Abstract
The carbon catabolite repressor protein 4 (Ccr4)–Negative on TATA (Not) complex controls gene expression at two levels. In the nucleus, it regulates the basal transcription machinery, nuclear receptor-mediated transcription and histone modifications. In the cytoplasm, the complex is required for messenger RNA (mRNA) turnover through its two associated deadenylases, Ccr4 and Caf1. Not1 is the largest protein of the Ccr4–Not complex and serves as a scaffold for other subunits of the complex. Here, we provide evidence that human Not1 in the cytoplasm associates with the C-terminal domain of tristetraprolin (TTP), an RNA binding protein that mediates rapid degradation of mRNAs containing AU-rich elements (AREs). Not1 shows extensive interaction through its central region with TTP, whereas binding of Caf1 is restricted to a smaller central domain within Not1. Importantly, Not1 is required for the rapid decay of ARE-mRNAs, and TTP can recruit the Caf1 deadenylase only in presence of Not1. Thus, cytoplasmic Not1 provides a platform that allows a specific RNA binding protein to recruit the Caf1 deadenylase and thereby trigger decay of its target mRNAs.
INTRODUCTION
Cells maintain tight control of gene expression by regulating rates of transcription, messenger RNA (mRNA) stability, protein translation and protein stability. Genes that are rapidly turned on and off frequently achieve such dynamic expression patterns by synthesizing short-lived mRNAs. Genome-wide studies in stimulated T cells, macrophages and fibroblasts revealed that mRNA half-life is a major determinant of gene expression patterns (1,2). An adenosine/uridine-rich element (ARE) is located in the 3′-untranslated region (UTR) of many short-lived mRNAs and mediates rapid mRNA degradation (3,4). Examples of mRNAs containing potent AREs include many cytokine transcripts such as tumor necrosis factor-α (TNF-α), granulocyte-macrophage colony-stimulating-factor (GM-CSF) and interleukin (IL)-3 mRNA as well as mRNAs encoding for transcription factors such as c-myc, c-fos and Ier3 (5).
Tristetraprolin (TTP) is an RNA binding zinc-finger protein required for ARE-mediated mRNA decay (AMD). The physiological role of TTP as an inhibitor of TNF-α expression was discovered by the analysis of TTP knock out mice (6). TTP was found to act at the post-transcriptional level by binding to the ARE of TNF-α mRNA and accelerating its degradation (7). TTP recognizes the ARE via its two cysteine–cysteine–cysteine–histidine (CCCH) zinc-finger domains. In addition to TNF-α, TTP was shown to mediate rapid degradation of the mRNAs encoding GM-CSF (8), IL-2 (9), IL-3 (10), IL-10 (11) and Ier3 (12). The TTP family of proteins comprises two additional paralogs, BRF1 and BRF2, which also promote AMD (13,14).
AREs destabilize mRNA by inducing a rapid shortening of the poly(A) tail (15). Subsequent degradation of the mRNA body occurs either from the 3′-end through the exosome (16–18) or from the 5′-end through decapping followed by Xrn1-mediated mRNA decay (19–21). Importantly, deadenylation is the first and rate-limiting step for both decay pathways. Several poly(A)-specific exoribonucleases in the cytoplasm share the job of shortening poly(A) tails. Whereas the Pan2–Pan3 complex was found to hydrolyse poly(A) in a distributive manner, the carbon catabolite repressor protein 4 (Ccr4)–Negative on TATA (Not) complex hydrolyses poly(A) in a processive manner and is responsible for rapid deadenylation (22). According to an early yet still valid classification, the highly potent class II AREs of cytokine mRNAs cause asynchronous deadenylation that reflects the activity of a processive deadenylase, whereas class I AREs, found e.g. in the 3′-UTR of the c-fos and c-myc transcription factors, cause synchronous deadenylation as the result of a distributive deadenylase activity. TTP typically interacts with class II AREs of TNFα, GM-CSF and IL-3 mRNA (7,8,23), and may thus recruit a processive deadenylase. In vitro decay studies suggested involvement of the poly-A ribonuclease (PARN) since recombinant TTP was found to stimulate PARN-induced deadenylation of an ARE-mRNA (24). Given that a direct interaction of TTP with PARN could not be observed, it is not clear how TTP activates or recruits PARN. In a different