapsid protein was tolerant for NaCl concentrations of up to 90 mM (10). Both Na+ and Mg2+ ions have been shown to compete with positively charged peptides for the interaction with RNA (54,55). Thus, the dependence of the peptide’s activity on the ion concentration hints at the importance of ionic interactions between the peptide’s basic amino acids and the negatively charged RNA backbone for the acceleration of annealing by Tat(44–61).
All basic amino acids are similarly important for the annealing activity of Tat(44–61)
The importance of basic amino acid stretches in nucleic acid annealing was reported by several groups (10,17–20,26,56,57) and is now generally accepted, although the specific role of the positive charges during the annealing process is under debate and might differ for each protein. The presence of eight basic residues in the 18 amino acid long sequence of Tat(44–61) supports the hypothesis of a mainly charge driven annealing acceleration process. The small size of the Tat peptide prompted us to explore the contribution of individual amino acids to its activity.
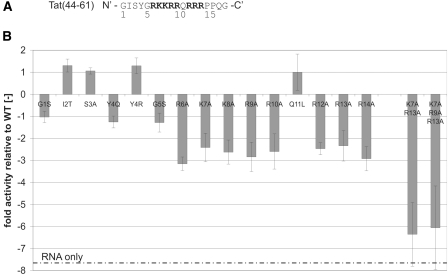
A set of peptides with single amino acid substitutions was therefore tested for their annealing activity in our FRET-based annealing assay (Figure 3, Supplementary Figure S7). Figure 3B shows the mutants’ kobs normalized to the wild-type kobs. Relative kobs values were confirmed for a few selected mutants using gel annealing assays (data not shown).
Figure 3.
The peptide’s basic amino acids are crucial for the acceleration of annealing. (A) Amino acid sequence of Tat(44–61) with the positively charged amino acids in bold. (B) Different single amino acid mutants and a double and triple amino acid mutant of Tat(44–61) were tested in our FRET-based annealing assay with the 21R RNA pair at 30°C (using 300 nM peptide). Normalized FRET-values were fitted to the monophasic second-order reaction equation yielding the kobs for each peptide mutant. The obtained kobs were used to calculate the mutant activities relative to the WT as follows. In case the kobs(mutant) was higher than the kobs(WT), kobs(mutant) was divided by kobs(WT) and plotted as a positive value. In case of a decreased mutant kobs relative to the WT kobs, kobs(WT) was divided by kobs(mutant) and plotted as a negative value. This calculation method was used in order to emphasize specifically kobs decreases. Notably, the activity of all peptides in which a basic arginine or lysine was exchanged against alanine was reduced by a factor of 2.5–3 relative to the wild-type peptide. The double and triple mutant ‘K7A R13A’ and ‘K7A R9A R13A’ did not accelerate annealing detectably at the applied conditions. Two peptide mutants (I2T and Y4R) showed a higher annealing activity than Tat(44–61).
Strikingly, each time a basic amino acid was replaced by an alanine the peptide’s activity was decreased by a factor of three. The position of the exchanged residue within the primary sequence did not seem to be important at all. Furthermore, the Y4R mutant in which a charge was added to the peptide was more active than the wild-type peptide while the Y4Q mutant did not accelerate annealing any different from Tat(44–61). The double and triple mutants (K7A R13A and K7A R9A R13A) did not accelerate annealing under the tested conditions. Only when we added mutant peptide amounts in the 10 µM range could we detect an activity similar to the activity of 300 nM wild-type peptide (Supplementary Figure S8).
The mutation of the polar amino acids serine (at position 3) and glutamine (position 11) that could theoretically interact with the RNA via hydrogen bonds or Van der Waals interactions did not show an effect. However, the exchange of isoleucine (position 2) for a threonine increased the peptide’s annealing activity. The additional threonine might either enhance the activity indirectly, for example by increasing the peptide’s stability and/or solubility. Alternatively, it might act directly by interacting with the RNA. Threonine has indeed often been shown to make contacts with phosphates of the nucleic acid backbone (58).
Together, these results demonstrate that the presence of a certain amount of basic residues is indispensable for the peptide to confer RNA annealing acceleration. Amino acids with the ability to hydrogen bond might support the annealing activity. This observation further bolsters the hypothesis of backbone phosphate charges conferring peptide/RNA binding. These (mainly ionic) interactions would explain the promiscuity of the peptide’s annealing activity towards different RNA substrates as well as DNA. The importance of the overall peptide charge can be partly explained by the peptide screening the nucleic acid surface similarly to metal ions. It is a well known fact that Na+ and Mg2+ stabilize RNA double strands thermodynamically by reducing the electrostatic stress arising from the small distances of the RNA backbone phosphates, with ions that have a higher charge density being more effective stabilizers (59). Thus, they accelerate annealing to a certain degree, especially at higher concentrations. Similarly, the multivalent Tat peptide might accelerate annealing partly because of this screening effect. However, as the magnitude of acceleration of annealing by Tat(44–61) can not be achieved by a simple increase of the salt concentration, there must be a different mechanism that is beyond charge neutralization.
The spatial arrangement of the amino acids in the Tat peptide is important for its activity
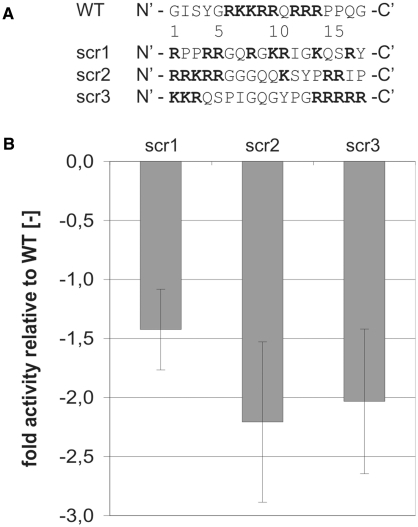
The results discussed in the previous section imply that the most important factor that confers annealing activity is the overall charge of the peptide. This could mean that the peptide acts as an octovalent ion. To assess the question as to whether a specific spatial arrangement of basic amino acids within the peptide is important we scrambled the peptide in three different ways (Figure 4A): (i) in the mutant scr1 the eight basic amino acids are distributed evenly over the peptide; (ii) scr3 contains two stretches of basic residues, as does the WT peptide, however these are separated by a larger amount of uncharged amino acids; (iii) scr2 contains only one basic stretch while the other basic residues are more distributed.
