Abstract
FACT complex is involved in elongation and ensures fidelity in the initiation step of transcription by RNA polymerase (pol) II. Histone variant H2A.Z is found in nucleosomes at the 5′-end of many genes. We report here H2A.Z-chaperone activity of the yeast FACT complex on the short, nucleosome-free, non-coding, pol III-transcribed yeast tRNA genes. On a prototype gene, yeast SUP4, chromatin remodeler RSC and FACT regulate its transcription through novel mechanisms, wherein the two gene-flanking nucleosomes containing H2A.Z, play different roles. Nhp6, which ensures transcription fidelity and helps load yFACT onto the gene flanking nucleosomes, has inhibitory role. RSC maintains a nucleosome abutting the gene terminator downstream, which results in reduced transcription rate in active state while H2A.Z probably helps RSC in keeping the gene nucleosome-free and serves as stress-sensor. All these factors maintain an epigenetic state which allows the gene to return quickly from repressed to active state and tones down the expression from the active SUP4 gene, required probably to maintain the balance in cellular tRNA pool.
INTRODUCTION
Eukaryotic genome is organized into transcriptionally inactive, condensed heterochromatin and transcriptionally active euchromatin in different epigenetic states. Chromatin formation, which results in repressive effect on elongation, can suppress non-specific initiations from the transcribed regions as well. A complex, facilitates chromatin transcription (FACT) with two subunits Spt16 and SSRP1 in humans, can counter this nucleosomal obstacle during elongation and even ensure the fidelity of transcription initiation by RNA polymerase (pol) II (1–3). FACT travels with pol II and removes/reinstates the H2A–H2B dimer from the nucleosome after passage of the pol II during elongation (4,5). Yeast FACT consists of three proteins: Spt16, Pob3 and a small, HMG box-containing, non-histone protein Nhp6 (6). Cells with deletion of both the copies of Nhp6 are viable (7), while Spt16/Pob3 are essential for cell viability (8), suggesting their functions are independent of each other.
Nucleosomal arrangement on pol II-transcribed genes follows a general pattern, wherein a nucleosome-free region (NFR) is flanked by two positioned nucleosomes (9). Several studies suggest that tDNAs are generally devoid of nucleosomes (9–11) and hence, refractory to chromatin mediated repression. Out of approximately 280 targets of pol III in the Saccharomyces cerevisiae genome (12,13), 275 genes code for different tRNAs. The most abundant ATP-dependent chromatin remodeler of yeast, RSC which interacts with all the three RNA polymerases, is found on most of the pol III-transcribed genes in yeast (14). Genome-wide localization studies of the chromatin modifying complexes also suggest that chromatin may be having an important regulatory role in expression of these genes (14–16). Recent studies have revealed that histone H2A variant H2A.Z (coded by Htz1 in S. cerevisiae) localizes to hundreds of repressed or basal, preferably TATA-less promoters of pol II (9). It acts as boundary mark for pol II-transcribed genes and is found around −200 and +200 bp from the transcription start site (TSS) of the tRNA genes (9). However, the role of H2A.Z in pol III transcription is largely an unexplored area. Earlier studies from our lab had shown that in vivo chromatin structure of a yeast pol III-transcribed gene, SNR6 is remodelled by RSC in response to nutritional stress but H2A.Z, present in the upstream, regulatory nucleosome is not required for its activation in vivo (17).
Much of the in vitro transcription by pol III has been studied using yeast SUP4, one of the tRNATyr gene family members. Like a typical pol III-transcribed gene, SUP4 has two internal promoter elements 55 bp apart, boxes A and B, but no upstream regulatory elements. The transcription complex formation is initiated with binding of the transcription factor (TF) IIIC to the boxes A and B, followed by the recruitment of the initiation factor IIIB, 30 bp upstream of TSS at +1 (18). Short length of genes facilitates terminator-dependent recycling of pol III which involves direct transfer of pol III to the TSS after termination in each round, giving several fold increase in transcription in vitro (19). Transcription by pol III is tightly regulated in response to any stress by a central regulator Maf1 (20,21). Nutrient deprivation like nitrogen and carbon starvation leads to immediate downregulation of transcription by pol III (22) and pol III is lost from its target genes under the conditions of repression (23).
In this study, we have shown the role of H2A.Z, RSC and FACT in modulating the transcription of the yeast tRNATyr gene. Working as H2A.Z chaperone, Spt16 with the help of SWR1 complex deposits as well as removes H2A.Z in the gene flanking nucleosomes. RSC promotes loading of FACT on the gene, maintains the transcribed region of SUP4 nucleosome-free and keeps a nucleosome close to the gene terminator region in vivo, which probably prevents the facilitated recycling of pol III, resulting in lower transcription from SUP4 in active state. These studies show a novel mechanism of transcriptional regulation of SUP4 by all these activities which together keep the SUP4 expression at a low level.
MATERIALS AND METHODS
Plasmid DNAs, primers, yeast strains and growth
Plasmid pLNwt containing the 256 bp DNA from the yeast SUP4 gene locus was gift. Sequences of the primers and list of the yeast strains used in this study are given under the Supplementary Tables S1 and S2, respectively. Yeast cultures were grown in YEP (yeast extract and peptone) media containing 2% glucose. Cells were nutritionally deprived by shifting to 0.15× YEP without any carbon source (23) and allowed to grow for further 1.5 h, unless otherwise stated. Yeast cultures of temperature sensitive mutants were rapidly shifted to 37°C by adding pre-warmed media to the cultures grown at 30°C.
In vitro chromatin assembly and transcription
Chromatin was assembled on pLNwt using S-190 extract of Drosophila embryos and in vitro transcription using pure TFIIIB, TFIIIC and pol III was performed as previously described (24). 6× His-tagged Nhp6A was purified from the overexpression clone (gift from Ian Willis). Spt16–Pob3 dimer was purified from strain carrying TAP-tagged Spt16 using standard tandem affinity purification protocol (25).
Antibodies
Antibodies against H3 (Ab46765), H2A.Z (Ab4626) and Myc (9E10) were purchased from Abcam. Anti-HA, anti-myc (05-904, 05-724MG; Millipore), IgG sepharose (GE Healthcare), FLAG M2 agarose (Sigma) were purchased while anti-Spt16 and anti-Pob3 (Tim Formosa) were gifts.
Chromatin structure, chromatin immunoprecipitation and RNA in vivo
Chromatin structure analysis by the IEL method, chromatin immunoprecipitation (ChIP) and real time PCR, RNA extraction and quantification were performed as described (17), and repeated at least three times for each experiment. Fold enrichments in ChIP and real time PCR assays were calculated as occupancy normalized against TELVIR region and average from three independent experiments with error bars are shown. For high-resolution footprinting, digestion of chromatin with MNase in vivo and sample preparations were as described for the IEL method. Samples were used for primer extension and products resolved on 8 M urea–6% polyacrylamide gel as described earlier (24). Further details for each method and primers can be found in Supplementary Data. Secondary digestion of DNA for IEL analysis was with a HindIII site present 860 bp downstream of the SUP4 gene and probe was 150 bp DNA abutting HindIII site.