study, TTP was found to interact through its N-terminal domain with the Ccr4 deadenylase (20), and more recently, TTP was shown to interact with Caf1 (25). Functional studies on the relative importance of Ccr4 and Caf1 deadenylases are challenging since multiple paralogs exist for both proteins in the human genome. Studies with dominant-negative mutants of Ccr4 and Caf1 suggested that Ccr4 may have a predominant role in the regular turnover of mRNAs lacking a dedicated destabilizing element (22,26). By knocking down simultaneously Ccr4a (CNOT6) and Ccr4b (CNOT6L) or Caf1a (CNOT7) and Caf1b (CNOT8, also CALIF or POP2) in human HT1080 cells, we could previously show that Caf1a/b are required for the degradation of a reporter mRNA containing the ARE of GM-CSF (a typical class II ARE), whereas the knock down of Ccr4a/b had no effect (27). In human, Ccr4 and Caf1 together are part of the large, 1.2 MDa Ccr4–Not complex that comprises Not1 (CNOT1), Not2 (CNOT2), Not3 (CNOT3), Caf40 (CNOT9 or Rcd1), CNOT10, TAB182 and C2ORF29 (28,29). In the nucleus, the Not1, Not2 and Caf40 subunits of the human Ccr4–Not complex play a role as regulators of transcription (30,31). Not1, e.g. binds to oestrogen receptor and thereby represses ligand-dependent transcriptional activation (31). In addition, studies in Saccharomyces cerevisiae showed that the Not subunits and Not4, in particular, are required to maintain normal levels of histone methylation (32,33). As for the cytoplasmic function of the Ccr4–Not complex, early studies in yeast identified Ccr4 and Caf1 (also termed Pop2) as major cytoplasmic deadenylases (34,35). In yeast, Ccr4 appears to be more active than Caf1 (36,37), whereas Caf1 seems to be the major deadenylase in Drosophila, trypanosomes and mammalian cells (27,38,39). Yeast studies further found that Not2 and Not5 enhance the deadenylation rate of cytoplasmic mRNAs and may thus serve as activators of Ccr4 or Caf1 (36). A detailed analysis in Drosophila showed that in addition to Caf1, Not1, Not2 and Not3 are critical for rapid deadenylation of both bulk and Hsp70 mRNA (38).
In this study, we purified proteins associated with TTP and found that TTP interacts with Not1. We show that Not1 is required for TTP-mediated mRNA decay, map the regions involved in the TTP–Not1 interaction and provide evidence that TTP recruits the Caf1 deadenylase via its interaction with Not1.
MATERIALS AND METHODS
Plasmid construction
Plasmids pcDNA3-TTP-mycHis (p2115), pTet-7B (p2254) and pTet-7B-ARE (p2260) have been described previously (40). For pTet-7B-MS2bs (p2255), six repeats of the improved MS2-binding site were amplified by PCR with primers G37 and G38 (Supplementary Table S1) from plasmid b-6bs (41) and ligated as a BamHI—BglII fragment into the BglII site of pTet-7B.
The Tet-inducible TOPuro (p2433) vector was created by cloning a PvuII—BamHI fragment from pPUR (Clontech) containing a puromycine resistence gene into the PvuII/BamHI sites of pcDNA4/TO (Invitrogen). For plasmid TOPuroGS (p2434) containing a protein-G and streptavidin-binding peptide tag for N-terminal fusions, the GS tag was amplified by PCR from plasmid pCeMM-NTAP(GS)-Gw (42) using primers G1050/G1051 and inserted into the KpnI/BamHI sites of TOPuro. For TOPuroGS-TTP-wt (p2436), the mouse TTP cDNA was amplified by PCR using primers G1052/G1053 from pcDNA3-TTPwt-mycHis (p2115) and ligated into the EcoRI/XhoI sites of TOPuroGS (p2434). To generate TOPuro-mycSG (p2484) for C-terminal fusions of a mycSG- tag, mycSG was amplified by PCR from pCeMM-CTAP(SG)-Gw (p2432) (42) using primers G1120/G1121 that contain a SalI and XhoI linker, respectively, and cloned into the XhoI site of TOPuro (p2433). Based on TOPuro-mycSG, TOPuro-Caf1a-mycSG (p2485) encoding human wt Caf1a and TOPuro-Caf1a-AA-mycSG (p2737) containing the D40A/E42A mutation were cloned as described previously (43).
For pcDNA3-YFP-TTP (p2789), pcDNA3-YFP-TTP-M1,2 (p2712), pcDNA3-YFP-TTP-N (p2790), pcDNA3-YFP-TTP-NZ (p2791) and pcDNA3-YFP-TTP-C (p2792), the TTP cDNA fragments were taken out of the corresponding pcDNA3-TTP-mycHis plasmids (40) and cloned into the EcoRI/XbaI sites of pcDNA3-YFP (p2168) described previously (43). The TTP-M1,2 mutant was kindly provided by Keith Blackwell (Joslin Diabetes Center, Boston, USA).