Figure 4.
The spatial arrangement of basic amino acids within the peptide is important for annealing activity. (A) Primary sequences of scrambled peptides with the same amino acid composition as the WT Tat(44–61) peptide. (B) The FRET-based annealing assay using 21R RNA was carried out at 30°C. The obtained reaction constant kobs of each peptide mutant was normalized to the WT kobs, yielding the ‘fold activity relative to the WT’. Interestingly, all scrambled peptides containing the same amount of basic amino acids as Tat(44–61) were less active than the WT.
Interestingly, all three scrambled peptides are less active than the wild-type peptide in our RNA annealing assay with scr1 being more active than scr2 and scr3 (Figure 4B). Thus, the overall charge [which is the same for scr1-3 and Tat(44–61)] alone is not sufficient to account for the extent of RNA annealing activity. In every charged molecule only a limited number of the charged amino acids can participate in the interaction with the binding partner due to structural reasons. Quantitatively, this is expressed by the effective charge Zeff as opposed to the overall charge (55). Changing Zeff by scrambling the Tat peptide sequence is likely, even though we expect a peptide of only 18 amino acids length to be extremely flexible which might result in a similar mean Zeff in the WT and scr1-3. We hypothesize that, apart from maintaining a certain effective charge, an annealing competent protein positions its positively charged residues in a well-defined (but still unknown) manner in space in order to facilitate rather defined interactions between amino acids and nucleic acids.
Binding strength and annealing activity do not correlate
We tested RNA binding of two different peptide mutants as well as of the wild-type peptide using filter binding assays. The RNA of interest was the single-stranded 21R+, one of the RNA strands used for the FRET-based annealing assay. We always recorded binding curves for the mutant peptide of interest in parallel with the wild-type. We thus obtained relative KD values and relative Hill coefficients (Table 1). The wild-type KD as measured in 16 independent experiments was 1110 ± 434 nM. The R13A mutant’s KD is slightly higher than the wild-type’s KD while the Hill factor remains unchanged. The K7A R13A mutant’s KD is double that of the wild-type’s binding constant. The different RNA binding strength of this mutant is also reflected in a slightly lower Hill coefficient. The loss of annealing activity of these two mutants is ∼70% (R13A) or complete (K7A R13A) at a peptide concentration of 300 nM (see Figure 3B). Thus, the mutants’ loss of activity is not reflected in their binding constants.
Table 1.
Binding constants for peptide-RNA binding of two different peptide mutants relative to the WT peptide
| R13A | K7A R13A | |
|---|---|---|
| kobs (mutant): kobs (WT) [−] | 0.43 ± 0.10 | 0.16 ± 0.04 |
| KD (mutant): KD (WT) [−] | 1.23 ± 0.13 | 2.27 ± 0.31 |
| h (mutant): h (WT) [−] | 1.07 ± 0.16 | 0.87 ± 0.10 |
Relative KD values and Hill factors h were calculated from three independent sets of filter binding experiments. For comparison, the kobs for mutant peptide accelerated annealing relative to the wild-type reaction constant is additionally listed.
We could therefore confirm the missing correlation between RNA annealing activity and RNA binding as described by several other groups (14,26,60–62). The results suggest that, besides RNA/peptide binding, another process (such as an RNA conformation change) occurs for which a certain amount of basic amino acids is necessary. Considering that the removal of one or two basic amino acids from Tat(44–61) will probably change the peptide’s conformation, the spatial arrangement of positive charges might also play a role in this process.
Tat(44–61) does not accelerate annealing through aggregation
One of the suggested mechanisms for protein accelerated annealing is the increase of the local nucleic acid concentration. Several studies have shown that HIV-1 nucleocapsid protein Ncp7 induces the sequence non-specific aggregation of nucleic acids which is considered to be a major element of its chaperone activity (46,63–67). Besides proteins, different multivalent cations such as spermidine and spermine compact and aggregate nucleic acids via electrostatic interactions (68,69) and thus it was suggested that highly positively charged annealing competent proteins in general accelerate annealing via this mechanism.
