Abstract
During cruciform extrusion, a DNA inverted repeat unwinds and forms a four-way junction in which two of the branches consist of hairpin structures obtained by self-pairing of the inverted repeats. Here, we use single-molecule DNA nanomanipulation to monitor in real-time cruciform extrusion and rewinding. This allows us to determine the size of the cruciform to nearly base pair accuracy and its kinetics with second-scale time resolution. We present data obtained with two different inverted repeats, one perfect and one imperfect, and extend single-molecule force spectroscopy to measure the torque dependence of cruciform extrusion and rewinding kinetics. Using mutational analysis and a simple two-state model, we find that in the transition state intermediate only the B-DNA located between the inverted repeats (and corresponding to the unpaired apical loop) is unwound, implying that initial stabilization of the four-way (or Holliday) junction is rate-limiting. We thus find that cruciform extrusion is kinetically regulated by features of the hairpin loop, while rewinding is kinetically regulated by features of the stem. These results provide mechanistic insight into cruciform extrusion and help understand the structural features that determine the relative stability of the cruciform and B-form states.
INTRODUCTION
A cruciform is a structure based on a DNA inverted repeat, and characterized by the presence of a four-way junction in which two of the branches are hairpin structures formed on each strand of the inverted repeat (Figure 1A). The bases located between the inverted repeats do not self-pair and instead form the apical loops of the hairpins. Although the four-way junction and the apical loops render the cruciform less stable than B-form DNA, negative DNA supercoiling has long been known to stabilize cruciform structures (1–4).
Figure 1.
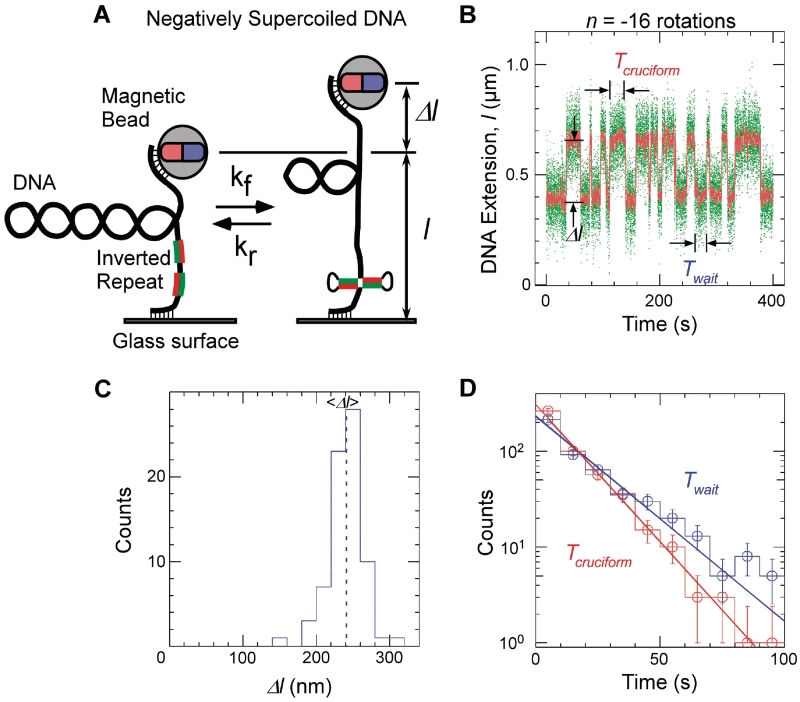
Single-molecule detection of cruciform extrusion. (A) Sketch of the experiment. A single DNA molecule containing an inverted repeat is tethered at one end to a glass surface and at the other end to a magnetic bead. A pair of magnets serves to control the vertical extending force, F, applied to the DNA and the number of rotations, n, imposed on the bead and hence the DNA supercoiling. The DNA end-to-end extension, l, is determined by measuring the position of the bead above the surface in real-time by computer-aided videomicroscopy. When the inverted repeat extrudes into a cruciform, unwinding of B-DNA titrates out negative supercoils and an increase, Δl, in DNA extension is observed. (B) Real-time data showing abrupt, reversible, changes in extension for Charomid 9-5 kb DNA (F ∼ 0.45 pN, n = −16 turns). Green points correspond to real-time data acquisition (60 Hz) while red points correspond to raw signal averaged over ∼1 s. Both structural (Δl) and kinetic (Twait and Tcruciform) features of the reaction can be obtained from these time-traces. (C) Histogram of transition amplitudes. The dashed line indicates the mean <Δl> = 241 ± 3 nm (n = 74 points). (D) Histograms of Twait (blue) and Tcruciform (red) and single-exponential fits to data. Mean lifetimes are <Twait> = 20.2 ± 1.2 s (n = 490 events) and <Tcruciform> = 13.5 ± 0.9 s (n = 489 events).
From the experimental standpoint, cruciform DNA has been extensively studied in vitro over the course of the past decades (5) using for instance gel-shift analysis of supercoiled inverted repeats to determine the equilibrium, and even the interconversion rates, between the B-DNA and extruded cruciform states (6–8). In such measurements, the B-DNA and cruciform states can be distinguished by their overall topology; indeed as discussed below, formation of the cruciform titrates out negative supercoils, typically causing the cruciform DNA to migrate more slowly than B-DNA during gel electrophoresis. In parallel, in vivo studies have suggested that the more structurally stable a cruciform, the more likely it is to be genetically unstable, for instance subject to excision as in bacteria (7,9) or even responsible for genetic rearrangements in eukaryotes (10–14). Theoretical, numerical and experimental studies (15,16) further indicate that negative DNA supercoiling in vivo may participate in modulating gene expression levels by driving DNA unwinding transitions in gene regulatory regions, and cruciform extrusion could participate in such processes.
On the other hand, many inverted repeats are obviously genetically stable (consider the DNA encoding tRNAs, microRNA precursors or riboswitches), and only a few clear examples of regulatory roles have been identified [e.g. promoters of the N4 virion (17,18)]. Thus, the role of cruciform extrusion in vivo remains elusive (19). To better understand how inverted repeats could behave under in vivo conditions of negative supercoiling, it is interesting to determine how their sequence and structural features contribute to the stability or instability of a cruciform structure. Here, we describe a single-molecule nanomanipulation procedure that allows us to directly monitor both cruciform extrusion and rewinding in real-time, and thus analyze cruciform stability in terms of the balance between the rates of extrusion and rewinding. This allows us to explore in detail how structural and sequence determinants of cruciforms contribute to their overall stability.