For pcDNA3-MS2cp-TTP-mycHis (p2200), the MS2cp was excised from pcNMS2 (41) using HindIII and ligated into the HindIII site of pcDNA3-mTTP-mycHis (p2115). pcDNA3-MS2cp-TTP-M1,2-mycHis (p2203) was generated in the same way using pcDNA3-TTP-M1,2-mycHis (p2117) as the vector. To clone pcDNA3-MS2cp-mycHis (p2205), pcDNA3-MS2cp-TTP-mycHis was opened with KpnI/XbaI to release TTP, and the vector was blunt-end religated. For pcDNA3-MS2cp-TTP-C-mycHis (p2808) and pcDNA3-MS2cp-TTP-N-mycHis (p2806), TTP was replaced in the pcDNA3-MS2cp-TTP-mycHis vector with the C- or N-terminus of TTP, respectively, by swapping with a ClaI—XbaI fragment from pcDNA3-TTP-C-mycHis (p2122) or pcDNA3-TTP-N-mycHis (p2123) (40).
pCMV-Flag-Not1 (p2556) encoding full-length Not1 has been described previously (31). For pCMV-Flag-Not1:1-700 (p2594), pCMV-Flag-Not1 was digested with NheI/XbaI and religated, thereby ejecting the region encoding amino acid 701–2376 of Not1. To generate pCMV-Flag-Not1:1-997, pCMV-Flag-Not1 was digested with XbaI and religated, thereby ejecting the region encoding aa 998–2376 of Not1. The empty vector pcDNA3-Flag (p2002) contains the sequence ACC ATG GAC TAC AAG GAC GAT GAC GAC AAG between the HindIII and BamHI sites of pcDNA3 (Invitrogen).
For pcDNA3-HA-Not1:727-1449 (p2642), pCMV-Flag-Not1 was digested with PvuII, and the 2172-bp fragment was ligated into the EcoRV site of pcDNA3-HA (p2003) described previously (43). Subsequently the HA tag was replaced by the YFP tag from plasmid pcDNA3-YFP by a HindIII/KpnI digest. From the same PvuII digest of pCMV-Flag-Not1, the 1911-bp fragment was cloned into the EcoRV site of pcDNA3-HA to generate pcDNA3-HA-Not1:1880-2376 (p2657). For pcDNA3-YFP-Not1:1880-2376 (p2693), the HA tag was then replaced by YFP via an SpeI/BamHI digest. For pcDNA3-HA-Not1:1330-1601 (p2673), the Not1 fragment 1330–1601 was amplified by PCR using primers G1653/G1654 and cloned into the BamHI/XhoI sites of pcDNA3-HA. Subsequently the HA tag was replaced by YFP via an SpeI/BamHI digest to generate pcDNA3-YFP-Not1:1330-1601 (p2694). For HA-tagged Ccr4a, the CNOT6 cDNA was cloned into the pMT2MS-HA vector.
Cell culture and transfection
HeLa and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% fetal calf serum (FCS) (Gibco), 2 mM l-glutamine, 100 U/ml penicillin and 0.1 mg/ml streptomycin (all PAN). T-REx-HeLa- cells (Invitrogen) were cultured in the same medium containing 10% tetracycline-negative FCS (PAA). All cells were grown at 37°C and 5% CO2.
Cells were transfected with DNA using polyethyleneimine (PEI) (Polysciences Europe; 1 mg/ml, pH 7.0) at a ratio of 1:2 (DNA:PEI) in serum-free DMEM without antibiotics. For transfection of siRNAs, Lipofectamine 2000 (Invitrogen) and Optimem (Gibco) were used according to the manufacturer's protocol.
Medium was changed to regular DMEM 4 h after transfection of DNA or siRNA.
TTP purification
T-REx-HeLa cells stably transfected with TOPuroGS or TOPuroGS-TTP-wt were treated with doxycycline (1 µg/ml) for 16 h to induce expression of the proteins. Cytoplasmic lysates were prepared from ∼2 × 108 cells using RNP lysis buffer (1% NP40, 150 mM NaCl, 50 mM Tris–HCl, pH 8.0, 1 mM MgCl2, 10% glycerol, 1 mM dithiothreitol, 1 mM Na2VO3, 50 mM NaF and 40 nM okadaic acid), and tagged proteins were bound to streptavidin sepharose (GE Healthcare). The beads were washed five times with RNP buffer and eluted by the addition of biotin (1 mM, pH 5.0). The eluate was concentrated by vacuum centrifugation, resolved on a 5–20% gradient polyacrylamide gel, and proteins were visualized by colloidal Coomassie staining.