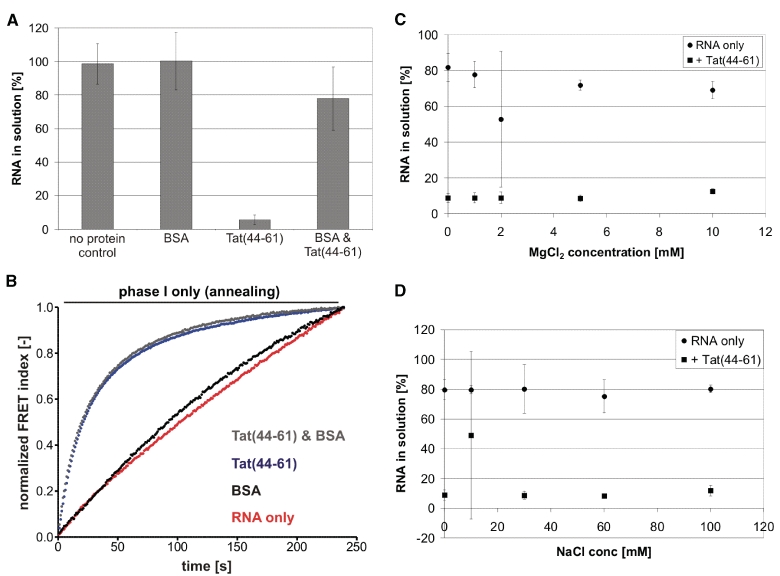
To address this hypothesis, we tested whether Tat(44–61) aggregates RNA with a simple centrifugation assay (46). We found the peptide to be positive in this assay using single-stranded 21R+ (Figure 5A) and double-stranded 21R RNA (data not shown). However, the addition of BSA to the reaction prevented Tat(44–61) caused depletion of RNA from the solution almost completely. Interestingly, BSA had no influence on the annealing activity of Tat(44–61) as assayed with the FRET-based annealing assay (Figure 5B). We therefore assume that the observed ‘aggregation’ was due to non-specific binding of the Tat peptide to test tube walls and that in contrast to Ncp7 no aggregates were formed. Further experiments supported this conclusion. MgCl2 and NaCl concentrations that inhibited annealing acceleration by the Tat peptide (Figure 2) did not affect the outcome of the aggregation assay (Figure 5C and D). In contrast, mono- and divalent ions have been shown to strongly influence Ncp7 induced nucleic acid aggregation (63). We could not detect a difference in RNA depletion for the three different peptide concentrations 100, 200 and 300 nM, which showed different magnitudes of annealing activity as well. Shorter centrifugation times resulted in less ‘aggregated’ RNA for all three concentrations without showing any significant discrimination between the three concentrations (Supplementary Figure S9).
Figure 5.
Aggregation and annealing acceleration by the Tat peptide do not correlate. (A) Aggregation assays with 21R+ ssRNA in the presence of 300 nM Tat(44–61), ∼0.1 µg/µl BSA or both proteins were carried out. While BSA did not aggregate the RNA, Tat(44–61) depleted >90% RNA from the solution. In the presence of BSA, however, Tat(44–61) had only a minor aggregation effect. (B) Interestingly, the same amount of BSA did not influence the RNA annealing activity of 300 nM of the peptide as determined using the FRET-based annealing assay. (C and D) RNA aggregation by 300 nM Tat(44–61) was assayed under different MgCl2 and NaCl concentrations that had been shown to inhibit the peptide’s annealing activity to a greater or lesser extent. However, no significant difference in measured aggregation was detectable.
We therefore conclude that nucleic acid aggregation is not the basis for Tat peptide accelerated RNA annealing.
Tat(44–61) increases the entropy of the annealing transition state
To learn more about the mechanism of Tat(44–61)-facilitated annealing we collected annealing rate constants at different temperatures using gel annealing assays (Figure 6A–C). From the temperature dependence of the rate constant k an Arrhenius plot was drawn (Figure 6D) and thermodynamic parameters of the transition state of annealing (Table 2) were calculated as described previously (29).
Table 2.
Thermodynamic data at 20°C derived from the Arrhenius plot in Figure 6
| K (M−1s−1) | Ea (kJ/mol) | ΔG≠ (kJ/mol) | ΔH≠ (kJ/mol) | ΔS≠ [J/(mol·K)] | |
|---|---|---|---|---|---|
| RNA only | 4.2 × 105 | 40 ± 1 | 40 ± 1 | 38 ± 1 | −8 ± 6 |
| Tat(44–61) 300 nM | 4.0 × 106 | 70 ± 6 | 35 ± 1 | 68 ± 6 | 112 ± 26 |
Parameters are as follows: k, RNA concentration corrected rate constant of the annealing reaction; Ea, Arrhenius activation energy; ΔG≠, free activation energy; ΔH≠, enthalpy; ΔS≠, entropy.
The relation between Gibbs free energy, enthalpy and the entropy term are described by the following equation:
| (7) |
The Tat peptide increases the activation energy Ea from 40 to 70 kJ/mol and concomitantly the transition enthalpy ΔH≠ of annealing by 30 kJ/mol which is detrimental to the reaction. Nevertheless, in the presence of the peptide the transition free energy ΔG≠ is lowered from 40 to 35 kJ/mol. This is also reflected by the increase of the annealing reaction constant by a factor of 9.5 under these conditions. Thus, the peptide-induced ΔG≠ decrease must be caused by entropic rather than enthalpic terms. Consistently, the transition entropy ΔS≠ is increased significantly in the presence of Tat(44–61) (Table 2). A similar thermodynamic effect on RNA annealing has so far been shown only for the chemical CTAB (29) and (with a not as pronounced magnitude) for the tumor suppressor protein p53 (17). The interaction of positively charged peptides with nucleic acids is generally accompanied by a counterion and water release which results in a considerable entropy increase (55,70). This entropy increase is driven solely by the net charge of the peptide, and not the amino acid arrangement (54). A very recent study on thermodynamics of binding of the Tat fragment Tat(46–60) to the TAR RNA confirmed an entropy increase of the system through RNA–peptide interaction (71). The calculated entropy increase through non-specific binding at 80 mM NaCl ΔSbinding was determined to be 63 ± 4 J/(mol K), which is in the same range as our calculated entropy increase for the transition state. We speculate that, also in the case of RNA/RNA annealing, ΔS≠ elevation is mainly driven by counterion and water release upon peptide binding. The question that remains unanswered is: What happens to the RNA upon peptide/RNA interaction that accelerates annealing? These processes might either support or counteract the entropic effect of counterion release.