Co-immunoprecipitations and pull-downs
Co-immunoprecipitation (IP) experiments (26) using yeast whole cell extract and in vitro pull downs to follow physical interactions of tagged H2A.Z and Spt16 proteins were performed as described under Supplementary Data.
RESULTS
We studied the role of chromatin in regulating SUP4 transcription, in vitro as well as in vivo. Recruitment of various activities to SUP4 in vivo under two different nutritional states in yeast was ascertained by ChIP and real time PCR assays.
Chromatin influences the transcription of SUP4 gene
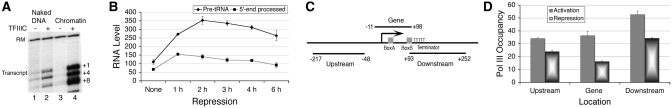
Naked DNA transcription of SUP4 is known to be TFIIIC-dependent (Figure 1A, lanes 1 and 2). Transcription from SUP4 using pure TFIIIC, pol III and recombinant TFIIIB gives two additional transcripts, corresponding to downstream initiations from the positions +4 and +8 with respect to the TSS at +1. Similar to SNR6 (24), addition of TFIIIC to the chromatin assembled on the plasmid pLNwt yields 5-fold more transcript (Figure 1A, lane 2 versus 4). Chromatin formation suppresses the non-specific transcription from the plasmid backbone (data not shown) which may contribute to the observed higher than naked DNA (lane 2) level of transcription from chromatin (lane 4). Unlike SNR6, TFIIIC addition does not result in a nucleosome positioning on the transcribed region of the gene (Supplementary Figure S1) suggesting absence of nucleosome on the gene may be due to the ATP-dependent chromatin remodelling activities present in the S-190 extract, which probably mobilize the nucleosome away from the gene region, making it transcriptionally active in the presence of TFIIIC. These in vitro observations suggest that chromatin may influence SUP4 expression in vivo as well.
Figure 1.
Transcription of SUP4 gene and chromatin. (A) Transcription from chromatin is higher than naked DNA in vitro. TFIIIC-dependent transcription of naked DNA (cf. lanes 1 and 2) and chromatin (cf. lanes 3 and 4) produces two additional transcripts due to downstream initiations from the positions +4 and +8 with respect to TSS at +1. RM represents the position of recovery marker in the gel. Transcript from +1 site is marked. (B) SUP4 RNA synthesis under nutritional stress. The total SUP4 transcript levels at different time points of repression are normalized against the U4 transcript used as an internal control. Average and scatter of RNA levels estimated from three independent experiments are plotted. (C) Schematic diagram showing the amplicon sizes and primer pairs used in real time PCR and chromatin immunoprecipitation studies. Positions of the gene sequence elements with respect to the three amplicons representing the upstream, gene and downstream regions of SUP4 gene are shown. Bent arrow represents TSS. For all ChIP data, averages from three independent experiments with error bars, normalized against TELVIR region, are calculated. (D) Relative occupancy of FLAG-tagged C160 subunit of pol III on SUP4 in both active and repressed conditions. Nutrient deprivation with 0.15× YEP medium lacking glucose was used to repress the pol III transcription for 80 min.
We measured SUP4 RNA levels under active and repressive conditions of pol III transcription (Figure 1B). Surprisingly, SUP4 pre-tRNA levels increase for the first 2 h while levels of 5′-end processed transcript do not change after 1 h of repression. Levels do not change much with prolonged starvation due to high stability of tRNAs. This suggests pre-tRNA levels increase after 1h of repression because processing stops but the increase during the first hour may be due to increased transcription. Starvation induced repression is accompanied by loss of pol III from the target genes (23). ChIP of Flag-RPC160 subunit using amplicons shown in the Figure 1C showed that after 80 min of nutritional stress, pol III levels on the gene drop by only 50% on the transcribed region (Figure 1D). This suggests that transcription from the gene may be continuing during initial repression and a change in the transcript level may be related to even chromatin structure around the gene.
A nucleosome-free gene region is flanked by two nucleosomes
Using micrococcal nuclease (MNase) digestion instead of sonication, we probed the SUP4 gene region (Figure 1C) for the presence of core histones by the ChIP and real time PCR method. As compared to nucleosome-dense TELVIR region, the gene region is found histone-depleted where levels appear close to background level (Figure 2A and B). Comparatively lower enrichment of H2B and H3 in downstream region as compared to upstream, suggests that the nucleosomes may be present on both sides of the SUP4 gene in vivo (−1 and +1, Figure 2C), but the +1 nucleosome is not well positioned. We confirmed the histone-depleted nature of the gene region in vivo by high-resolution MNase footprinting. As compared to naked DNA, chromatin shows MNase hypersensitivity at the position from +93 to +96 in the gene region (Figure 2D), while the region downstream of the terminator is protected (lanes 3, 4 versus 1, 2), suggesting presence of nucleosomes. The MNase digestion profile of rest of the gene sequence is same for both chromatin and naked DNA, suggesting the observed hypersensitivity probably represents the 5′-boundary of the +1 nucleosome, downstream of the gene.
Figure 2.
Effect of Repression and H2A.Z deposition on the SUP4 gene locus in vivo. Chromatin organization at the SUP4 locus is probed by ChIP/Real Time PCR and footprinting methods. (A) Relative occupancy of FLAG-tagged H2B. (B) Relative occupancy of Myc-tagged H3. (C) Nucleosomes flank the histone-free SUP4 gene: Cartoon showing positions of the nucleosomes −1 and +1, relative to the cis elements of the gene, as marked in the panel 1C. (D) High-resolution MNase footprinting in vivo. MNase digested chromatin and naked DNA samples were used for extension with a primer which hybridizes to the bottom strand, 13 bp downstream of the TSS. Lane M shows a 50 bp ladder used as molecular size marker while GATC represent the sequencing reaction over genomic DNA. Two levels of MNase digestions are shown for SUP4 as naked DNA (lanes 1 and 2) and chromatin (lanes 3 and 4). Positions of the box B and terminator are marked while grey ovals represent nucleosomes. Position of MNase hypersensitivity immediate upstream of the terminator is marked with a short vertical bar in the left-hand side. (E) Low-resolution chromatin structure analysis by the IEL method. Grey ovals denote the nucleosomal size protections and arrows mark the MNase cut sites in the chromatin. Gene region is marked with a rectangle. All numbers represent bp with respect to TSS at +1. Numbers on the left-hand side mark the MNase cuts seen on the naked DNA in lanes 1 and 2. Numbers on the right-hand side mark the MNase cuts seen on chromatin and −1, +1 mark the gene flanking nucleosomes. MNase cleavage pattern of wild type (lanes 3–6) and Htz1Δ cells (lanes 7–10) without (lanes 3, 4 and 7, 8) or with nutritional stress (lanes 5, 6 and 9, 10) for 2 h is shown. (F) H2A.Z deposition in the nucleosomes around the SUP4 gene in vivo. Relative occupancies of FLAG-tagged H2A and H2A.Z on SUP4 against TELVIR region are shown. H2A.Z levels increase with repression.