MATERIALS AND METHODS AND DATA ANALYSIS
In these experiments, an individual DNA molecule is torsionally constrained by attaching both strands at one end of the DNA to a treated glass coverslip, and by attaching both strands at the other end to a 1-µm magnetic bead, as previously described (20,21). To accomplish this, the DNA is multiply labeled at one end with digoxigenin and at the other end with biotin. The labeled DNA is then allowed to bind to 1-µm streptavidin-coated magnetic beads. The bead-DNA solution is then deposited on an antidigoxigenin-coated glass surface, and when the DNA binds to the glass surface a tethered DNA-bead system forms. The sample is placed on an inverted microscope through which bead images can be viewed and captured by videomicroscopy. The position of the bead in the three spatial directions is tracked at 30 Hz and with ∼5 nm accuracy using the PicoTwist software package. From the position of the bead above the surface, we determine the end-to-end extension, l, of the DNA.
A pair of permanent magnets mounted on translation and rotation stages is located above the sample and generates a magnetic field that imposes a vertical extending force on the bead and sets the bead’s orientation. The extending force is increased (decreased) by translating the magnets closer to (farther away from) the sample. Rotating the magnets causes the bead to rotate in a perfectly synchronous fashion about the axis defined by the extended DNA, allowing for quantitative supercoiling of the DNA. Both extending force and DNA supercoiling can therefore be controlled in a fully reversible and quantitative fashion (20). The extending force is calibrated using Brownian motion analysis of the tethered bead, and determination of the DNAs elastic behavior confirms that a single DNA molecule tethers the bead to the surface (20).
Indeed, the mechanical and topological properties of a single supercoiled DNA molecule have been extensively studied experimentally (20,22–25) and theoretically (26–28). Briefly, DNA topology is described by three numbers that reflect the number of times the two strands of DNA wind about one another (29). The twist number, Tw, counts the number of times the two strands wrap one around the other in the double-helix structure; in B-DNA, there is one unit of twist for every 10.5 bp. The writhe, Wr, counts the number of times the axis of the DNA crosses itself, in particular in the looped structures (also termed ‘plectonemes’) that arise as a result of DNA supercoiling and that resemble the tangles observed on a twisted phone cord. Finally, the linking number Lk is the sum total of crossings in the DNA, either by twist or by writhe: Lk = Tw + Wr. In a topologically constrained system (i.e. a closed circular plasmid or a DNA torsionally constrained in a magnetic trap), Lk is a constant, and any change in Tw must be compensated by an equal but opposite change in Wr. By rotating the magnets and thus the bead, we directly modify Lk.
Supercoiling a DNA molecule subjected to a very low extending force (F = 0.45 pN) rapidly causes it to form loops, or writhe, as a means of storing the topological constraint (20). It thereby adopts a compact conformation with low extension (see sketch in leftmost panel of Figure 1A). Loops, and therefore writhe, can be calibrated by measuring the changes in DNA extension that take place upon supercoiling of the molecule (30,31) (see Supplementary Figure S1 for an example of such calibration curves). From these curves, we determine that adding or removing a supercoil changes the DNA extension by an amount δ = 74 nm for the experimental conditions used here.
Thus for a negatively supercoiled DNA containing an inverted repeat, cruciform extrusion is expected to result in a large increase in DNA end-to-end extension (Figure 1A). This strategy is formally identical to that used in standard electrophoresis assays to measure cruciform extrusion, and has previously been used in this single-molecule assay to study DNA unwinding in real-time, for instance during transcription by Escherichia coli RNA polymerase (32,33). Indeed the change in twist ΔTw that occurs during cruciform extrusion must be compensated by an equal but opposite change in the writhing number, Wr, of the DNA: ΔWr = −ΔTw (34). Conversion of Ncruciform bp of B-DNA to cruciform DNA leads to a decrease in twist ΔTw = −Ncruciform/h = −ncruciform and an equal increase in Wr (h, the DNA pitch, is 10.5 bp/turn). For negatively supercoiled DNA, this results in a loss of negatively signed loops, and thus an increase in DNA extension of δ·ncruciform or ∼7 nm/bp in the cruciform. It is important to note that this is a very large increase, far greater than the 2 Å decrease in DNA extension that results from removal of the base pair from the end-to-end extension of the DNA so that it may participate in the cruciform (lbp, the extension of a base pair of DNA at this low extending force, is only ∼2 Å).
The DNA constructs studied in this article include the Charomid 9-5 kb plasmid (35) (for studies of the Charomid X sequence, see ‘Results’ section) and the 3.7 kb pColIR315 plasmid (36) (for studies of the ColE1 inverted repeat and its derivatives). Experiments with Charomid DNA were carried out in SB solution containing 10 mM potassium phosphate buffer at pH 8.0, 0.1% Tween-20 and 0.1 mg/ml BSA, and for experiments with pColIR315 the potassium phosphate concentration was 11 mM. The extending force acting on the DNA was F = 0.5 pN for experiments performed in 10 or 11 mM phosphate buffer and F = 0.8 pN for experiments performed in the presence of 50 mM NaCl. The experimental system was thermally regulated to 26 ± 0.1°C using Pelletier modules as the rates of cruciform extrusion and rewinding were found to be highly temperature sensitive.
Data analysis
Raw data traces were filtered at 1–2 s and analyzed using in-house software to determine where transitions between the B-DNA and extruded cruciform state occurred. The low signal-to-noise ratio of the data, which results from the weak stiffness of supercoiled DNA under the experimental conditions used, can limit the effectiveness of automatic analysis, and so results of this processing were then manually verified. Typically, the mean rates were determined from analysis of at least 300 transitions (providing for a standard error of about 6% on the mean rates), and transition amplitude histograms contain at least 50 points (smaller error was achieved here due to the lower variance of the observed distributions).