Tat(44–61) favors an annealing-competent RNA structure
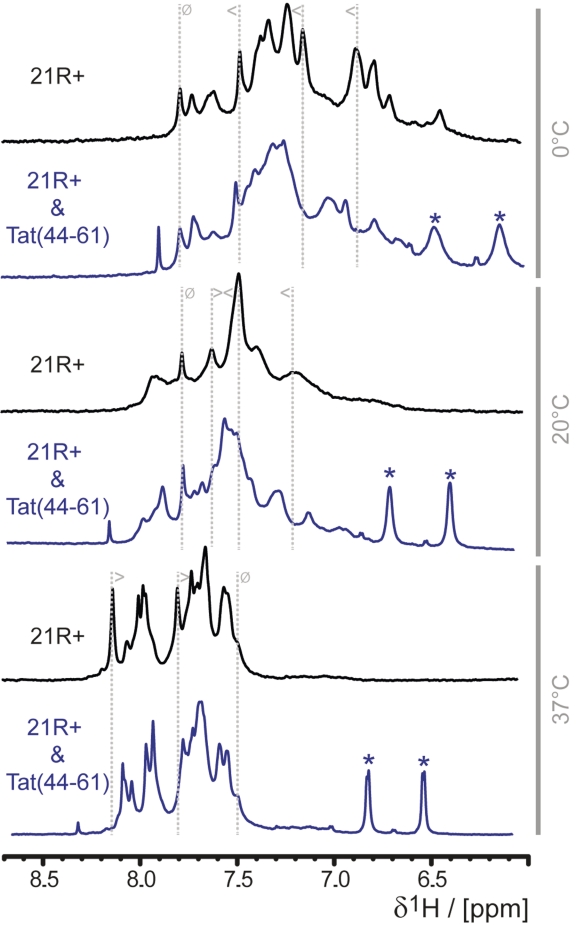
In order to learn more about the structural influence of Tat(44–61) on its RNA substrate we recorded 1D 1H spectra of the 21R + RNA in the absence or presence of different amounts of Tat peptide and at different temperatures (Figure 7).
Figure 7.
Tat(44–61) selects a specific RNA structure. 1H 1D NMR spectra of 21R+, a 21nt single stranded RNA, in the absence (black) or the presence (blue) of two equivalents of Tat(44–61) at different temperatures (indicated in gray at the right panel of the figure) were recorded. The chemical shifts of the aromatic protons did not overlap with the peptide chemical shifts and serve thus as an indicator for the peptide’s influence on the RNA conformation: the only visible peaks of the peptide in this spectral region are stemming from the aromatic protons of Y47 and are marked with asterisks. To follow changes in the spectrum upon addition of peptide gray dotted lines are added exemplarily, the symbol indicate if peaks are shifting upon addition of the peptide (less than and greater than symbol) or show no change (struck through circle).
The 1D 1H NMR spectra of the 21R+ RNA recorded in 9/1 H2O/D2O-buffer did not show any signals in the imino-proton region at a temperature range of 0–47°C (data not shown). Therefore we conclude that under our experimental conditions the RNA molecule does not exhibit any secondary-structure elements that involve base-pair interactions and truly represents an ‘unfolded’ RNA chain. Changing the solvent from H2O to D2O allows recording of the spectra without the deterring effect from water suppression. The same signal pattern of non-exchangeable protons is found, indicating that changing the solvent has no effect on the conformation of the RNA.
The NMR spectra of the 21R+ RNA alone show a strong temperature-dependent behavior in the range between 0 and 47°C (Figure 7). At low temperatures, such as 0°C, distinct signals are detectable and show a reasonable chemical shift dispersion for an RNA void of any canonical structural elements. As displayed in Figure 7 (and Supplementary Figure S10), the peaks of the aromatic protons spread over a spectral region of ∼1.3 ppm. As the temperature is increased the dispersion of the signals is lost and the spread of the aromatic protons is narrowed down to ∼0.7 ppm at 37°C. Furthermore, the shape of the peaks shows a marked temperature dependence. At temperatures ∼20–25°C, the peaks become broad but the line-shape narrows down again when the temperature is further increased to 37°C or higher (Supplementary Figure S10). This behavior is not only observed in the aromatic spectral region, but for all RNA peaks in the whole spectrum (the line-width of signals from buffer molecules stay constant within error over the whole temperature scale). The changes in the spectrum are reversible and multiple cycles of cooling and heating over the indicated temperature range always produces the same spectra. The integrity of the RNA was confirmed by biochemical methods before and after NMR measurements.
For a more analytical evaluation of the experimental spectra, simulated NMR spectra of the 21R+ RNA were generated with the NUCHEMICS software. The simulations were based on two structures of the 21R+ RNA: one representing the single strand in A-form conformation and a second representing a structurally randomized conformation. Although the exact experimental spectrum could not be reproduced by the simulation, the chemical shift dispersion behavior could be reproduced (Supplementary Figure S11). For the aromatic region, the simulated spectra show a dispersion of signals over ∼1.8 ppm based on the A-form conformation, whereas the randomized conformation leads to a reduction of chemical shift dispersion down to ∼0.8 ppm and an overlap of peaks. Generally, the dispersion in RNA spectra (especially for the aromatic protons in the nucleo-bases) derives from the stacking behavior of the RNA chain. Therefore, the absence of helicality in the RNA reduces the proportion of stacked nucleotides and the effect on the chemical shifts in the NMR spectrum is a reduced chemical shift dispersion.