Nucleosome positions near the gene were further confirmed by analysing the chromatin structure of SUP4 tDNA locus in vivo using the indirect-end-labelling (IEL) approach (Figure 2E). As compared to MNase digestion pattern of naked DNA, the chromatin structure in vivo shows presence of an array of positioned nucleosomes upstream of the gene sequence (cf. lanes 1, 2 and 3, 4). The transcribed region of the gene is not cut well by MNase, but the broad band around +98 bp, downstream of the box B at +80 bp, shows a MNase hypersensitivity of the gene, not seen on naked DNA. Thus, considering the results from the Figure 2A and B, and the low-resolution nature of the IEL mappings, the region from −47 to +98 (±20) bp encompassing the SU4 transcribed region is devoid of a nucleosome. Protection of naked DNA sites at −151 and −63 bp positions shows the presence of the −1 nucleosome positioned between −242 and −47 bp giving a median at −140 bp as opposed to −130 bp mapped by the ChIP-seq method (http://h2az.atlas.bx.psu.edu) for the same nucleosome. The protection of naked DNA sites, including the +192 bp position but absence of any strong boundaries between +100 and +350 bp positions (cf. lanes 3, 4 and 1, 2) suggests that as compared to the −1 nucleosome, the boundaries of +1 nucleosome are comparatively diffused. The span of this nucleosome matches well with the position of a fuzzy nucleosome between +90 and +300 bp (http://h2az.atlas.bx.psu.edu). Fuzzy nature of the +1 nucleosome may also be the reason that H2B and H3 levels in the downstream region are lower as compared to the upstream region in active state of the gene (Figure 2A and B). Increase in H2B levels only in +1 nucleosome under repression (Figure 2A) suggests a differential histone dynamics in both the nucleosomes.
Higher association of H2A.Z with SUP4 gene under repressed state
Nucleosome mobility and occupancy on the promoter regions are often associated with the state of activity of a gene (11). As above data show absence of nucleosomes on the SUP4 in its active state, we followed the nucleosome dynamics on the SUP4 gene locus under repression conditions using IEL method. No change in the arrangement of nucleosomes upstream or downstream of the gene after repression (Figure 2D, lanes 5 and 6) is seen. But, the MNase sensitivity of the gene at +98 bp is reduced, suggesting a change in +1 nucleosome region, probably resulting in further terminator exposure and higher transcription in the repressed state (Figure 1B).
Nucleosomes flanking the tRNA genes (9) are reported to have H2A.Z. We looked for the presence of H2A and its only variant in the budding yeast, H2A.Z, in both the nucleosomes. As opposed to negligible H2A levels, H2A.Z shows significant enrichment in both, especially the −1 nucleosome (Figure 2F, Supplementary Figure S2). This profile of H2A.Z is similar to the general pattern of H2A.Z-containing nucleosomes flanking an NFR around the TSS, reported for active pol II-transcribed genes (10). However, IEL analysis shows that similar to SNR6 (17); the chromatin structure remains unperturbed in the absence of H2A.Z, even under repression (Figure 2D, lanes 7–10), supporting a previous report showing no role of H2A.Z in general nucleosome positioning or maintenance of nucleosomal organization (27). H2A.Z associates with both active and inactive genes (28). On SUP4, H3 levels do not change (Figure 2B) but H2A.Z (Figure 2F) levels increase under repression. As compared to wild-type cells, higher SUP4 RNA levels found in htz1Δ cells (Figure 3A), suggest its repressive role in SUP4 expression. Decrease in occupancy upon activation of SUP4 is consistent with the reports showing that H2A.Z is lost during the active transcription of a gene (29,30).
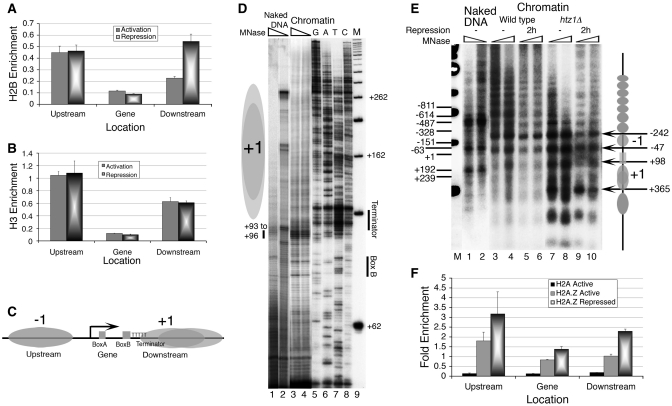
Figure 3.
RSC has a repressive role in SUP4 expression. (A) RNA was extracted from different mutants under active transcription conditions and SUP4 levels estimated as given under the ‘Material and Methods’ section. U4 was used as the internal control to normalize the SUP4 transcript levels. Average and standard deviations calculated from three independent isolations are plotted. (B) Relative enrichment of myc-tagged Sth1 subunit of RSC complex on SUP4 in both active and repressed conditions. (C) IEL analysis of the chromatin structure in RSC mutant and wild-type yeast cells. Ovals represent the nucleosomes; arrow and rectangle mark the gene position. Bent arrow marks the TSS, asterisk marks the MNase hypersensitive site on the gene region. Digestions with two MNase levels are shown for each strain. (D) Comparison of the digestion profiles of the similarly digested DNA in lanes 2 and 4 from the panel C. Profiles were generated using the PhosphorImager and the Image Gauge (Fuji) software. Asterisk marks the MNase hypersensitive site on the gene region while short arrow marks the direction of nucleosome shift. Box marks the gene region, bent arrow the TSS and Term the gene terminator. Light and dark ovals represent the positioned nucleosomes in wild-type and mutant cells, respectively. (E) Relative enrichment of myc-tagged Nhp6 in rsc4-Δ4 mutant in active condition. (F) High-resolution MNase footprinting. Samples were prepared and primer extension reactions carried out as described under the legends for the Figure 2D. The primer hybridizes to the bottom strand, 92 bp downstream of the TSS. Nucleosomes in rsc4-Δ4 mutant are restricted to downstream of the terminator. Three levels of MNase digestions are shown for wild type (lanes 1–3) and mutant (lanes 4–6) cells. Dark grey oval represents the downstream nucleosome in mutant while light grey ovals represent fuzzy nucleosomes in wild-type condition. Numbers in the right hand side denote bp with respect to TSS while short vertical bar marks the position of MNase hypersensitivity in lanes 4–6.
Shift in the position of +1 nucleosome modulates SUP4 transcription
In vitro results from the Figure 1A suggest involvement of ATP-dependent chromatin remodelers in SUP4 transcriptional activation. Therefore, we monitored the SUP4 tRNA levels in cells mutated for a subunit of the major chromatin remodelling complexes of yeast. Deletion of Isw2 or Chd1 does not have any effect on SUP4 levels (Figure 3A), and both of them did not show enrichment near the gene (data not shown). In comparison, RSC localizes to the region (Figure 3B) and an increase in RNA level is found (Figure 3A) in cells with an Rsc4 mutation known to cause a loss of interaction with pol III (31). Similar to repressed state (Figure 1B), higher RNA levels in both H2A.Z and RSC mutants could be due to higher transcription as well as defective processing. Decrease in occupancy of RSC (Figure 3B) along with increase of H2B (Figure 2A) under repression, only in the downstream region, suggests that the repressive effect of RSC on SUP4 expression may also be due to a remodelling of +1 nucleosome by RSC.