RESULTS
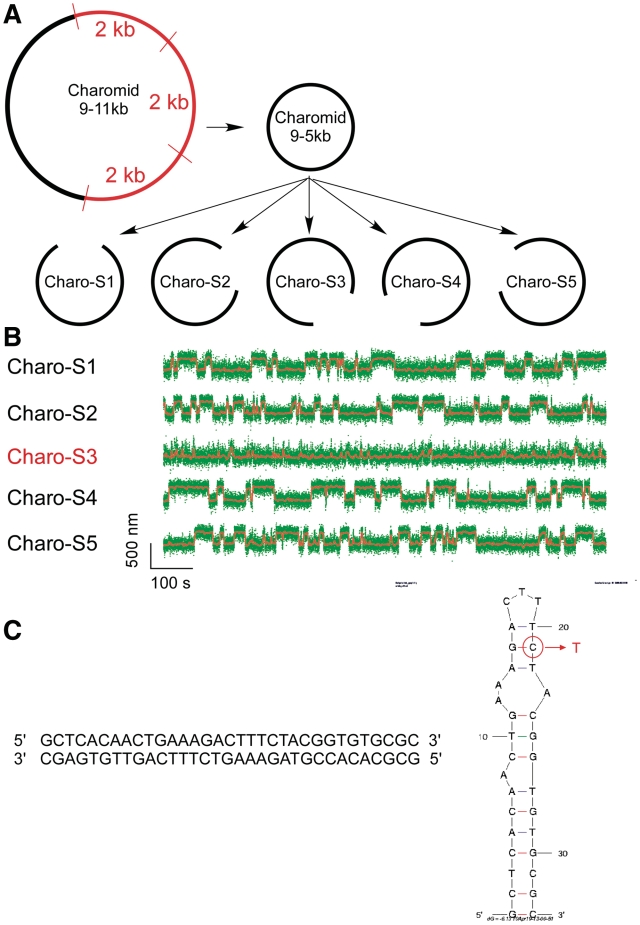
In Figure 1B, we present a real-time measurement of the change in DNA extension resulting from reversible extrusion of the Charomid X imperfect inverted repeat under conditions of negative supercoiling. The time-trace shows the 5-kb DNA undergoing abrupt transitions (‘hopping’) between a low-extension state and a high-extension state. The Charomid X sequence responsible for these fluctuations was identified by selectively removing different regions of the Charomid 9-5 kb DNA and screening the resulting constructs for the slow, large-scale fluctuations (Figure 2). The low- and high-extension states we observe are separated by a remarkably large change in DNA extension of <Δl> = 241 ± 3 nm (Figure 1C), nearly 15% of the full contour length of the DNA. No such slow, large-scale fluctuations are observed if the Charomid X sequence is positively supercoiled (Supplementary Figure S1). We thus estimate that when the DNA hops from the low-extension state to the high-extension state, <Δl>/δ = 3.3 units of writhe are titrated out, consistent with formation of a cruciform involving Ncruciform = 34 bp. This is in agreement with the expected size of the cruciform based on the Charomid X quasipalindromic sequence (33 bp, see Figure 2C). The Charomid X sequence still undergoes these fluctuations if separated from its original flanking sequences (Supplementary Figure S2).
Figure 2.
Single-molecule screen of Charomid 9-5 kb is used to identify the Charomid X quasipalindrome. (A) Screen strategy. Charomid 9-5 kb is derived from Charomid 9-11 kb by removing the 6 kb region of the plasmid, corresponding to three consecutive repeats of a 2-kb fragment (35). Then, five constructs are derived from Charomid 9-5 kb using the PCR reaction, each construct lacking a different 1 kb segment of the plasmid. (B) Time-traces for the five constructs described above: first level of screening. DNA constructs were unwound by n = −12 turns using the magnetic trap, and their extension monitored in real-time. The Charomid 9-5 kb displays the same behavior as the Charomid 9-11 kb plasmid, indicating that the repeat region is not responsible for the observed fluctuations. The Charo-S3 construct is the only one of the five constructs to lack the canonical fluctuations observed on the original plasmid, indicating that the sequence responsible for this behavior is located in the corresponding 1 kb of DNA, which is specifically lacking in the construct. Fast residual fluctuations indicate denaturation or cruciform extrusion on secondary sequences in the plasmid (C) Screen results. The above process was reiterated two times to narrow down the sequence responsible for the observed fluctuations. The 33-bp Charomid X sequence responsible for the observed fluctuations is a quasipalindrome, which extrudes to form a complex and unstable cruciform. A putative minimum energy fold derived from mfold for high-salt conditions (0.5 M NaCl) is presented; in the lower salt conditions initially used here, the 3-bp separating the two apical loops are unlikely to be stable at room temperature, implying that the apical loop is in fact the sum of the two smaller loops. This is consistent with the extensive amount of unwinding estimated to be present in the transition state. The mutation made to increase the loop size involved converting the G circled in red into a T, disrupting a crucial base pair for high-salt conditions but not low-salt conditions (Table 1).
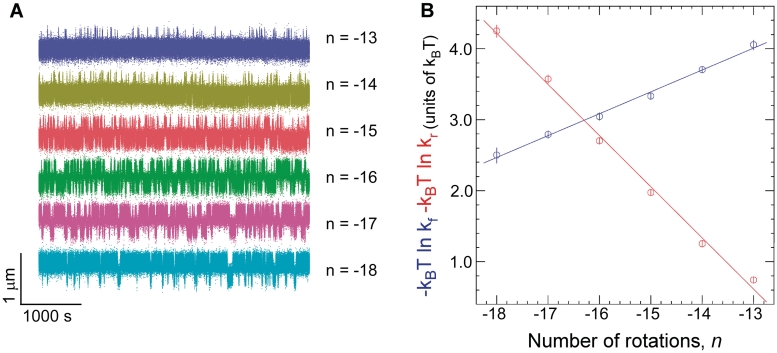
The time elapsed before an extrusion event, Twait, and the lifetime of the extruded state, Tcruciform, can be directly determined from the time-traces (Figure 1B). Histograms of Twait and Tcruciform display single-exponential lifetime distributions (Figure 1D), suggesting that at the temporal resolution of the experiment (∼1 s) a single energy barrier dominates the transition between the B-DNA and cruciform states. These measurements were then repeated on DNA for different levels of negative supercoiling (Figure 3A). The data show that as the DNA is progressively unwound, the cruciform state becomes more likely as a result of an increase in the rate of cruciform extrusion, kf = 1/<Twait>, and a concomitant decrease in the rate of cruciform rewinding, kr = 1/<Tcruciform>. More precisely, kf increases exponentially while kr decreases exponentially with negative supercoiling as per an Arrhenius law (Figure 3B). At the same time, we find that ncruciform remains essentially constant, indicating that the size of the cruciform does not increase with negative supercoiling (Supplementary Figure S3). These results imply that the cruciform is fully formed upon extrusion, and that the hairpin branches cannot be extended beyond the inverted repeats. Instead, the cruciform becomes progressively more stable with negative supercoiling, consistent with mechanical stabilization of the extruded cruciform by the torque generated by negative supercoiling.