In-gel tryptic digestion and LC-MS/MS analysis
Mass spec analyses were carried out at the core facility for mass spectrometry and proteomics of the Center for Molecular Biology at the University of Heidelberg (ZMBH). Proteins in the gel were stained with colloidal Coomassie, and the entire lane was cut into five gel slices with a scalpel. Gel slices were transferred to a 96-well plate and reduced, alkylated and digested with trypsin (44) using a Digest pro MS liquid handling system (Intavis). Following digestion, tryptic peptides were extracted from the gel pieces with 50% acetonitrile/0.1% trifluoroacetic acid (TFA), concentrated nearly to dryness in a vacuum centrifuge and diluted to a total volume of 30 µl with 0.1% TFA; 25 µl of the sample was analyzed by a nano-HPLC system (Eksigent 1D plus) coupled to a ESI LTQ Orbitrap mass spectrometer (Thermo Fisher). Sample was loaded on a C18 trapping column (Inertsil, LC Packings) with a flow rate of 10 µl/min 0.1% TFA. Peptides were eluted and separated on an analytical column (75 µm × 150 mm) packed with Inertsil 3 µm C18 material (LC Packings) with a flow rate of 200 nl/min in a gradient of buffer A (0.1% formic acid) and buffer B (0.1% formic acid, acetonitrile): 0–6 min: 3% B; 6–60 min: 3–40% B; 60–65 min: 60–90% B. The column was connected with a nano-ESI emitter (New Objectives), and 1500 V was applied via liquid junction. One survey scan (resolution: 60000) was followed by five information-dependent product ion scans in the LTQ. Only doubly and triply charged ions were selected for fragmentation. All MS/MS samples were analyzed using Mascot (Matrix Science; version 2.2.03). The NCBInr database (taxonomy: homo sapiens, 204 932 entries) was used with the following search parameters: cleavage with trypsin, parent ion tolerance of 4.0 ppm and fragment ion mass tolerance of 0.20 Da. Iodoacetamide derivative of cysteine was specified in Mascot as a fixed modification. Deamidation of asparagine and oxidation of methionine were specified as variable modifications.
Immunoprecipitation and western blot analysis
Cells were transiently transfected 24–48 h prior to lysis. When Tet-inducible expression vectors were used, cells were treated over night with 1 µg/ml doxycycline (Sigma). Typically, cells from a 10-cm dish were solubilized in 400 µl RNP lysis buffer and nuclei were removed by centrifugation. YFP-tagged proteins were purified using the GFP-binder as described in Ref. (45). Cytoplasmic lysates were allowed to bind affinity matrices for 2–4 h at 4°C prior to washing. For IPs with specific antibodies, cytoplasmic lysates were pre-cleared by the addition of 30 µl protein A/G agarose beads (Thermo Scientific) for 1 h at 4°C, incubated with 1 µg of antibody for 2 h and 30 µl of protein A/G beads for additional 2 h before washing. Three to five washes were carried out using RNP lysis buffer. From the GFP-binder and the protein A/G agarose beads, proteins were eluted with 1% SDS-containing sample buffer. Where indicated, RNase A (Genomed) was added during the IP at a concentration of 1 µg/ml. RNase I (Ambion) was used at a concentration of 500 U/ml. RNA was extracted from the unbound fraction using TriFast (Peqlab) according to manufacturer’s protocol.
Proteins were resolved on 5–20% gradient polyacrylamide gels and transferred onto 0.2 µm pore size nitrocellulose membranes (Peqlab) for western blotting. Horseradish peroxidase-coupled secondary antibodies (Jackson Immunoresearch) in combination with Western Lightning-enhanced chemiluminescence substrate (Perkin Elmer) were used for detection on chemiluminescence film from GE Healthcare.
Antibodies
The following antibodies were used for IP and western blot analysis: Mouse monoclonal anti-HA (HA.11, Covance), anti-myc 9E10 (MMS-150P, Covance), anti-Flag (M2, Sigma Aldrich), rabbit polyclonal anti-GFP (Abcam ab290) and anti-14-3-3 K-19 (Sant Cruz, sc-629). Anti-Caf1a antibody was kindly provided by Ann-Bin Shyu (University of Texas, Houston, TX, USA).
Northern blot analysis
For RNA decay experiments, HeLa cells were transiently transfected with pTet-Off (Clontech) and a pTet-7B reporter plasmid; 1 µg/ml doxycycline (Sigma) was added to cell cultures for indicated time periods and total RNA was extracted using the Genematrix RNA purification kit (Eurx, Roboklon). To generate deadenylated mRNA, the 20 µg of total RNA was annealed with 1 nmol/µl oligo dT18 in presence of 1 mM EDTA for 15 min at 26°C. RNA:DNA hybrids were digested using 5 U/µl RNase H (NEB) for additional 15 min at 37°C. RNA was extracted with phenol:chloroform:isoamylalcohol (50:50:1) and precipitated; 5–15 µg of RNA was resolved by 1.1% agarose/2% formaldehyde/MOPS gel electrophoresis and blotted over night with 8 × saline-sodium citrate (SSC) buffer onto Hybond-N+ Nylon membranes (GE Healthcare). Membranes were hybridized overnight at 55°C with digoxigenin-labeled RNA probes synthesized in vitro using Sp6 polymerase (Fermentas). Membranes were washed twice with 2× SSC/0.1% SDS for 5 min, and twice with 0.5× SSC/0.1% SDS for 20 min at 65°C. Alkaline phosphatase-coupled anti-digoxigenin Fab fragments and CDP-Star substrate (both Roche) were used for detection according to the manufacturer’s instructions. The following primers were used to generate templates for Sp6 probes by PCR: G1000 and G1001 for the probe against exons 1 and 2 of rabbit β-globin; G83 and G1009 for the probe against human nucleolin.