We used IEL method, which can map the nucleosomes with fair accuracy (32), to see the remodelling, suggested by above data. Digestion pattern of chromatin in rsc4-Δ4 mutant does not show any apparent changes in the chromatin structure (Figure 3C) except a reduced MNase sensitivity (cf. lanes 1–4) near the box B (asterisk), which could be either due to loss of nucleosome and its boundary or further encroachment by a nucleosome from the downstream region. Comparison of the profiles of similarly digested DNA in lanes 2 and 4 (Figure 3D) shows a change further downstream, showing two positioned nucleosomes with new boundaries (right-side cartoon, Figure 3C; and darker ovals, Figure 3D), in place of a fuzzy +1 nucleosome. Physical and functional interaction of a subunit of yeast FACT, Nhp6 with RSC helps its loading onto the nucleosome in vitro (33). Figure 3E shows that the Nhp6 shows comparatively higher enrichment in the upstream region of SUP4 from where it is lost in rsc4-Δ4 mutant, suggesting RSC may help recruit Nhp6 on SUP4. Chromatin shows MNase hypersensitivity in the presence of Nhp6 in vitro (Supplementary Figure S3A) as Nhp6 binding changes the conformation of DNA (34). Thus, loss of MNase hypersensitivity of the gene region in the rsc4-Δ4 mutant (Figure 3D) may be due to loss of Nhp6 as well as the shift of boundary of the +1 nucleosome.
To resolve this further, we used the high-resolution MNase footprinting in vivo (Figure 3F). The presence of a strong cut around +160 bp position in the mutant as compared to the wild-type condition, suggests the presence of the common boundary of the two positioned nuclesomes at ∼50 bp downstream of the terminator. Out of the two, positioning of the gene-proximal nucleosome places its terminator at the dyad axis, though its footprint is not easy to ascertain since the chromatin-specific cut in the gene region is broad and diffuse. Therefore, the remodelling activity of RSC in the wild type cells must be keeping the gene region nucleosome-free and +1 nucleosome fuzzy, making it encroach the second terminator of SUP4, T2 at +108 bp (35) in active state.
SUP4 gene in vivo is flanked by a Ty element in upstream and an ORF of unknown gene in the downstream region. The TSS of both the divergent genes are found two nucleosomes away from the SUP4. We did not find either transcript of the downstream gene or effect of repression or RSC mutation on the upstream transcript (data not shown), confirming the observed changes in the gene downstream region are related directly to SUP4 expression.
Nhp6 regulates the transcription initiation from the SUP4 chromatin
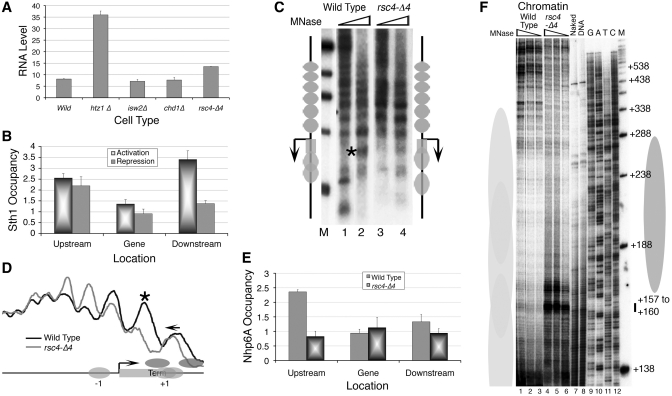
Nhp6 is reported to suppress the downstream initiations from naked SUP4 DNA in vitro (36). In a titration experiment (Figure 4A), addition of purified, recombinant Nhp6A to chromatin also abolishes downstream initiations and the right SUP4 transcript increases with increasing Nhp6 (lanes 1–3). However, in contrast to steady increase in naked DNA transcription with addition of upto ∼300 ng Nhp6 (data not shown), transcription from chromatin drops after the saturation levels of Nhp6 at 60 ng (lanes 4–10, Figure 4A). Expecting a similar effect of Nhp6 in vivo, we used an intron-specific primer which also detects transcripts from two isogenes of SUP4, all differing from each other by 1 or 2 bp in TSS selection (36). As marked in the Figure 4B, −1 represents barely visible SUP2 transcript; SUP3 and SUP4 appear together at +1; while +2 and +3 represent downstream initiated transcripts from SUP3 and SUP2, respectively. Deletion of Nhp6 is found to reduce transcription from the right positions in all three genes (−1 and +1 bands in lanes 3, 4 versus lanes 1, 2). In Nhp6ΔΔ cells, the downstream initiated transcripts from +2 and +3 (transcripts from SUP3 and SUP2 respectively), show an increase and a transcript initiated from +8 position, probably from SUP4, is also found, as seen in vitro (cf. Figures 1A and 4B).
Figure 4.
FACT has inhibitory effects on SUP4 transcription. (A) Effect of Nhp6 on SUP4 transcription in vitro. Transcription of S-190 assembled chromatin over SUP4 was followed using pure yeast pol III transcription machinery in the absence (lane1) and presence (lanes 2–10) of increasing amounts (36, 60, 72, 84, 99, 108, 120, 132 and 144 ng) of pure Nhp6A. Transcript and RM are as given for the Figure 1A. (B) Nhp6 confers the fidelity of transcription in vivo. Total RNA was isolated and used to generate extension products using the intron-specific primer of SUP4. The three products obtained from the two yeast strains using 5 or 10 µg total RNA are resolved on a gel as described under methods. Arrow marks the major SUP4 transcript initiating from the +1 bp position. Numbers on the right-hand side denote the positions of all the transcripts with respect to +1 site of SUP4, as explained in the text. (C) Relative enrichment of myc-tagged Nhp6A in both active and repressed states. (D) Relative enrichment of myc-tagged Spt16 in active and repressed states. (E) Relative enrichment of TAP-tagged Pob3 in both active and repressed states. (F) Effect of FACT on the chromatin transcription of SUP4 in vitro. Transcription of S-190 assembled chromatin over SUP4 was followed using pure yeast pol III transcription machinery. Pure FACT components in excess were added as indicated.
Nhp6 can recruit Spt16 and Pob3 to nucleosome to form the stable FACT complex (6). Accordingly, presence of Nhp6 specially in the upstream region of SUP4 is matched with levels of other two FACT subunits Spt16 and Pob3 in active state (Figure 4C–E) and its increase in all three regions under repression (Figure 4C) is in agreement with its inhibitory role in SUP4 transcription in vitro, at higher levels. However, unlike Nhp6, Spt16 and Pob3 show loss from the repressed gene (Figure 4D and E) and mutations in Spt16 and Pob3 did not affect the fidelity of pol III (not shown), suggesting the functions of Nhp6 and Spt16/Pob3 are independent of each other. In agreement to this, when purified Spt16 and Pob3 are added for in vitro transcription, downstream-initiated transcripts from chromatin are not abolished (lane 3 versus 1, Figure 4F). This result is in contrast to reported association of Spt16 with the increase in transcription fidelity for many of the pol II-transcribed genes (5,37). Being in excess, Nhp6, wherever added, shows increase in fidelity (lanes 2 and 4) but decrease in total transcript, whether added alone (lane 2) or with Spt16/Pob3 (lane 4). These results show that all the three components of yFACT occupy the SUP4 gene simultaneouly in vivo but the ability to ameliorate the downstream initiated transcripts resides with Nhp6 alone.