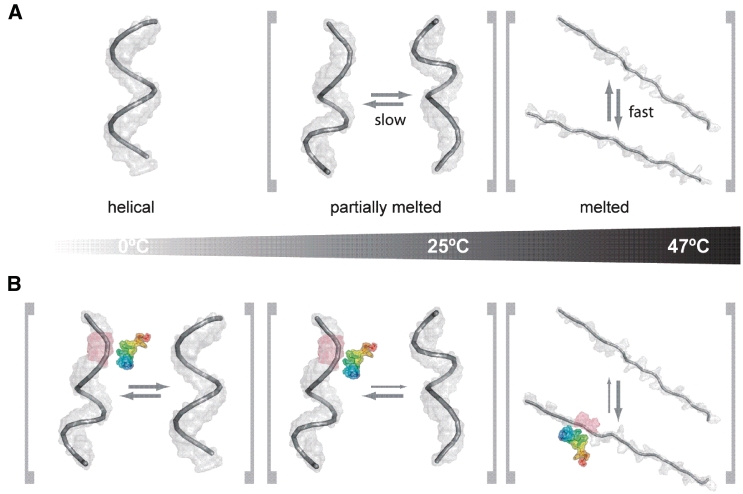
We conclude that at low temperatures a distinct, probably helical conformation is adopted by the single-stranded 21R+ RNA (Figure 8A). When the temperature is raised, this conformation starts to melt. Around 20–25°C the molecule is no longer best described by a single static conformation but by an ensemble of interconverting partially melted ‘helices’. The temperature increase to 47°C converts the 21R+ RNA into similar, unstructured (and most probably unstacked) conformations that give rise to the low dispersion but narrow line-shape signature of the peaks. This observation is in accordance with CD-spectroscopic melting experiments (Supplementary Figure S12). When the CD-temperature row is analyzed for a two-state unfolding process, the melting temperature is determined to be Tm = 38°C meaning that at this particular temperature the RNA is in an equilibrium of 50% unfolded and folded conformations.
Figure 8.
Schematic representation of peptide-induced RNA structural changes. (A) RNA only, (B) RNA in the presence of Tat(44–61) with its N-terminus in blue and its C-terminus in red. At low temperature, the interaction of the peptide with the RNA induces changes in the conformational equilibrium in a way that not only a single helical conformation is present but the helical-conformation is locally disrupted or partially melted. At intermediate temperatures (close to the ‘melting point’) the RNA is anyhow in a conformational equilibrium between helical and partially melted conformations. The addition of peptide to this pool of conformers results in a partial stabilization of the partially melted RNA conformations, which are probably the ones where RNA duplex formation initiates. At high temperatures, Tat(44–61) will stabilize not the completely randomized unfolded form of the RNA, but intermediates that are prone to interaction with a complementary strand of RNA.
The addition of peptide to the 21R+ changed the RNA spectra in a temperature dependent way (Figure 7). A control spectrum of peptide alone demonstrated that RNA aromatic hydrogen shifts did not overlap with peptide peaks (Supplementary Figure S13). Thus, the chemical shift changes upon addition of peptide could be attributed directly to structural changes within the RNA, assuming that shift changes in the peptide do not lead to an overlap in the NMR spectra of the complex. The addition of peptide Tat(44–61) to the RNA induces chemical shift changes in the RNA and leads to a broadening of the signals at low (0°C) and high (37°C) temperatures. At these temperatures, the interaction of the RNA with Tat(44–61) leads to an equilibrium between the selected conformation and the ground-state conformation on a timescale turning the peaks in the spectrum broad. At intermediate temperatures (20°C) (when the signals are already broad and the chemical shift dispersion is limited), the spectrum gets more disperse and more distinct signals and are visible upon addition of peptide. This is due to the selection of a certain conformation from the pool of slowly interconverting RNA conformations (Figure 8).
This conformation selection might be achieved by true selective binding of already formed RNA conformations which relocates the equilibrium between different RNA structures. Another possible reason for the population change is the modulation of RNA structure upon peptide binding. Indeed, it has been shown by other groups that arginine/lysine oligopeptides are able to change DNA conformations by tilting the bases or altering the winding angle (70). Regarded on a molecular level, a lot of encounters between RNA molecules turn out to be fruitless due to unfavorable RNA conformations and/or orientations. Switching the RNA molecules to a favorable conformation and/or orientation would thus increase the probability of procession from an encounter to the transition state of duplex formation. At the level of the RNA molecule ensemble, this activity would have the propensity to accelerate the overall annealing reaction. Thus, we suggest that Tat(44–61) shifts the RNA structure into an annealing-competent form which might be a partly helical and partly melted conformation as indicated by our data.
CONCLUSIONS
HIV-1 Tat as well as its fragment Tat(44–61) have potent nucleic acid annealing activity but do not catalyze strand displacement and are thus nucleic acid annealers. The activity of the model peptide Tat(44–61) is based on ionic interactions between peptide and the RNA backbone. Therefore, positively charged amino acids play a dominant role in annealing acceleration while other amino acids are not important. Both the net charge of the peptide, as well as an exact arrangement of basic amino acids within the peptide, determine the magnitude of the annealing activity. Thermodynamically, Tat(44–61) increases the entropy of the annealing transition state, thereby decreasing the Gibbs free energy. According to previous studies, the peptide-conferred entropy increase is caused by a counterion release upon peptide binding (55,70). The actual acceleration of annealing, however, is probably mainly caused by a conversion of the RNA into an annealing prone conformation. This RNA conformational change in the presence of peptide must be detrimental to the reaction’s entropy but its contribution is apparently too small to compensate the counterion release effect. These findings also explain the equal importance of both the peptide’s overall charge and the basic amino acid distribution: while the peptide net charge dictates RNA/peptide binding and entropy increase (54), the basic amino acid distribution probably mediates the RNA conformational change. We found no evidence for the contribution of the proposed annealing mechanism (A) (active increase of the local RNA concentration, see ‘Introduction’ section) to the annealing activity of the Tat peptide. The potential annealing mechanism (B) (stabilization of the annealing transition state by shielding the negative RNA backbone charges) can be excluded for Tat(44–61) since it would demand a decrease of the activation energy Ea. Considering the promiscuous annealing acceleration of RNAs with different sequences, it seems obvious that Tat(44–61) does not form strong complexes with its substrates. Thus, binding constants might not be of significance when discussing RNA annealers and chaperones. In fact, the interaction of Tat(44–61) and RNA are most probably of a very transient nature which we believe to be a key element to annealing acceleration (72). Although CTAB-, Ncp7- and p53-facilitated annealing apparently underlie different mechanisms, we think that our findings might apply to other facilitators of annealing that are rich in basic amino acids, such as Escherichia coli StpA, ribonucleoprotein hnRNP A1 and guideRNA binding protein gBP21. In each unique case other amino acids (like hydrogen bond-forming ones) might contribute to the activity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of Vienna (to M.D.); Fonds zur Förderung der wissenschaftlichen Forschung (FWF) through a Lise Meitner-Position (M1157-B12 to B.F.), RNA Biology Doktoratskolleg (W1207 to M.D. and R.S.), (F1703 to R.S). Funding for open access charge: Austrian Science Fund FWF.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Peter Steinlein for the fast synthesis of peptides and to Michael Jantsch for help concerning the conduction of the filter binding assays. We would like to thank all members of the Schroeder Lab for helpful discussions on the topic of RNA chaperones. We are indebted to Christina Waldsich, Brooke Morriswood and Jennifer L. Boots for critical reading of the article and helpful comments.