Figure 3.
Kinetic analysis of cruciform extrusion and rewinding for Charomid 9-5 kb DNA. (A) As negative DNA supercoiling increases, progressively less time is spent in the B-DNA state and more time is spent in the extruded-cruciform state. Experiments were performed at F = 0.45 pN. (B) Free energy barrier to cruciform extrusion (blue) and rewinding (red) as a function of negative supercoiling. The barrier to cruciform extrusion decreases linearly with negative supercoiling at a rate of 0.31 ± 0.015 kBT/turn (B-DNA is destabilized by unwinding), while the barrier to rewinding increases linearly by 0.71 ± 0.014 kBT/turn (cruciform DNA is stabilized by unwinding). At least ∼300 events were measured at each rotation point to determine each mean transition rate.
This can be seen by calculating the difference in free energy between the two states, ΔG = −kBT ln kf/kr, as a function of supercoiling. In Figure 3B, this corresponds to the difference between the blue and red curves: thus ΔG grows linearly with negative DNA supercoiling at a rate of about −1 kBT/turn. We conclude that in this regime torque increases slowly and linearly with negative supercoiling, even after the buckling transition. Indeed, ΔG = ΔGcruciform + ΔGsupercoiling and ΔGcruciform is constant. Thus, the increase in free energy between the B-DNA and cruciform states can only be due to the increase in mechanical work done on the system by torque Γ during the unwinding of ncruciform turns of the double helix: ΔGsupercoiling = Γ·2πncruciform. As ncruciform does not depend on supercoiling, this implies that Γ increases with supercoiling. Note that this increase in torque is quite small, on the order of 0.05 kBT for every turn added. Although this slow increase in torque after the formation of DNA plectonemes has already been observed (25), there are currently no theoretical models of DNA statistical mechanics to explain this weak increase in torque occurring after the formation of DNA writhe (24).
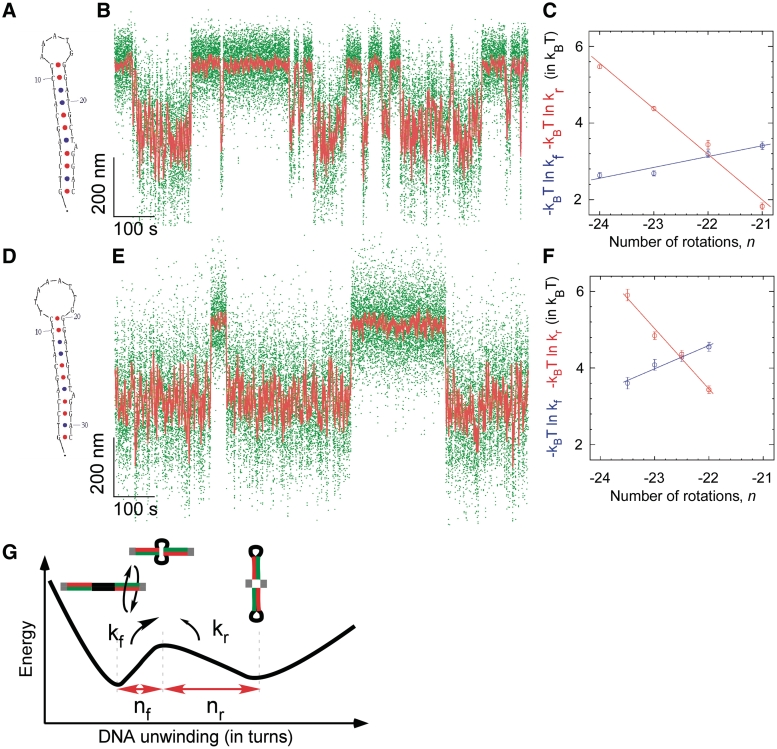
As can be done with force spectroscopy (37,38), the supercoiling dependence of kf and kr can be quantitatively analyzed to determine the ratio nr/nf, where nf is the amount of DNA unwinding separating the B-DNA state from the transition state, nr is the amount of DNA unwinding required to complete extrusion, i.e. to go from the transition state to the cruciform state and nf + nr = ncruciform is well-determined experimentally (for details, see Supplementary Data and Figure 5E). We thus find for the Charomid X quasipalindrome nr/nf = 2.3 ± 0.1. Using nr + nf = ncruciform = 34 ± 1 bp, we interpret this as meaning that in the transition state 10 ± 1 bp of DNA are unwound. Note that the Charomid X cruciform is predicted to have a large, weakly structured and presumably degenerate ∼12 base loop region at its apex (Figure 2C). [We further note that this result is independent of the extending force used during the measurement, as in the range of forces tested increasing the force leaves the relative position of the transition state essentially unchanged (Supplementary Figure S4).] Based on the simple two-state model the data permit for, these experiments suggest that the transition-state intermediate to cruciform extrusion corresponds to a state where the DNA is unwound only in the apical loop region of the cruciform.
Figure 5.