Repressive role of FACT in tDNA transcription
Apart from being a part of the FACT complex, Spt16 has a specific role in chromatin organization on certain pol II-transcribed genes (38). The IEL analysis of Nhp6 as well as Spt16 mutants did not show any change in gross chromatin structure around SUP4 (Supplementary Figure S3B), suggesting their association with SUP4 locus may have effect at some other level.
Spt16 is known to differentially affect transcription of the pol II-transcribed genes (38). The SUP4 RNA levels in cells defective in either Spt16 or Pob3 (Figure 5A) show an increase while pol II-transcribed genes CMD1 and U4 do not show change in their transcript levels in Spt16 mutant cells (Supplementary Figure S4). In contrast to roles of FACT subunits in pol II transcription activation (5,6,39,40), the higher SUP4 levels in Spt16 and Pob3 mutants suggest a repressive role for them in SUP4 expression.
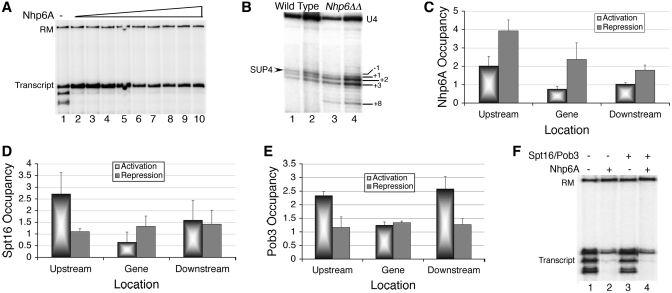
Figure 5.
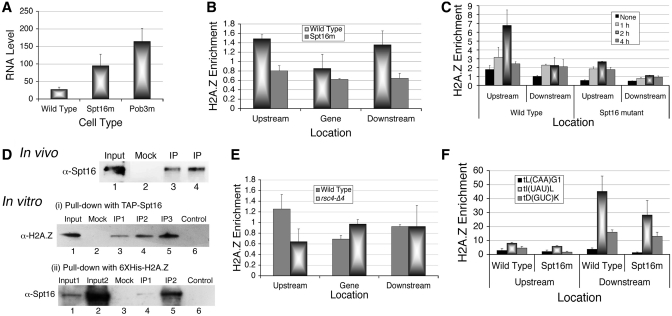
FACT is a H2A.Z chaperone. (A) In vivo analysis of SUP4 expression in mutants of Spt16 and Pob3. Total RNA was isolated and intron-specific primer extension products of SUP4 were visualized by phosphorImaging of the gel as described under methods. RNA levels were normalized against U4 and average from three independent experiments with scatter are shown. (B) Relative enrichment of HA-tagged H2A.Z in the absence of functional Spt16 in active condition. Comparative levels of H2A.Z are low in both the nucleosomes in Spt16 mutant cell. (C) HA-H2A.Z levels around the SUP4 gene in wild type cells change with time in parrallel to transcriptional repression under nutrient starvation. (D) Co-immunoprecipitation of H2A.Z and Spt16 in vivo and in vitro. Mock was as described under Supplementary Data. Input lanes are loaded with one-third (in vivo) or one-fourth (in vitro) of the sample as compared to IP (immunoprecipitate) lanes. Spt16 was at increasing amounts in IP1, 2 and 3. Upper panel: In vivo, H2A.Z-HA is immunoprecipitated with anti-HA antibody and IP is probed with anti-Spt16 antibody. Lanes 3 and 4 are the duplicates of the immunoprecipitation (IP). Lower panel: (i) and (ii) show in vitro pull-downs, using purified Spt16/Pob3 or 6XHis-tagged H2A.Z; probed with anti-H2A.Z or anti-Spt16 antibodies, respectively. Control shows the immobilized bait protein alone on beads while mock lanes show incubation of the prey with bare beads. (E) Relative enrichment of HA-tagged H2A.Z in rsc4-Δ4 mutant in active condition. (F) Relative enrichment of H2A.Z-HA on tDNA loci present on three different chromosomes.
Spt16 is required for H2A.Z deposition on SUP4 locus
Spt16 has been shown to work as chaperone for H2A (4) and H2A.X (41). It co-purifies with H2A.Z in the absence of Nap1 and H2A.Z-specific chaperone Chz1 and helps in SWR1-mediated exchange of H2A.Z–H2B dimer in vitro (42). We found a loss of H2A.Z levels on SUP4 in the absence of Chz1 (data not shown) and Swr1 (Supplementary Figure S2) suggesting Swr1 and Chz1 are required for H2A.Z deposition at SUP4 locus in vivo. A loss of Spt16/Pob3 (Figure 4D and E) but increase of H2A.Z (Figure 2F) under repression suggest Spt16 may be required for H2A.Z removal from nucleosomes. Similarly, a loss of H2A.Z in the absence of functional Spt16 (Figure 5B) indicates an involvement of Spt16 in the deposition of H2A.Z.
Spt16 is known to interact with histone H3. In the absence of Spt16, a rapid loss of H3 is seen on both active and inactive genes (43). This suggests that the observed H2A.Z dynamics on SUP4 could be indirectly due to H3/nucleosome dynamics and may not have any direct correlation with Spt16. However, total H3 as well as H2A.Z levels in mutant and wild type cells did not show any significant difference (Supplementary Figure S5A), suggesting lack of Spt16 activity may be directly responsible for the lower H2A.Z levels in nucleosmes flanking the SUP4 (Figure 5B). Mutant cells showed a differential behaviour of H3 in −1 and +1 nucleosomes (Supplementary Figure S5B), again suggesting both the nucleosomes follow different dynamics under repression, and may be targets of different chromatin modifiers. Preferential occupancy of FACT in the upstream region and NFR on the transcribed region of SUP4 advocate for requirement of FACT activity only in the −1 nucleosome, which shows enrichment of H2A.Z.
While general pol III transcription is repressed within minutes of nutrient deprivation, the SUP4 transcript (Figure 1B) and H2A.Z levels (Figure 5C) show steady increase till 2 h. The increase probably reflects the initial cellular response to the stress wherein H2A.Z plays a positive role as discussed later. The H2A.Z shows significantly higher level in the −1 nucleosome after 2 h repression. Levels drop after 2 h, in parallel to transcription levels but remain at higher than active state in the later stages of repression, suggesting its loss may be necessary for activation. Unlike H3, H2A.Z levels in both nucleosomes are comparatively lower in Spt16 mutant cells (Figure 5C) but time course of H2A.Z dynamics looks similar in both types of cells. While a slight increase in H3 could be the reason behind the small increase of H2A.Z in the −1 nucleosome in mutant cells (Supplementary Figure S5B) under repression, it could also be due to activity of the H2A.Z specific chaperone Chz1 (42). These differences in dynamics of the two suggest that H2A.Z does not follow H3 dynamics on SUP4. While Chz1 may be an alternate chaperone, Spt16 is required on SUP4 for H2A.Z deposition during active state and removal in repressed state. Thus, working as H2A.Z chaperone along with Chz1, Spt16 may deposit as well as remove H2A.Z from the SUP4 flanking nucleosomes. This is not surprising, as most of the histone chaperones show broad specificity and often work in redundant manner in vivo (44). Therefore, H2A.Z levels at any point of time may represent the outcome of the removal/deposition by Spt16 and Chz1.