REFERENCES
- 1.Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 2.Uhlenbeck OC. Keeping RNA happy. RNA. 1995;1:4–6. [PMC free article] [PubMed] [Google Scholar]
- 3.Semrad K, Schroeder R. A ribosomal function is necessary for efficient splicing of the T4 phage thymidylate synthase intron in vivo. Genes Dev. 1998;12:1327–1337. doi: 10.1101/gad.12.9.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schroeder R, Grossberger R, Pichler A, Waldsich C. RNA folding in vivo. Curr. Opin. Struct. Biol. 2002;12:296–300. doi: 10.1016/s0959-440x(02)00325-1. [DOI] [PubMed] [Google Scholar]
- 5.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Bläsi U, Schroeder R. RNA Chaperones, RNA Annealers and RNA Helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 6.Cristofari G, Darlix JL. The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:223–268. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- 7.Rajkowitsch L, Schroeder R. Dissecting RNA chaperone activity. RNA. 2007;13:2053–2060. doi: 10.1261/rna.671807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 9.Valdez BC, Henning D, Perumal K, Busch H. RNA-unwinding and RNA-folding activities of RNA helicase II/Gu–two activities in separate domains of the same protein. Eur. J. Biochem. 1997;250:800–807. doi: 10.1111/j.1432-1033.1997.00800.x. [DOI] [PubMed] [Google Scholar]
- 10.Lapadat-Tapolsky M, Pernelle C, Borie C, Darlix JL. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 1995;23:2434–2441. doi: 10.1093/nar/23.13.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang ZS, Chen AY, Wu HN. Characterization and application of the selective strand annealing activity of the N terminal domain of hepatitis delta antigen. FEBS Lett. 2004;578:345–350. doi: 10.1016/j.febslet.2004.11.043. [DOI] [PubMed] [Google Scholar]
- 12.Stewart-Maynard KM, Cruceanu M, Wang F, Vo MN, Gorelick RJ, Williams MC, Rouzina I, Musier-Forsyth K. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J. Virol. 2008;82:10129–10142. doi: 10.1128/JVI.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopkins JF, Panja S, McNeil SA, Woodson SA. Effect of salt and RNA structure on annealing and strand displacement by Hfq. Nucleic Acids Res. 2009;37:6205–6213. doi: 10.1093/nar/gkp646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayer O, Rajkowitsch L, Lorenz C, Konrat R, Schroeder R. RNA chaperone activity and RNA-binding properties of the E. coli protein StpA. Nucleic Acids Res. 2007;35:1257–1269. doi: 10.1093/nar/gkl1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S, Marians KJ. DNA and RNA-DNA annealing activity associated with the tau subunit of the Escherichia coli DNA polymerase III holoenzyme. Nucleic Acids Res. 1995;23:1374–1379. doi: 10.1093/nar/23.8.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Portman DS, Dreyfuss G. RNA annealing activities in HeLa nuclei. EMBO J. 1994;13:213–221. doi: 10.1002/j.1460-2075.1994.tb06251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nedbal W, Frey M, Willemann B, Zentgraf H, Sczakiel G. Mechanistic insights into p53-promoted RNA-RNA annealing. J. Mol. Biol. 1997;266:677–687. doi: 10.1006/jmbi.1996.0813. [DOI] [PubMed] [Google Scholar]
- 18.Muller UF, Lambert L, Goringer HU. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J. 2001;20:1394–1404. doi: 10.1093/emboj/20.6.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller UF, Goringer HU. Mechanism of the gBP21-mediated RNA/RNA annealing reaction: matchmaking and charge reduction. Nucleic Acids Res. 2002;30:447–455. doi: 10.1093/nar/30.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee CG, Zamore PD, Green MR, Hurwitz J. RNA annealing activity is intrinsically associated with U2AF. J. Biol. Chem. 1993;268:13472–13478. [PubMed] [Google Scholar]
- 21.Akhmedov AT, Bertrand P, Corteggiani E, Lopez BS. Characterization of two nuclear mammalian homologous DNA-pairing activities that do not require associated exonuclease activity. Proc. Natl Acad. Sci. USA. 1995;92:1729–1733. doi: 10.1073/pnas.92.5.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baechtold H, Kuroda M, Sok J, Ron D, Lopez BS, Akhmedov AT. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J. Biol. Chem. 1999;274:34337–34342. doi: 10.1074/jbc.274.48.34337. [DOI] [PubMed] [Google Scholar]
- 23.Altmann M, Wittmer B, Methot N, Sonenberg N, Trachsel H. The Saccharomyces cerevisiae translation initiation factor Tif3 and its mammalian homologue, eIF-4B, have RNA annealing activity. EMBO J. 1995;14:3820–3827. doi: 10.1002/j.1460-2075.1995.tb00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skabkin MA, Evdokimova V, Thomas AA, Ovchinnikov LP. The major messenger ribonucleoprotein particle protein p50 (YB-1) promotes nucleic acid strand annealing. J. Biol. Chem. 2001;276:44841–44847. doi: 10.1074/jbc.M107581200. [DOI] [PubMed] [Google Scholar]
- 25.Kwon SH, Lee IH, Kim NY, Choi DH, Oh YM, Bae SH. Translation initiation factor eIF1A possesses RNA annealing activity in its oligonucleotide-binding fold. Biochem. Biophys. Res. Commun. 2007;361:681–686. doi: 10.1016/j.bbrc.2007.07.084. [DOI] [PubMed] [Google Scholar]