Kinetic and structural analysis of cruciform extrusion in mutColE1 inverted repeats bearing (A–C) a 5 base loop or (D–F) an 8 base loop. (A) The mutColE1-5b inverted repeat is seen (B) to extrude reversibly as shown previously in Figure 4. (C) The free energy barrier to cruciform rewinding (red) is more sensitive to torque than the free energy barrier to cruciform extrusion (blue), d ln kf/d ln kr = 4.2 ± 0.6. (D) The mutColE1-8b inverted repeat with an 8 base loop is seen (E) to extrude with slower kinetics than the 5-base loop. (F) The free energy barrier to rewinding is still more sensitive to torque than the free energy barrier to extrusion, but less so: d ln kf/d ln kr = 2.6 ± 0.5. (G) Two-state model for torque-induced cruciform extrusion. The transition-state intermediate to cruciform extrusion is formed by unwinding B-DNA by nf turns, and for complete formation of the cruciform the B-DNA must thereafter be unwound by an additional nr turns, with nf + nr = ncruciform. The relative sensitivity of transition rates kf and kr to supercoiling predicts the position of the transition state between the two states, d ln kf/d ln kr = nf/nr.
To test this, we determined the effect of a single base mutation designed to disrupt the stem and increase the size of the loop (see Figure 2C). However in the salt conditions previously used (10 mM potassium phosphate buffer, pH 8.0), we do not find any effect for this mutation (Table 1), suggesting that under these salt conditions the loop is already large enough to encompass the targeted base. Upon addition of 50 mM NaCl, however, analysis of the mutant becomes possible (Table 1). First, we find that the wild-type cruciform becomes slightly more structured. Indeed here the ratio nr/nf increases, implying proportionally less unwinding in the transition state and consistent with an increase in stem length at the cost of a decrease in loop length, for a constant sum of stem and loop length. This is expected for ionic conditions that stabilize base pairing. In these higher salt conditions, the mutation now causes a large decrease in the ratio nr/nf, consistent with an increase in loop size and a decrease in stem length resulting from this disruptive mutation. These experiments support the notion that disrupting base pairs involved in closing the loop affects the transition state intermediate.
Table 1.
Compilation of transition amplitudes and torque dependency of cruciform extrusion and refolding rates for the inverted repeats studied in this article
| DNA | Ionic conditions | Ncruciform (bp) | d(ln kr)/dn (kBT/turn) | d(ln kf)/dn (kBT/turn) | d(ln kr)/d (ln kf) = nr/nf | DNA unwinding in TS (bp) |
|---|---|---|---|---|---|---|
| WT Charomid X | 10 mM Phosphate buffer | 34 ± 1 (74) | 0.71 ± 0.014 | 0.31 ± 0.015 | 2.3 ± 0.1 | 10 ± 1 |
| 10 mM Phosphate Buffer + 50 mM NaCl | 36 ± 1 (58) | 0.30 ± 0.01 | 0.10 ± 0.01 | 2.9 ± 0.6 | 9 ± 1 | |
| Mutated Charomid X | 10 mM Phosphate buffer | 34 ± 1 (51) | 0.91 ± 0.02 | 0.43 ± 0.02 | 2.1 ± 0.11 | 11 ± 1 |
| 10 mM Phosphate buffer + 50 mM NaCl | 36 ± 1 (65) | 0.11 ± 0.02 | 0.11 ± 0.02 | 1.0 ± 0.25 | 18 ± 3 | |
| ColE1-5b | 11 mM Phosphate buffer | 30 ± 1 | 0.39 ± 0.05 | 1.68 ± 0.05 | 4.3 ± 0.5 | 5.8 ± 0.8 |
| ColE1-8b | 11 mM Phosphate buffer | 32 ± 1 | 0.61 ± 0.13 | 1.57 ± 0.13 | 2.6 ± 0.5 | 9.1 ± 1.5 |
For unwinding amplitudes, the number in parenthesis is the number of events used to calculate the average.
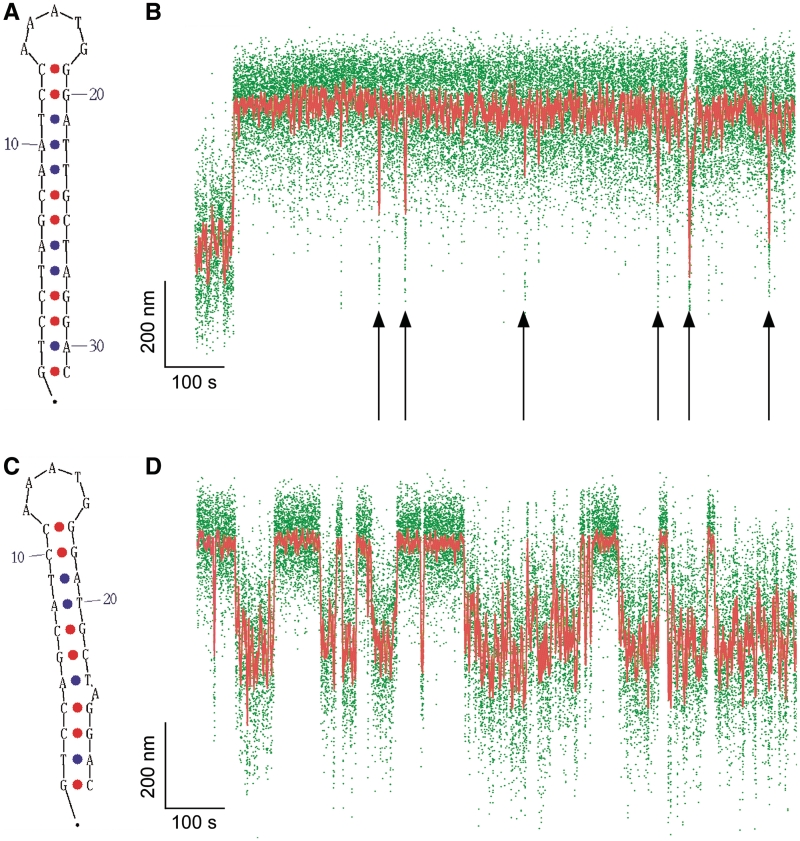
To further test our model, we performed another series of experiments investigating the kinetics of extrusion and rewinding for the canonical 31 bp ColE1 perfect inverted repeat, a cruciform characterized by a 5-base apical loop and perfectly paired 13 bp stem (Figure 4A) (1). We selected this sequence based on its perfect pairing and its reported propensity for irreversible extrusion, as it could potentially provide a basis on which to test modifications leading to reversible cruciform extrusion. Single-molecule analysis of the wtColE1 inverted repeat subjected to negative supercoiling indeed showed that cruciform extrusion was essentially irreversible, with a large increase in DNA extension during cruciform extrusion which never fully reversed. Nevertheless, the data indicate that the extruded cruciform can make attempts at refolding (Figure 4B). During such attempts, the extension of the system can nearly return to that of the native low-extension state. However, these attempts are unsuccessful and do not correspond to full rewinding of the cruciform, as their lifetime, on the order of seconds, is much shorter than the lifetime of the true B-DNA state (410 ± 100 s, see Supplementary Figure S5). In the context of our two-state model describing the relative position of the transition state, these results suggest that the transition state to rewinding lies very close to the native state for this perfect inverted repeat. Further experiments (Supplementary Figure S6) show that rewinding of the cruciform can occur when the DNA is returned to n = −9 turns but that this is still a slow process. Thus under the conditions in which this perfect inverted repeat extrudes, it is thereafter kinetically trapped and thus not a good thermodynamic system.