Higher RNA and lower H2A.Z levels in Spt16 mutant and higher H2A.Z but lower Spt16 in repressed wild-type cells suggest a close relation between H2A.Z dynamics, FACT and transcription of SUP4. Unlike SUP4, H2A.Z on CMD1 and U4, two Spt16-independent genes transcribed by pol II (6, Supplementary Figure S4) shows lower levels under active as well as repressed states in mutant cells (Supplementary Figure S5C). Therefore, Spt16 is required for the deposition of H2A.Z in active state although the transcription of these genes does not require Spt16. As Spt16 is a known chaperone for other H2A variants, loss of the variant H2A.Z from the genes in the mutant cells reflects a direct outcome of defect in H2A.Z deposition by Spt16 in active state.
FACT is a chaperone of H2A.Z for pol III-transcribed genes
A chaperone role would require direct interaction between H2A.Z and Spt16. Using yeast cells carrying HA-tagged H2A.Z, we found that Spt16 co-immunoprecipitates with H2A.Z in vivo (Figure 5D, upper panel), while an N-terminal mutant of Spt16 could not pull down H2A.Z (Supplementary Figure S6A), re-enforcing the possibility of Spt16 working as H2A.Z chaperone in vivo. For confirming direct interaction of the two in vitro, we purified TAP-tagged Spt16-Pob3 heterodimer from yeast cells and 6XHis-tagged recombinant yeast H2A.Z from E. coli cells (Supplementary Figure S6B). We used the tagged proteins immobilized on Calmodulin sepharose or Ni-NTA agarose beads, respectively, and added the other partner as pure protein (Figure 5D, lower panel). We found that both Spt16 and H2A.Z can efficiently pull-down each other, demonstrating a direct physical interaction between the two. As Nhp6/FACT are enriched specifically in the upstream region (Figure 4C and D), H2A.Z levels also show larger changes only in the −1 nucleosome (Figures 2F and 5D). All these results support a H2A.Z chaperone role for FACT on SUP4 locus. In agreement with Nhp6 requirement for Spt16/Pob3 binding to nucleosome (45) and loss of Nhp6 in rsc4-Δ4 mutant; H2A.Z is specifically lost from the −1 nucleosome in this mutant (Figure 5E), suggesting Nhp6/FACT may be a link between RSC and H2A.Z. Nucleosomes flanking NFRs containing tDNA sequences (9) are reported to have H2A.Z and recently human FACT was shown to associate with some pol III-transcribed genes (46). We monitored H2A.Z levels in the absence of functional Spt16 on some more tRNA genes (Figure 5F). Similar to SUP4, H2A.Z levels in the mutant cells show significant loss on all these genes, suggesting FACT may have a general role in H2A.Z dynamics on pol III-transcribed genes.
Taken together, results presented in this study show while Nhp6 is required for fidelity; loss of RSC activity, H2A.Z or the FACT components results in higher SUP4 RNA levels in active state, suggesting they may be working in the same pathway, probably to reduce the SUP4 expression. This may be a novel mode of gene regulation whereby all chromatin related activities target the gene to repress it rather than activate in active state. Importantly, regulation of H2A.Z levels in −1 nucleosome by FACT as H2A.Z chaperone and shift in position of +1 nucleosome by RSC in direct response to transcription repression show a differential role of both the gene flanking nucleosomes in the gene expression.
DISCUSSION
According to the general belief, the tRNA genes are largely devoid of well-defined, positioned nucleosomes obviating the need for epigenetic modes of regulation. In the genome-wide sequence-directed nucleosome position predictions (http://genie.weizmann.ac.il/software/nucleo_prediction.html), the SUP4 gene sequence is predicted to exclude nucleosomes from the gene region. Actively transcribed SUP4 gene on a multicopy plasmid, was shown to resist incorporation into a positioned nucleosome in vivo (47). Using salt-dilution chromatin assembly method, we found that the SUP4 gene excludes nucleosome deposition over itself in vitro while a single nucleosome is positioned in the upstream region (not shown). Therefore, sequence-directed nucleosomal absence near the SUP4 tDNA in vivo could potentially exclude role of all epigenetic mechanisms in regulating SUP4 transcription. Results presented in this study have shown a central role for H2A.Z, RSC and FACT in tRNA transcription and suggest a complex interplay of a number of epigenetic processes in regulating SUP4 gene.
H2A.Z as stress-receptor in SUP4 expression
Presence of H2A.Z may have important biological implications (48). Yeast H2A.Z, present in many loci throughout the genome, is implicated in locus-specific activation and repression of genes (48–50). The initial increase of SUP4 transcription under repression is similar to induction of many stress-response genes shortly after stress signal is received (51,52). During first 2 h, when pol III levels drop, increase in Swr1 levels and loss of Spt16 may increase the H2A.Z levels in −1 nucleosome. The drop in H2A.Z levels after prolonged repression probably poises the gene for the activation process through a quick H2A.Z-independent trigger. H2A.Z presence in the nucleosome reduces its mobilization by a chromatin remodeller (27). Thus, increase in H2A.Z under repressed condition may help keep the −1 nucleosome in its place while loss of H2A.Z and increase of RSC in active state would delocalize the downstream nucleosomes making the +1 nucleosome fuzzy. Thus, H2A.Z confers differential behaviour on these nucleosomes in response to repression while its dynamics on SUP4 is not related to active transcription. These observations suggest H2A.Z serves as a starvation-respone factor for SUP4, similar to its recently suggested thermosensory role (53). Significantly, on two of pol II-transcribed genes as well, which show a delayed repression, late increase of H2A.Z levels is seen (Supplementary Figures S4 and S5C), suggesting H2A.Z may be a general stress-response factor.
Role of FACT components on SUP4 gene
Our results suggest that FACT may be responsible for the reported presence of H2A.Z near tDNAs in NFR-flanking nucleosomes (9), a role not attributed to the FACT till now. Different core histone dimers can be exchanged by more than one histone chaperone in the chromatin assembly/disassembly reactions in vivo on different genes, suggesting a broad range of carriers for them (44). Events like histone exchange or nucleosome loss on the transcribed or promoter regions of the genes may be either the cause or the consequence of transcription. Nevertheless, these events are neither spontaneous nor autonomous and require the assistance of a chromatin remodeler and chaperone. Moreover, transcription-associated loss/exchange of histones due to passage of the elongating RNA polymerase II takes place with the help of FACT only in the transcribed region of a gene (2–5). The N-terminal domain of Spt16 is known to interact with H3 and in its absence a rapid loss of H3 is seen on both the active and inactive genes (43). Thus, on SUP4, the observed effect of Spt16 on the dynamics of H2A.Z in the flanking nucleosomes, which are regulatory and not structural in nature, need not be transcription process-associated, but very well be due to the Spt16 chaperone activity. Spt16 was shown to co-purify with H2A.Z in the absence of Nap1 and H2A.Z-specific chaperone Chz1 and help in SWR1-mediated exchange of H2A.Z–H2B dimer in vitro (42). We have shown that FACT interacts with H2A.Z both in vitro and in vivo. Thus, FACT may be a chaperone for the dimer of H2B with H2A or its major variants on different genes which may or may not require FACT for their expression.