- 26.Hitti E, Neunteufl A, Jantsch MF. The double-stranded RNA-binding protein X1rbpa promotes RNA strand annealing. Nucleic Acids Res. 1998;26:4382–4388. doi: 10.1093/nar/26.19.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schumacher MA, Karamooz E, Zikova A, Trantirek L, Lukes J. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell. 2006;126:701–711. doi: 10.1016/j.cell.2006.06.047. [DOI] [PubMed] [Google Scholar]
- 28.Zikova A, Kopecna J, Schumacher MA, Stuart K, Trantirek L, Lukes J. Structure and function of the native and recombinant mitochondrial MRP1/MRP2 complex from Trypanosoma brucei. Int. J. Parasitol. 2008;38:901–912. doi: 10.1016/j.ijpara.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nedbal W, Homann M, Sczakiel G. The association of complementary ribonucleic acids can be strongly increased without lowering Arrhenius activation energies or significantly altering structures. Biochemistry. 1997;36:13552–13557. doi: 10.1021/bi970459e. [DOI] [PubMed] [Google Scholar]
- 30.Jones KA, Peterlin BM. Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 31.Brigati C, Giacca M, Noonan DM, Albini A. HIV Tat, its TARgets and the control of viral gene expression. FEMS Microbiol. Lett. 2003;220:57–65. doi: 10.1016/S0378-1097(03)00067-3. [DOI] [PubMed] [Google Scholar]
- 32.SenGupta DN, Berkhout B, Gatignol A, Zhou AM, Silverman RH. Direct evidence for translational regulation by leader RNA and Tat protein of human immunodeficiency virus type 1. Proc. Natl Acad. Sci. USA. 1990;87:7492–7496. doi: 10.1073/pnas.87.19.7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennasser Y, Jeang KT. HIV-1 Tat interaction with Dicer: requirement for RNA. Retrovirology. 2006;3:95. doi: 10.1186/1742-4690-3-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 35.Lin J, Cullen BR. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J. Virol. 2007;81:12218–12226. doi: 10.1128/JVI.01390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMillan NA, Chun RF, Siderovski DP, Galabru J, Toone WM, Samuel CE, Mak TW, Hovanessian AG, Jeang KT, Williams BR. HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virology. 1995;213:413–424. doi: 10.1006/viro.1995.0014. [DOI] [PubMed] [Google Scholar]
- 37.Cai R, Carpick B, Chun RF, Jeang KT, Williams BR. HIV-I TAT inhibits PKR activity by both RNA-dependent and RNA-independent mechanisms. Arch. Biochem. Biophys. 2000;373:361–367. doi: 10.1006/abbi.1999.1583. [DOI] [PubMed] [Google Scholar]
- 38.Chiu YL, Ho CK, Saha N, Schwer B, Shuman S, Rana TM. Tat stimulates cotranscriptional capping of HIV mRNA. Mol. Cell. 2002;10:585–597. doi: 10.1016/s1097-2765(02)00630-5. [DOI] [PubMed] [Google Scholar]
- 39.Harrich D, Ulich C, Garcia-Martinez LF, Gaynor RB. Tat is required for efficient HIV-1 reverse transcription. EMBO J. 1997;16:1224–1235. doi: 10.1093/emboj/16.6.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kameoka M, Morgan M, Binette M, Russell RS, Rong L, Guo X, Mouland A, Kleiman L, Liang C, Wainberg MA. The Tat protein of human immunodeficiency virus type 1 (HIV-1) can promote placement of tRNA primer onto viral RNA and suppress later DNA polymerization in HIV-1 reverse transcription. J. Virol. 2002;76:3637–3645. doi: 10.1128/JVI.76.8.3637-3645.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Apolloni A, Hooker CW, Mak J, Harrich D. Human immunodeficiency virus type 1 protease regulation of tat activity is essential for efficient reverse transcription and replication. J. Virol. 2003;77:9912–9921. doi: 10.1128/JVI.77.18.9912-9921.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuciak M, Gabus C, Ivanyi-Nagy R, Semrad K, Storchak R, Chaloin O, Muller S, Mely Y, Darlix JL. The HIV-1 transcriptional activator Tat has potent nucleic acid chaperoning activities in vitro. Nucleic Acids Res. 2008;36:3389–3400. doi: 10.1093/nar/gkn177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fanales-Belasio E, Moretti S, Nappi F, Barillari G, Micheletti F, Cafaro A, Ensoli B. Native HIV-1 Tat protein targets monocyte-derived dendritic cells and enhances their maturation, function, and antigen-specific T cell responses. J. Immunol. 2002;168:197–206. doi: 10.4049/jimmunol.168.1.197. [DOI] [PubMed] [Google Scholar]
- 44.Rajkowitsch L, Schroeder R. Coupling RNA annealing and strand displacement: a FRET-based microplate reader assay for RNA chaperone activity. Biotechniques. 2007;43:304. doi: 10.2144/000112530. 306, 308 passim. [DOI] [PubMed] [Google Scholar]
- 45.Hallegger M, Taschner A, Jantsch MF. RNA aptamers binding the double-stranded RNA-binding domain. RNA. 2006;12:1993–2004. doi: 10.1261/rna.125506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vo MN, Barany G, Rouzina I, Musier-Forsyth K. Effect of Mg(2+) and Na(+) on the nucleic acid chaperone activity of HIV-1 nucleocapsid protein: implications for reverse transcription. J. Mol. Biol. 2009;386:773–788. doi: 10.1016/j.jmb.2008.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atkins P, Höpfner A, Schleitzer A, Bär M. Physikalische Chemie. 1996. VCH, Weinheim. [Google Scholar]