Real-time PCR
cDNA was synthesized from 5 µg of total RNA using oligo-dT18 (Invitrogen) and M-MLV H(–) reverse transcriptase (Promega). PCRs were carried out with 1:40 of a cDNA reaction, 200 nM target-specific primers and SYBR Green Mastermix (Roche) in a total volume of 10 µl on a Lightcycler 480 (Roche). The following primers were used: G1578/G1579 for Not1, and G1576/G1577 for nucleolin as internal control.
siRNAs
Cells were transfected twice with siRNA at a final concentration of 100 nM over a period of 4 days using Lipofectamine 2000 (Invitrogen). The siRNA concentration was 50 nm each in cases where two siRNAs were transfected simultaneously. Where indicated, plasmid DNA was added to the second siRNA transfection. siRNAs were synthesized by Ambion and correspond to the following sequences (sense strand):
D0/S015 (control), 5′-GCAUUCACUUGGAUAGUAAdTdT-3′; C2/S014 (control), 5′-GCAUUCACUUGGAUAGUAAdTdT-3′; U0 (control), 5′-GAAUGCUCAUGUUGAAUCAdTdT-3′ S021 (Not1), 5′-GGAACUUGUUUGAAGAAUAdTdT-3′; S034 (Not1), 5′-GAGGAUGACAAUCGAGAAAdTdT-3′.
RESULTS
Not1 interacts with TTP
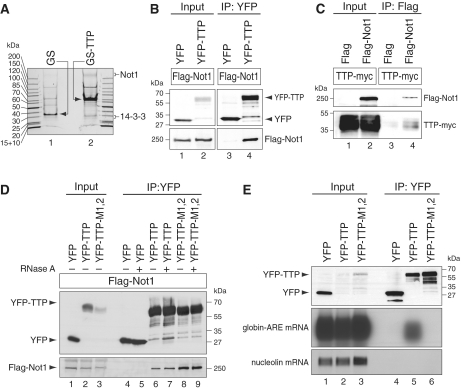
In order to identify proteins that associate with TTP, we generated a Tet-inducible vector expressing GS-tagged TTP. This tag encodes for protein G and a streptavidin-binding peptide (42). Stably transfected HeLa T-REx cells were generated with inducible expression of GS-TTP, and the fusion protein was affinity-purified on streptavidin sepharose. Due to difficulties with TEV cleavage, we could not perform a two-step purification as intended initially. Upon elution with biotin, co-purifying proteins were visualized by Coomassie staining and compared to proteins co-purifying with the GS tag alone (Figure 1A). Proteins from both purifications were identified by mass spectrometry. If a protein was identified by three or more peptides, and if the peptide number was ≥3-fold higher in the GS-TTP purification compared with the GS purification, it was considered a potential candidate associated with TTP (Supplemental Table S2). Among previously known interacting partners (46), we found 14-3-3 adaptor proteins (40,47) to co-purify with wild-type TTP (Figure 1A), but not with a non-phosphorylatable S52A/S178A mutant of TTP (data not shown). Interestingly, five of the seven isoforms of 14-3-3 could be identified in our mass spec analysis, suggesting that the interaction of phospho-TTP with 14-3-3 is not isoform-specific.
Figure 1.
TTP interacts with Not1 independently of RNA. (A) T-REx-Hela cells were stably transfected with either the GS tag alone or GS-TTP, and treated with doxycycline for 16 h to induce expression of the tagged proteins. Cytoplasmic lysates were prepared from ∼2 × 108 cells, and tagged proteins were purified using streptavidin sepharose. Proteins were eluted with biotin, resolved on a 5–20% gradient polyacrylamide gel and visualized by colloidal Coomassie staining. (B) HEK293 cells were transiently transfected with Flag-Not1 together with either YFP or YFP–TTP. After 1 day, cytoplasmic lysates (input) were prepared for IP with GFP-binder. Western blot analysis was carried out with antibodies against the Flag and YFP tags. (C) HEK293 cells were transiently transfected with TTP-myc together with either Flag alone or Flag-Not1. Cytoplasmic lysates were used for IP with Flag antibody, and western blots analysis was carried out with antibodies against the myc and Flag tags. (D) HEK293 cells were transiently transfected with Flag-Not1 together with YFP, YFP–TTP or the zinc-finger mutant YFP–TTP-M1,2. IP and western blot analysis was carried out as in panel B, except that RNase A was added during IP where indicated. (E) HeLa cells were transiently transfected with a β-globin reporter gene containing the ARE of TNFα in its 3′-UTR. In addition, cells were transfected with YFP, YFP–TTP or the zinc-finger mutant YFP–TTP–M1,2. RNA and protein were extracted from both the input and IP samples. Western blot analysis was carried out with an antibody against YFP, and globin-ARE as well as nucleolin mRNA were detected by northern blot analysis.