Nhp6 stimulates the transcription of pol III-transcribed U6snRNA, while the deletion of both the copies of Nhp6 abolishes the TFIIIB footprint in the TATA box region (54). Although de novo synthesis of bulk of tRNA is not affected in Nhp6ΔΔ strain (55), we have found multiple effects of Nhp6 on SUP4. It physically interacts with RSC and blocks remodelling action of RSC in vitro (33). As it is required to load Spt16/Pob3 (45), mutation in RSC or loss of Nhp6, results in probably the same effect; loss of Spt16/Pob3 recruitment, and hence, H2A.Z. Therefore, increase in SUP4 RNA levels in RSC, Spt16 and Pob3 mutants could be due to the loss of H2A.Z in these strains, a condition similar to htz1Δ cells.
RSC plays multiple roles in SUP4 gene expression
Loss of RSC is reported to shrink NFR (56) on pol II-transcribed genes and increase the nucleosome density on pol III-transcribed genes (57). We have shown that RSC is required to keep the nucleosome immediate downstream of the SUP4 terminator, fuzzy in vivo (Figure 6A). It changes the local density of nucleosmes near the SUP4 gene (Figure 6B and C), maintains nucleosome positions to suppress the transcription in active state, helps loading Nhp6/FACT for transcriptional fidelity and differential marking with H2A.Z according to the expression state. While leaving the +1, RSC persists on the −1 nucleosome under repression, keeping the gene poised for activation under repressed state.
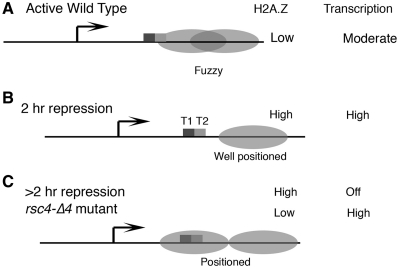
Figure 6.
Relationship of transcription from SUP4 with H2A.Z level and nucleosome positions. Ovals represent nucleosomes, boxes and T1, T2 denote two terminators, bent arrow marks the TSS. Only downstream nucleosomes are shown with H2A.Z levels and transcriptional state of the gene shown on the right-hand side. (A) In wild-type cells, active state, nucleosmes are fuzzy and block T2. (B) In wild-type cells, following 2 h repression, +1 nucleosome is positioned at a short distance from the terminators. (C) In wild-type cells, after repression longer than 2 h, two nucleosomes may be positioned similar to that seen in rsc4-Δ4 cells. Both terminators are near the dyad axis of the gene-proximal nucleosome.
Loss of Sth1, the catalytic subunit of RSC, from the downstream region of SUP4 within first hour during repression, may result in a nucleosome positioning similar to that seen in the rsc4-Δ4 mutant (which shows loss of remodelling activity on SNR6, 17). On some of pol III-transcribed genes, loss of Sth1 activity was earlier found to result in gradual increase of histone density but as RSC was not lost under repression, histone density on these genes did not show any change (57). However, despite an increase in histone density near the gene terminator at +100 bp (Figure 2A) we could not see a similar nucleosomal protection in the gene proximal region after 2 h of repression. As Sth1 is lost from the downstream of SUP4, a gradual increase in histone density in the gene region may take place with further repression. Accordingly, a nucleosome was seen to encroach the gene after 4 hr repression (not shown), suggesting an organization similar to rsc4-Δ4 cells may be generated after prolonged satrvation.
Regulation of SUP4 from downstream
Absence of the transcription terminator of pol III-transcribed genes results in loss of the facilitated recycling of pol III and transcript yield (19). As the cartoon in the Figure 6 depicts, due to its fuzzy nature, the +1 nucleosome encroaches the gene terminator T2 at +108 bp (Figure 6A). This may allow transcription termination only at T1, ensuring utilization of only one gene terminator in vivo; but interfere with terminator-directed recycling of pol III (19), resulting in reduced transcription rate in active state. Under initial hours of repression, +1 nucleosome positioning away from the gene would make both the terminators equally exposed (Figure 6B) and the resultant increase in the transcription rate would give higher RNA levels from SUP4 as observed under repression for some time (Figure 1B). Interestingly, presence of the gene terminators at the dyad axis of a positioned nucleosme in the rsc4-Δ4 mutant, similar to that seen on the SNR6 gene in active state (17), may allow the terminator-dependent recycling of pol III, giving higher transcription rate. In comparison, prolonged stress may completely abolish the transcription despite the same positioning in the wild type cells. This may be related to different signals from H2A.Z dynamics in both conditions (Figures 5C and F, and 6C).
Thus, a fine tuning of the +1 nucleosome position by RSC along with H2A.Z as the stress-sensor, probably regulates the gene output under different states from pol III-transcribed genes. These observations suggest SUP4 may be an example where diverse chromatin related activities come together and co-ordinately keep the gene expression low in active state generating a milieu in which gene remains in a poised state for release from the repressed state.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Council of Scientific and Industrial Research (CSIR); Department of Science and Technology, Govt. of India, research grant (SR/SO/BB-71/2003 to P.B.); CSIR senior research fellowship (to S.M. and P.S.D.). Funding for open access charge: Waived by Oxford University Press.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tim Formosa, Phillipp Korber, Hiten Madhani, Sebastian Chavez, Frank Pugh, David Stillman, Kevin Struhl, Toshio Tsukiyama, Michel Werner, Ian Willis and Jerry Workman for kind gifts of some of the yeast strains used in this study.