- 48.Laidler K, King M. Development of transition-state theory. J. Phys. Chem. 1983;87:2657–2664. [Google Scholar]
- 49.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 50.Wijmenga S, Kruithof M, Hilbers C. Analysis of 1 H chemical shifts in DNA: assessment of the reliability of 1 H chemical shift calculations for use in structure refinement. J. Biomol. NMR. 1997;10:337–350. doi: 10.1023/A:1018348123074. [DOI] [PubMed] [Google Scholar]
- 51.Cromsigt JA, Hilbers CW, Wijmenga SS. Prediction of proton chemical shifts in RNA. Their use in structure refinement and validation. J. Biomol. NMR. 2001;21:11–29. doi: 10.1023/a:1011914132531. [DOI] [PubMed] [Google Scholar]
- 52.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Semrad K, Green R, Schroeder R. RNA chaperone activity of large ribosomal subunit proteins from Escherichia coli. RNA. 2004;10:1855–1860. doi: 10.1261/rna.7121704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mascotti DP, Lohman TM. Thermodynamics of single-stranded RNA binding to oligolysines containing tryptophan. Biochemistry. 1992;31:8932–8946. doi: 10.1021/bi00152a033. [DOI] [PubMed] [Google Scholar]
- 55.Record MT, Jr, Lohman ML, De Haseth P. Ion effects on ligand-nucleic acid interactions. J. Mol. Biol. 1976;107:145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- 56.Huang ZS, Wu HN. Identification and characterization of the RNA chaperone activity of hepatitis delta antigen peptides. J. Biol. Chem. 1998;273:26455–26461. doi: 10.1074/jbc.273.41.26455. [DOI] [PubMed] [Google Scholar]
- 57.Croitoru V, Semrad K, Prenninger S, Rajkowitsch L, Vejen M, Laursen BS, Sperling-Petersen HU, Isaksson LA. RNA chaperone activity of translation initiation factor IF1. Biochimie. 2006;88:1875–1882. doi: 10.1016/j.biochi.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 58.Draper DE. Themes in RNA-protein recognition. J. Mol. Biol. 1999;293:255–270. doi: 10.1006/jmbi.1999.2991. [DOI] [PubMed] [Google Scholar]
- 59.Draper DE. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munroe SH, Dong XF. Heterogeneous nuclear ribonucleoprotein A1 catalyzes RNA.RNA annealing. Proc. Natl Acad. Sci. USA. 1992;89:895–899. doi: 10.1073/pnas.89.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cobianchi F, Calvio C, Stoppini M, Buvoli M, Riva S. Phosphorylation of human hnRNP protein A1 abrogates in vitro strand annealing activity. Nucleic Acids Res. 1993;21:949–955. doi: 10.1093/nar/21.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ammerman ML, Fisk JC, Read LK. gRNA/pre-mRNA annealing and RNA chaperone activities of RBP16. RNA. 2008;14:1069–1080. doi: 10.1261/rna.982908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Le Cam E, Coulaud D, Delain E, Petitjean P, Roques BP, Gerard D, Stoylova E, Vuilleumier C, Stoylov SP, Mely Y. Properties and growth mechanism of the ordered aggregation of a model RNA by the HIV-1 nucleocapsid protein: an electron microscopy investigation. Biopolymers. 1998;45:217–229. doi: 10.1002/(SICI)1097-0282(199803)45:3<217::AID-BIP4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 64.Tsuchihashi Z, Brown PO. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mely Y, de Rocquigny H, Sorinas-Jimeno M, Keith G, Roques BP, Marquet R, Gerard D. Binding of the HIV-1 nucleocapsid protein to the primer tRNA(3Lys), in vitro, is essentially not specific. J. Biol. Chem. 1995;270:1650–1656. doi: 10.1074/jbc.270.4.1650. [DOI] [PubMed] [Google Scholar]
- 66.al-Ghusein H, Ball H, Igloi GL, Gbewonyo A, Coates AR, Mascagni P, Roberts MM. Chemically synthesised human immunodeficiency virus P7 nucleocapsid protein can self-assemble into particles and binds to a specific site on the tRNA(Lys,3) primer. Biochem. Biophys. Res. Commun. 1996;224:191–198. doi: 10.1006/bbrc.1996.1006. [DOI] [PubMed] [Google Scholar]
- 67.Cruceanu M, Urbaneja M, Hixson C, Johnson D, Datta S, Fivash M, Stephen A, Fisher R, Gorelick R, Casas-Finet J. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006;34:593. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen T, Rouzina I, Shklovskii B. Reentrant condensation of DNA induced by multivalent counterions. J. Chem. Phys. 2000;112:2562. [Google Scholar]
- 69.Raspaud E, Olvera de la Cruz M, Sikorav JL, Livolant F. Precipitation of DNA by polyamines: a polyelectrolyte behavior. Biophys J. 1998;74:381–393. doi: 10.1016/S0006-3495(98)77795-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Helene C, Maurizot JC. Interactions of oligopeptides with nucleic acids. CRC Crit. Rev. Biochem. 1981;10:213–258. doi: 10.3109/10409238109113600. [DOI] [PubMed] [Google Scholar]
- 71.Suryawanshi H, Sabharwal H, Maiti S. Thermodynamics of peptide-RNA recognition: the binding of a Tat peptide to TAR RNA. J. Phys. Chem. B. 2010;114:11155–11163. doi: 10.1021/jp1000545. [DOI] [PubMed] [Google Scholar]
- 72.Schreiber G, Haran G, Zhou HX. Fundamental aspects of protein-protein association kinetics. Chem. Rev. 2009;109:839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.