We further noticed that a large protein >200 kDa in size co-purified with GS-TTP (Figure 1A), and this protein was identified as Not1 (Supplementary Table S2). Not1 is the scaffold protein of the Ccr4–Not complex that regulates transcription in the nucleus and contains two major cytoplasmic deadenylases, Ccr4 and Caf1 (48). Since Ccr4 and Caf1 are required for turnover of eukaryotic mRNAs (27,34,35,39,49), we further explored the interaction of Not1 with TTP. By transient transfection in HEK293 cells, we found that yellow fluorescent protein (YFP)-tagged TTP co-IPs with Flag-tagged Not1, whereas YFP alone did not (Figure 1B, lanes 3 and 4). By inverse IP we confirmed that Flag-Not1 co-IPs with myc-tagged TTP, whereas the Flag tag alone does not (Figure 1C, lanes 3 and 4). We then expressed the TTP mutant M1,2 (50), which does not bind RNA because of CCCH to SSCH mutations in both of its zinc fingers. IP analysis showed that Flag-Not1 was efficiently co-immunoprecipitated with both wt and mutant TTP (Figure 1D, lanes 6 and 8). Likewise, addition of RNase A during the IP did not abolish the interaction between YFP-TTP and Flag-Not1 (Figure 1D, lanes 7 and 9), suggesting that the interaction between TTP and Not1 is RNA-independent. Lack of RNA binding by TTP-M1,2 was confirmed by RNA-IP: whereas wt TTP efficiently co-purified with a globin-ARE reporter mRNA (Figure 1E, lane 5) and the zinc-finger mutant TTP-M1,2 did not (lane 6).
Not1 is required for TTP-mediated mRNA deadenylation and decay
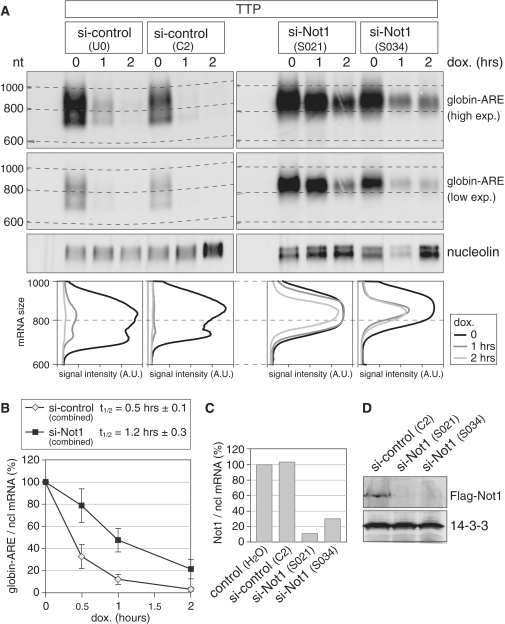
Next, we determined whether Not1 is required for AMD. We first reduced Not1 expression by transfection of two different siRNAs in HeLa cells, and subsequently transfected TTP together with a β-globin reporter gene that contains the ARE of TNFα in its 3′-UTR and is driven by a tetracycline-sensitive promoter. Degradation of the reporter mRNA was measured after blocking its transcription with doxycycline. In the presence of two different control siRNAs, the globin-ARE mRNA showed very rapid deadenylation and subsequent degradation (Figure 2A, left panels). In contrast, both siRNAs directed against Not1 caused a strong stabilization of the globin-ARE mRNA (right panels). By analyzing the reporter mRNA size, we noticed that the globin-ARE mRNA remained above the 800 nt mark after Not1 knock down, whereas it was shortened to ∼700 nt in the control conditions. A lower exposure of the globin-ARE mRNA signal in the second panel of Figure 2A clearly shows the deadenylated mRNA species that is present in the control knock downs, but absent in the Not1 knock downs. The difference was visualized by plotting the signal intensity as a function of mRNA size in the panels below of the northern blots of Figure 2A. This result indicated that Not1 is primarily required for full deadenylation of the reporter mRNA. Quantification of the overall mRNA signals in three biological repeat experiments showed that knocking down Not1 increases the half-life of globin-ARE mRNA from 0.5 to 1.2 h (Figure 2B). By quantitative PCR, we determined that the two siRNAs reduced Not1 mRNA expression levels to ∼10% and 30%, respectively (Figure 2C). Knock down efficiency was also confirmed at the protein level, as both siRNAs reduced Flag-Not1 levels below the detection limit of our western blot analysis (Figure 2D). Taken together, we concluded from these experiments that Not1 is required for AMD of a reporter mRNA containing a class II ARE.
Figure 2.