Figure 4.
Irreversible and reversible cruciform extrusion on the wtColE1 inverted repeat and its derivative mutColE1-5b. (A) The wtColE1 inverted repeat forms a perfect cruciform (B) in an irreversible fashion under conditions of negative supercoiling (n = −21). Immediately after the extrusion event, supercoiling is returned to n = −19 rotations to favor rewinding. Despite this no complete rewinding is observed, although unsuccessful attempts at refolding are regularly observed (down arrows). (C) The mutColE1-5b inverted repeat extrudes into an imperfect cruciform (D) in a reversible fashion, allowing for statistical analysis of the kinetics of cruciform extrusion and rewinding. The increased level of noise in the extension of the B-DNA state is due to the presence of A + T-rich regions flanking the inverted repeat (see Supplementary Figure S7).
So as to perform kinetic and structural analysis of this inverted repeat, we first sought to introduce limited changes to the sequence that would convert the cruciform into a thermodynamically reversible system. We accomplished this by removing three bases from each stem so as to shorten each stem by a full AT base pair and generate a single base adenine or thymine bulge on each stem (Figure 4C). Thanks to these modifications in the stem region, the resulting inverted repeat, mutColE1-5b, was now observed to extrude and rewind reversibly (Figure 4D).
Structural analysis of the extrusion transition for this inverted repeat gives <Δl> = 213 ± 4 nm, corresponding to Ncruciform = 30 ± 1 bp (Supplementary Figure S7), in close agreement with the expected length of the inverted repeat (28 bp). From a kinetic standpoint, with this cruciform kr is again significantly more sensitive to changes in supercoiling than kf, and analysis of the rates as above gives nr/nf = 4.2 ± 0.6 (Figure 5C and Table 1). We infer that in the transition state the inverted repeat is unwound by 5.8 ± 0.8 bp. In agreement with the simple two-state model for cruciform extrusion, this amount of unwinding in the transition state is just enough to accommodate formation of the five base apical loop of the cruciform. Note that, in agreement with previous findings, this family of inverted repeats requires the presence of A + T-rich flanking regions to extrude (see Supplementary Figure S8); transient unwinding in these regions is the cause of the increase in noise level in the B-DNA state and presumably allows for attempts at cruciform folding.
Furthermore, using a second cruciform made reversible by a separate set of destabilizing changes in the stem region, we again find the same position for the transition state intermediate (Supplementary Figure S9). Thus in further agreement with the simple two-state model, we find that the position of the transition state is unaffected by the position of destabilizing changes in the stem.
To see if we could quantitatively predict the structure of the transition state intermediate from the minimal two-state model, we derived the mutColE1-8b inverted repeat, for which the apical loop is three bases larger than for the mutColE1-5b inverted repeat (Figure 5D–F). Structural analysis of the unwinding transition gives <Δl> = 233 ± 3 nm (Supplementary Figure S7B), corresponding to Ncruciform = 32 ± 1 bp, again in close agreement with the expected length of the inverted repeat (31 bp). For this construct, torque spectroscopy yielded nr/nf = 2.6 ± 0.5 (Figure 5F and Table 1). We infer that in the transition state, 9.1 ± 1 bases of the inverted repeat are unwound, in agreement with the predicted size of the cruciform’s apical loop. We conclude that in the transition state to cruciform extrusion only the apical loop is unwound; presumably, the stem is then poised to zip up via base pairing as in the Holliday junction (39,40).
DISCUSSION
Although single-molecule approaches to studying DNA cruciforms have already been described (41,39), these prior studies focused on the behavior of a preexisting cruciform and did not analyze the nucleation process itself. Here, we show that it is also possible to observe in real-time the process of cruciform extrusion, as well as the reverse rewinding process. Cruciform size can be measured to within base pair accuracy, and kinetics determined to ∼1 s. The ColE1 and Charomid X cruciforms presumably begin extrusion by different mechanisms: the ColE1 cruciform requires flanking A + T-rich regions to initiate cruciform extrusion, but not the Charomid X cruciform (Supplementary Figures S8 and S3, respectively). Thus, extrusion of the ColE1 cruciform likely depends on unwinding fluctuations in the flanking A + T-rich regions, while the A + T-rich content of the Charomid X cruciform loop region suggests that unwinding fluctuations required to nucleate the apical loop are localized within the apical loop itself. This is consistent with past studies of cruciform extrusion (8) in which the ColE1 cruciform was classified as C-type, i.e. requiring unwinding of a large flanking region to initiate extrusion (42). We here propose that the Charomid X behaves more like the S-type cruciform, as it does not require unwinding of a large flanking region to initiate extrusion.
Despite the difference in how DNA unwinding in the apical loop initiates, we find that a similar transition state rate-limits the extrusion of both Charomid X and ColE1 cruciforms (Figure 5G). Although these cruciforms may depend on DNA unwinding fluctuations to initiate extrusion, it appears that in negatively supercoiled DNA this is not the rate-limiting step of extrusion. For instance, although the ColE1 palindrome requires large-scale unwinding to extrude (as evidenced by its dependence on A + T-rich flanking regions), the rate-limiting step of extrusion is the formation of an intermediate with unwinding only equal to that of the final apical loop. This supports the proposal that after initial unwinding most of the DNA reanneals but for the apical loop, and it is the subsequent formation of a four-way junction intermediate that is rate-limiting and which obligatorily precedes zipping of the stem (6,43). Indeed in the high-energy state of the four-way junction intermediate, bases are unstacked and little or no base pairing takes place in the stem yet. Once the bases between the inverted repeats are unwound and the apical loop stabilized by pairing at its base, pairing in the stem can proceed and supercoiling torque drives full extrusion of the cruciform.