REFERENCES
- 1.Biswas D, Yu Y, Prall M, Formosa T, Stillman DJ. The yeast FACT complex has a role in transcriptional initiation. Mol. Cell. Biol. 2005;25:5812–5822. doi: 10.1128/MCB.25.14.5812-5822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 3.Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 4.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 5.Mason PB, Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 2003;23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Formosa T, Eriksson P, Wittmeyer J, Ginn J, Yu Y, Stillman DJ. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 2001;20:3506–3517. doi: 10.1093/emboj/20.13.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costigan C, Kolodrubetz D, Snyder M. NHP6A and NHP6B, which encode HMG1-like proteins, are candidates for downstream components of the yeast SLT2 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 1994;14:2391–2403. doi: 10.1128/mcb.14.4.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wittmeyer J, Formosa T. The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol. Cell. Biol. 1997;17:4178–4190. doi: 10.1128/mcb.17.7.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL. Global nucleosome occupancy in yeast. Genome Biol. 2004;5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 12.Harismendy O, Gendrel C-G, Soularue P, Gidrol X, Sentenac A, Werner M, Lefebvre O. Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J. 2003;22:4738–4747. doi: 10.1093/emboj/cdg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Ng HH, Robert F, Young RA, Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 2002;16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 2005;19:942–954. doi: 10.1101/gad.1298905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guffanti E, Percudani R, Harismendy O, Soutourina J, Werner M, Iacovella MG, Negri R, Dieci G. Nucleosome depletion activates poised RNA polymerase III at unconventional transcription sites in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:29155–29164. doi: 10.1074/jbc.M600387200. [DOI] [PubMed] [Google Scholar]
- 17.Arimbasseri AG, Bhargava P. Chromatin structure and expression of a gene transcribed by RNA polymerase III are independent of H2A.Z deposition. Mol. Cell. Biol. 2008;28:2598–2607. doi: 10.1128/MCB.01953-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kassavetis GA, Geiduschek EP. Transcription factor TFIIIB and transcription by RNA polymerase III. Biochem. Soc. Trans. 2006;34:1082–1087. doi: 10.1042/BST0341082. [DOI] [PubMed] [Google Scholar]
- 19.Dieci G, Sentenac A. Facilitated recycling pathway for RNA polymerase III. Cell. 1996;84:245–252. doi: 10.1016/s0092-8674(00)80979-4. [DOI] [PubMed] [Google Scholar]
- 20.Geiduschek EP, Kassavetis GA. Transcription: adjusting to adversity by regulating RNA polymerase. Curr. Biol. 2006;16:R849–R851. doi: 10.1016/j.cub.2006.08.071. [DOI] [PubMed] [Google Scholar]
- 21.Willis IM, Moir RD. Integration of nutritional and stress signaling pathways by Maf1. Trends Biochem. Sci. 2007;32:51–53. doi: 10.1016/j.tibs.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Clarke EM, Peterson CL, Brainard AV, Riggs DL. Regulation of the RNA polymerase I and III transcription systems in response to growth conditions. J. Biol. Chem. 1996;271:22189–22195. doi: 10.1074/jbc.271.36.22189. [DOI] [PubMed] [Google Scholar]
- 23.Roberts DN, Stewart AJ, Huff JT, Cairns BR. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc. Natl Acad. Sci. USA. 2003;100:14695–14700. doi: 10.1073/pnas.2435566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shivaswamy S, Kassavetis GA, Bhargava P. High-level activation of transcription of the yeast U6 snRNA gene in chromatin by the basal RNA polymerase III transcription factor TFIIIC. Mol. Cell. Biol. 2004;24:3596–3606. doi: 10.1128/MCB.24.9.3596-3606.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 26.Martin DG, Grimes DE, Baetz K, Howe L. Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin. Mol. Cell. Biol. 2006;26:3018–3028. doi: 10.1128/MCB.26.8.3018-3028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl Acad. Sci. USA. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:2100–2110. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soutourina J, Bordas-Le Floch V, Gendrel G, Flores A, Ducrot C, Dumay-Odelot H, Soularue P, Navarro F, Cairns BR, Lefebvre O, et al. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol. Cell. Biol. 2006;26:4920–4933. doi: 10.1128/MCB.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vinayachandran V, Pusarla RH, Bhargava P. Multiple sequence-directed possibilities provide a pool of nucleosome position choices in different states of activity of a gene. Epigenetics Chromatin. 2009;2:4. doi: 10.1186/1756-8935-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szerlong H, Saha A, Cairns BR. The nuclear actin-related proteins Arp7 and Arp9: a dimeric module that cooperates with architectural proteins for chromatin remodeling. EMBO J. 2003;22:3175–3187. doi: 10.1093/emboj/cdg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhoades AR, Ruone S, Formosa T. Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol. Cell. Biol. 2004;24:3907–3917. doi: 10.1128/MCB.24.9.3907-3917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chedin S, Riva M, Schultz P, Sentenac A, Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998;16:3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kassavetis GA, Steiner DF. Nhp6 is a transcriptional initiation fidelity factor for RNA polymerase III transcription in vitro and in vivo. J. Biol. Chem. 2006;281:7445–7451. doi: 10.1074/jbc.M512810200. [DOI] [PubMed] [Google Scholar]
- 37.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 38.Jimeno-Gonzalez S, Gomez-Herreros F, Alepuz PM, Chavez S. A gene-specific requirement for FACT during transcription is related to the chromatin organization of the transcribed region. Mol. Cell. Biol. 2006;26:8710–8721. doi: 10.1128/MCB.01129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahata S, Yu Y, Stillman DJ. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol. Cell. 2009;34:405–415. doi: 10.1016/j.molcel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ransom M, Williams SK, Dechassa ML, Das C, Linger J, Adkins M, Liu C, Bartholomew B, Tyler J. FACT and the proteasome promote promoter chromatin disassembly and transcriptional initiation. J. Biol. Chem. 2009;284:23461–23471. doi: 10.1074/jbc.M109.019562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, Lieber MR, Yang AS, An W. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol. Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 42.Luk E, Vu ND, Patteson K, Mizuguchi G, Wu WH, Ranjan A, Backus J, Sen S, Lewis M, Bai Y, et al. Chz1, a nuclear chaperone for histone H2AZ. Mol. Cell. 2007;25:357–368. doi: 10.1016/j.molcel.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 43.Jamai A, Puglisi A, Strubin M. Histone chaperone Spt16 2005 promotes redeposition of the original H3-H4 histones evicted by elongating RNA Polymerase. Mol. Cell. 2009;35:377–383. doi: 10.1016/j.molcel.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Koning LD, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nat. Struct. Mol. Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 45.Ruone S, Rhoades AR, Formosa T. Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and to reorganize nucleosomes. J. Biol. Chem. 2003;278:45288–45295. doi: 10.1074/jbc.M307291200. [DOI] [PubMed] [Google Scholar]
- 46.Birch JL, Tan BCM, Panov KI, Panova TB, Andersen JS, Hughes TAO, Russell J, Lee SC, Zomerdijk JCBM. FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J. 2009;28:854–865. doi: 10.1038/emboj.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morse RH, Roth SY, Simpson RT. A transcriptionally active tRNA gene interferes with nucleosome positioning in vivo. Mol. Cell. Biol. 1992;12:4015–4025. doi: 10.1128/mcb.12.9.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pusarla RH, Bhargava P. Histones in functional diversification. Core histone variants. FEBS J. 2005;272:5149–5168. doi: 10.1111/j.1742-4658.2005.04930.x. [DOI] [PubMed] [Google Scholar]
- 49.Santisteban MS, Kalashnikova T, Smith MM. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- 50.Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16:166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 51.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 52.Uffenbeck SR, Krebs JE. The role of chromatin structure in regulating stress-induced transcription in Saccharomyces cerevisiae. Biochem. Cell Biol. 2006;84:477–489. doi: 10.1139/o06-079. [DOI] [PubMed] [Google Scholar]
- 53.Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2009;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 54.Lopez S, Livingstone-Zatchej M, Jourdain S, Thoma F, Sentenac A, Marsolier MC. High-mobility-group proteins NHP6A and NHP6B participate in activation of the RNA polymerase III SNR6 gene. Mol. Cell. Biol. 2001;21:3096–3104. doi: 10.1128/MCB.21.9.3096-3104.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kruppa M, Moir RD, Kolodrubetz D, Willis IM. Nhp6, an HMG1 protein, functions in SNR6 transcription by RNA polymerase III in S. cerevisiae. Mol. Cell. 2001;7:309–318. doi: 10.1016/s1097-2765(01)00179-4. [DOI] [PubMed] [Google Scholar]
- 56.Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parnell TJ, Huff JT, Cairns BR. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J. 2007;27:100–110. doi: 10.1038/sj.emboj.7601946. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.