Not1 is required for ARE-mediated mRNA deadenylation and decay. (A) HeLa cells were transfected with two independent control siRNAs or two siRNAs targeting Not1. After 48 h, cells were transfected again with the same siRNAs together with a plasmid encoding TTP, a pTet-Off-driven β-globin reporter gene containing the ARE of TNF-α in its 3′ UTR, and the Tet-Off transactivator. Reporter gene transcription was blocked specifically by addition of doxycycline 24 h after the second transfection, and RNA was isolated after the time intervals indicated. Globin-ARE and nucleolin mRNA were detected by northern blot analysis; a high and low exposure is provided for the globin-ARE signal. The size of the mRNA was determined by comparison to an RNA marker. In the bottom panel, deadenylation was visualized by quantifying the signal intensity of globin-ARE mRNA along the length of the signal and plotting it as a function of mRNA size. (B) Quantification of the globin-ARE reporter mRNA decay in panel A and repeat experiments. Signal intensities of globin-ARE mRNA were normalized to nucleolin mRNA and represented as % of the initial value. Results from the two control and the two Not1 siRNA transfections were combined. The graph shows average values ± SE from eight (si-control) and six (si-Not1) biological repeat experiments. (C) HeLa cells were transfected twice with siRNAs as in panel A. Not1 mRNA levels were quantified by real-time PCR using nucleolin mRNA for normalization. (D) HEK293 cells were first transfected with Flag-Not1, and 1 day later with the siRNAs indicated. After one additional day, cytoplasmic lysates were prepared and analyzed by western blotting using antibodies against Flag and 14-3-3.
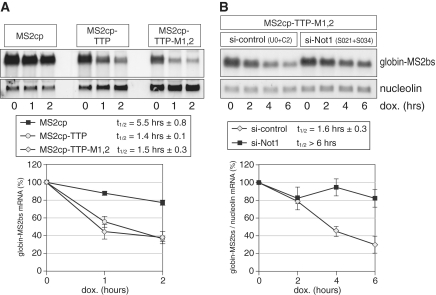
Since several ARE-binding proteins including TTP, BRF1, KSRP and AUF1 participate in AMD (51), we then tested whether an mRNA whose decay is mediated by TTP alone would also require Not1 for degradation. For this purpose, we chose a tethering approach whereby TTP is forced to bind an mRNA lacking an ARE through the tight interaction of the MS2 bacteriophage coat protein (cp) with its cognate binding site (bs) in the MS2 RNA. MS2cp-TTP fusion proteins were expressed in HeLa cells together with a β-globin reporter mRNA containing six repeats of the MS2bs in its 3′-UTR. Figure 3A shows that tethering of MS2cp-TTP causes rapid degradation of the mRNA, whereas the mRNA remains stable after tethering of MS2cp alone. Tethering of the TTP-M1,2 mutant, which does not bind RNA, was as efficient as tethering of wt TTP (Figure 3A). This confirms that TTP functions by recruiting components of the RNA decay machinery to the bound RNA (16,20). We then tested the effect of knocking down Not1 by transfecting two siRNAs (S021 and S034) simultaneously. As seen with the ARE-containing mRNA above, we observed that the mRNA tethered to MS2cp-TTP-M1,2 was stabilized by knock down of Not1 (Figure 3B). Quantification of four biological repeat experiments showed an increase in the mRNA half-life from 1.6 h to >6 h. Thus, mRNA decay mediated by binding of TTP alone is clearly dependent on the deadenylase scaffold protein Not1.
Figure 3.
Not1 is required for mRNA decay induced by tethering of TTP. (A) HeLa cells were transiently transfected with MS2cp, MS2cp-TTP or the zinc-finger mutant MS2cp-TTP-M1,2 that does not bind mRNA by itself. In addition, a pTet-Off-driven β-globin reporter gene that contains six MS2-binding sites (bs) in its 3′ UTR together with the Tet-Off transactivator were transfected into all cells. Transcription of the reporter gene was turned off by treating cells with doxycycline. RNA was isolated after the indicated time intervals. Globin-MS2bs and nucleolin mRNA levels were detected by northern blot analysis. The bottom graph shows quantification of the globin-MS2bs mRNA. Average values ± SE from three biological repeat experiments are represented. (B) HeLa cells were transfected simultaneously with either two control siRNAs or two siRNAs targeting Not1. After 48 h, cells were transfected with the same siRNAs again together with MS2cp-TTP-M1,2, the pTet-Off-globin-MS2bs reporter gene and the Tet-Off transactivator; 24 h later, cells were treated with doxycycline and total RNA was isolated after the indicated time intervals. Globin-MS2bs and nucleolin mRNA levels were detected by northern blot analysis. For quantification, signal intensities of globin-MS2bs mRNA were normalized to nucleolin mRNA and represented as percentage of the initial value. The bottom graph shows average values ± SE from four biological repeat experiments.