According to this model, the rate of cruciform extrusion is set principally by the size of the unpaired loop, while the rate of cruciform rewinding is set principally by the size and structure of the stem. For instance, the position of the transition state intermediate close to the native B-DNA state helps to explain the apparently irreversible extrusion of perfect stem–loop structures with small loops (1): these are in fact kinetically trapped.
In particular, these results help explain why certain inverted repeats can lead to genetic instability (10,11,14), while those that underlie functional RNA structures such as microRNA precursors or riboswitches do not. Genetically unstable inverted repeats are characterized by short loops and long stems (10,11), making their extrusion frequent and their rewinding highly unlikely. The high probability of the resulting cruciform facilitates its targeting by excision and repair machineries. On the other hand, the apical loops in, for instance, structured RNA precursors are typically (but not always) quite large, on the order of >10 bases. For the dsDNA encoding the precursor, this corresponds to a large amount of DNA to unwind, making the barrier to cruciform extrusion too high and too far along the reaction coordinate to be crossed via thermal fluctuations. This is important as the long length of the DNA inverted repeats encoding miRNA precursors (typically 20–25 bp) is likely to forbid rewinding of an extruded cruciform in the underlying dsDNA. In agreement with this, we were unable to detect cruciform extrusion in DNA inverted repeats encoding the miRNA precursors BRF1-1, BRF1-2, HSA-mir-379 and HSA-mir-1225b (data not shown).
Finally, we note that the inverted repeats studied in this work underwent cruciform extrusion at levels of negative supercoiling no more than half of what is expected in vivo. As we find that the rates of cruciform extrusion and rewinding vary exponentially with supercoiling, it is therefore likely that supercoiling in vivo may in certain cases regulate cruciform extrusion. Thus, these results may help better understand the ‘genome rules’ that dictate the way in which ubiquitous inverted repeats in dsDNA can evolve to generate a novel function rather than be targeted for repair.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cold Spring Harbor Fellows program; CNRS ATIP program; University of Paris 7 – Diderot; EURYI grant from the European Science Foundation (to T.R.S.); R.S. acknowledges the support of Cold Spring Harbor Laboratory. Funding for open access charge: CNRS core funding.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Lilley D, Palecek E. The supercoil-stabilized cruciform of ColE1 is hyper-reactive to osmium tetroxide. EMBO J. 1984;3:1187–1192. doi: 10.1002/j.1460-2075.1984.tb01949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murchie A, Bowater R, Aboul-ela F, Lilley DM. Helix opening transitions in supercoiled DNA. Biochim. Biophys. Acta. 1992;1131:1–15. doi: 10.1016/0167-4781(92)90091-d. [DOI] [PubMed] [Google Scholar]
- 3.Benham C, Savitt A, Bauer W. Extrusion of an imperfect palindrome to a cruciform in superhelical DNA: complete determination of energetics using a statistical mechanical model. J. Mol. Biol. 2002;316:563–581. doi: 10.1006/jmbi.2001.5361. [DOI] [PubMed] [Google Scholar]
- 4.Dawid A, Guillemot F, Brème C, Croquette V, Heslot F. Mechanically controlled DNA extrusion from a palindromic sequence by single molecule micromanipulation. Phys. Rev. Lett. 2006;96:188102. doi: 10.1103/PhysRevLett.96.188102. [DOI] [PubMed] [Google Scholar]
- 5.Lilley DM. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc. Natl Acad. Sci. USA. 1980;77:6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizuuchi K, Mizuuchi M, Gellert M. Cruciform structures in palindromic DNA are favored by DNA supercoiling. J. Mol. Biol. 1982;156:229–243. doi: 10.1016/0022-2836(82)90325-4. [DOI] [PubMed] [Google Scholar]
- 7.Courey AJ, Wang JC. Cruciform formation in a negatively supercoiled DNA may be kinetically forbidden under physiological conditions. Cell. 1983;33:817–829. doi: 10.1016/0092-8674(83)90024-7. [DOI] [PubMed] [Google Scholar]
- 8.Lilley DM. The kinetic properties of cruciform extrusion are determined by DNA base-sequence. Nucleic Acids Res. 1985;13:1443–1465. doi: 10.1093/nar/13.5.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinden RR, Zheng GX, Brankamp RG, Allen KN. On the deletion of inverted repeated DNA in Escherichia coli: effects of length, thermal stability, and cruciform formation in vivo. Genetics. 1991;129:991–1005. doi: 10.1093/genetics/129.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collick A, Drew J, Penberth J, Bois P, Luckett J, Scaerou F, Jeffreys A, Reik W. Instability of long inverted repeats within mouse transgenes. EMBO J. 1996;15:1163–1171. [PMC free article] [PubMed] [Google Scholar]
- 11.Lobachev KS, Shor BM, Tran HT, Taylor W, Keen JD, Resnick MA, Gordenin DA. Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics. 1998;148:1507–1524. doi: 10.1093/genetics/148.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waldman AS, Tran H, Goldsmith EC, Resnick MA. Long inverted repeats are an at-risk motif for recombination in mammalian cells. Genetics. 1999;153:1873–1883. doi: 10.1093/genetics/153.4.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rattray A. A method for cloning and sequencing long palindromic DNA junctions. Nucleic Acids Res. 2004;32:e155–e162. doi: 10.1093/nar/gnh143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narayanan V, Mieczkowski P, Kim H-M, Petes T, Lobachev K. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell. 2006;125:1283–1296. doi: 10.1016/j.cell.2006.04.042. [DOI] [PubMed] [Google Scholar]
- 15.Hwa T, Marinari E, Sneppen K, Tang L-H. Localization of denaturation bubbles in random DNA sequences. Proc. Natl Acad. Sci. USA. 2003;100:4411–4416. doi: 10.1073/pnas.0736291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Opel M, Aeling K, Holmes W, Johnson R, Benham C, Hatfield G. Activation of transcription initiation from a stable RNA promoter by a Fis protein-mediated DNA structural transmission mechanism. Mol. Microbiol. 2004;53:665–674. doi: 10.1111/j.1365-2958.2004.04147.x. [DOI] [PubMed] [Google Scholar]
- 17.Dai X, Greizerstein M, Nadas-Chinni K, Rothman-Denes L. Supercoil-induced extrusion of a regulatory DNA hairpin. Proc. Natl Acad. Sci. USA. 1997;94:2174–2179. doi: 10.1073/pnas.94.6.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai X, Kloster M, Rothman-Denes L. Sequence-dependent extrusion of a small DNA hairpin at the N4 virion RNA polymerase promoters. J. Mol. Biol. 1998;283:43–58. doi: 10.1006/jmbi.1998.2096. [DOI] [PubMed] [Google Scholar]
- 19.Strawbridge E, Benson G, Gelfand Y, Benham C. The distribution of inverted repeat sequences in the Saccharomyces cerevisiae genome. Curr. Genet. 2010;56:321–340. doi: 10.1007/s00294-010-0302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strick T, Allemand J, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 21.Revyakin A, Ebright R, Strick T. Single-molecule DNA nanomanipulation: improved resolution through use of shorter DNA fragments. Nat. Methods. 2005;2:127–138. doi: 10.1038/nmeth0205-127. [DOI] [PubMed] [Google Scholar]
- 22.Strick T, Croquette V, Bensimon D. Homologous pairing in stretched supercoiled DNA. Proc. Natl Acad. Sci. USA. 1998;95:10579–10583. doi: 10.1073/pnas.95.18.10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Léger J, Romano G, Sarkar A, Robert J, Bourdieu L, Chatenay D, Marko J. Structural transitions of a twisted and stretched dna molecule. Phys. Rev. Lett. 1999;83:1066–1069. [Google Scholar]
- 24.Forth S, Deufel C, Sheinin M, Daniels B, Sethna J, Wang M. Abrupt buckling transition observed during the plectoneme form of individual molecules. Phy. Rev. Lett. 2008;100:148301–148305. doi: 10.1103/PhysRevLett.100.148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mosconi F, Allemand J-F, Bensimon D, Croquette V. Measurement of the torque on a single stretched and twisted dna using magnetic tweezers. Phys. Rev. Lett. 2009;102:078301. doi: 10.1103/PhysRevLett.102.078301. [DOI] [PubMed] [Google Scholar]
- 26.Bouchiat C, Mezard M. Elasticity model of a supercoiled DNA molecule. Phys. Rev. Lett. 1998;80:1556–1559. [Google Scholar]
- 27.Moroz J, Nelson P. Entropic elasticity of twist-storing polymers. Macromolecules. 1998;31:6333–6347. [Google Scholar]
- 28.Marko J. Torque and dynamics of linking number relaxation in stretched supercoiled DNA. Phys Rev. E. 2007;76:021926. doi: 10.1103/PhysRevE.76.021926. [DOI] [PubMed] [Google Scholar]
- 29.White J. Self linking and the gauss integral in higher dimensions. Am. J. Math. 1969;91:693–728. [Google Scholar]
- 30.Strick T, Allemand J-F, Bensimon D, Croquette V. The behavior of supercoiled DNA. Biophys. J. 1998;74:2016–2028. doi: 10.1016/S0006-3495(98)77908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charvin G, Allemand J-F, Strick T, Bensimon D, Croquette V. Twisting DNA: single molecule studies. Contemp. Phys. 2004;45:385–403. [Google Scholar]
- 32.Revyakin A, Ebright R, Strick T. Promoter unwinding and promoter clearance by rna polymerase: detection by single-molecule DNA nanomanipulation. Proc. Natl Acad. Sci. USA. 2004;101:4776–4780. doi: 10.1073/pnas.0307241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Revyakin A, Liu C-Y, Ebright R, Strick T. Abortive initiation and productive initiation by rna polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White J, Bauer W. Superhelical DNA with local substructures. A generalization of the topological constraint in terms of the intersection number and the ladder-like correspondence surface. J. Mol. Biol. 1987;195:205–213. doi: 10.1016/0022-2836(87)90337-8. [DOI] [PubMed] [Google Scholar]
- 35.Saito I, Stark G. Charomid: cosmid vectors for efficient cloning and mapping of large or small restriction fragments. Proc. Natl Acad. Sci. USA. 1986;83:8664–8668. doi: 10.1073/pnas.83.22.8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lilley D. Hairpin-loop formation by inverted repeats in supercoiled DNA is a local and transmissible property. Nucleic Acids Res. 1981;9:1271–1289. doi: 10.1093/nar/9.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liphardt J, Dumont S, Smith S, Tinoco I, Bustamante C. Equilibrium information from nonequilibrium measurements in an experimental test of Jarzynski’s equality. Science. 2002;296:1832–1835. doi: 10.1126/science.1071152. [DOI] [PubMed] [Google Scholar]
- 38.Larson M, Greenleaf W, Landick R, Block S. Applied force reveals mechanistic and energetic details of transcription termination. Cell. 2008;132:971–982. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hohng S, Zhou R, Nahas M, Yu J, Schulten K, Lilley D, Ha T. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the Holliday junction. Science. 2007;318:279–283. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mikheikin A, Lushnikov A, Lyubchenko Y. Effect of DNA supercoiling on the geometry of Holliday junctions. Biochemistry. 2006;45:12998–13006. doi: 10.1021/bi061002k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dawid A, Croquette V, Grigoriev M, Heslot F. Single-molecule study of RuvAB-mediated Holliday-junction migration. Proc. Natl Acad. Sci. USA. 2004;101:11611–11616. doi: 10.1073/pnas.0404369101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowater R, Aboul-ela F, Lilley DM. Large-scale stable opening of supercoiled DNA in response to temperature and supercoiling in (A + T)-rich regions that promote low-salt cruciform extrusion. Biochemistry. 1991;30:11495–11506. doi: 10.1021/bi00113a003. [DOI] [PubMed] [Google Scholar]
- 43.Gellert M, O’Dea MH, Mizuuchi K. Slow cruciform transitions in palindromic DNA. Proc. Natl Acad. Sci. USA. 1983;80:5545–5549. doi: 10.1073/pnas.80.18.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.