Caf1 is required for TTP-induced mRNA deadenylation
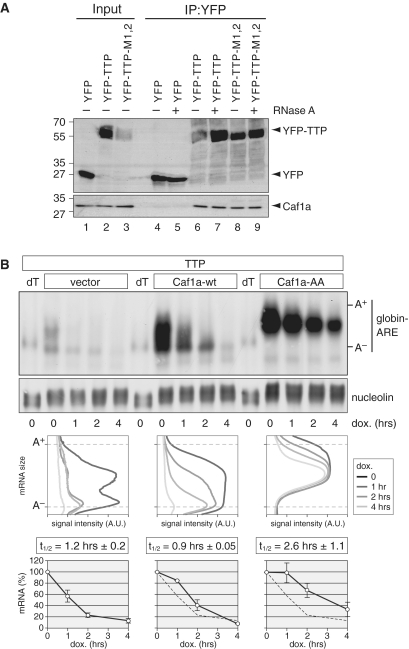
In yeast, Ccr4 and Caf1 were found to be the major cytoplasmic deadenylases (35), whereas in trypanosomes, Drosophila and human cells, Caf1 appears to be the predominant deadenylase (27,49). Previously, we have shown that in human HT1080 fibrosarcoma cells, Caf1 is of particular importance for the rapid degradation of an ARE-containing mRNA (27). To further establish the link between Caf1 and AMD, we examined whether TTP could co-IP with endogenous Caf1a in HEK293 cells. Indeed, we found that YFP-tagged TTP associates with Caf1a (Figure 4A, lane 6), whereas YFP alone did not (lane 4). Importantly, addition of RNase A to the IP did not reduce the interaction of YFP-TTP with Caf1a (lane 7). Given that YFP-TTP-M1,2 also co-IPs with Caf1a (lanes 8 and 9), we concluded that TTP associates with the Caf1 deadenylase in an RNA-independent manner.
Figure 4.
Caf1 interacts with TTP and mediates deadenylation of globin-ARE-mRNA. (A) HeLa cells were transiently transfected with YFP, YFP–TTP or TTP–M1,2. Cytoplasmic lysates (input) were prepared after 24 h for IP with GFP-binder. Inputs were divided and one half was treated with RNase A during IP. Western blot analysis was carried out with antibodies against YFP and endogenous Caf1a. (B) HeLa cells were transiently transfected with TTP, the Tet-Off transactivator and a pTet-Off-driven β-globin reporter gene containing the ARE of TNF-α in its 3′-UTR. Cells were co-transfected with either empty vector, Caf1a-wt or dominant negative Caf1a-AA. Reporter gene transcription was blocked specifically by addition of doxycycline 20 h after transfection, and total RNA was isolated after the indicated time intervals. Globin-ARE and nucleolin mRNA were detected by northern blot analysis. dT indicates that the RNA was treated with oligo-dT and RNase H to generate deadenylated mRNA. In the middle panel, deadenylation was visualized by quantifying the signal intensity of globin-ARE mRNA along the length of the signal and plotting it as a function of mRNA size. In the bottom panel, the overall signal intensity of globin-ARE mRNA was quantified and normalized to nucleolin mRNA. Average values ± SE were obtained from three biological repeat experiments and plotted as % of the initial time point.
We then tested whether Caf1 is important for TTP-induced mRNA degradation. Since knocking down both Caf1a and Caf1b together is not very efficient (27), we made use of a dominant-negative mutant of Caf1a, Caf1a-AA (D40A/E42A), which can efficiently prevent deadenylation (43). TTP was expressed together with a globin-ARE reporter mRNA in order to accelerate AMD (Figure 4B). Compared with vector control, co-expression of Caf1a-wt had only a small effect on the deadenylation pattern and decay rate of globin-ARE mRNA. Conversely, co-expression of dominant-negative Caf1a-AA strongly reduced both deadenylation and decay of the ARE-containing reporter mRNA. Figure 4B shows that only about half of the poly(A) tail is removed in the presence of Caf1a-AA. This is in line with the notion that Caf1 takes part in the second, rapid phase of deadenylation, as pointed out by Yamashita et al. (22). The effect on deadenylation was visualized by plotting the mRNA signal intensity along the length of the mRNA signal (Figure 4B, middle panel). Quantification of the mRNA decay rate from three biological repeat experiments (Figure 4B, bottom panel) showed that Caf1a-AA increased the mRNA half-life by >2-fold.
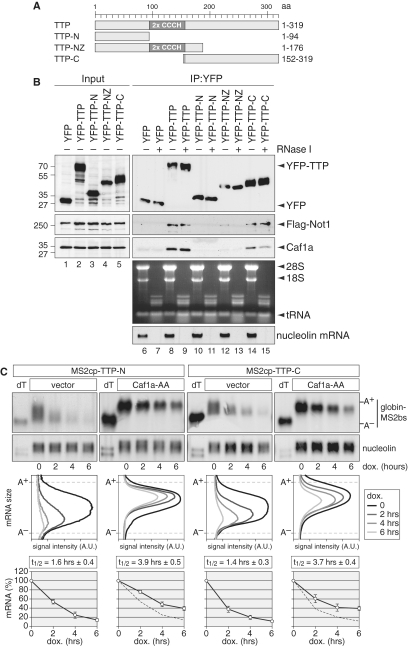
The C-terminal domain of TTP is critical for recruitment of Not1 and Caf1
TTP contains in its central part a tandem CCCH zinc-finger domain that is required for binding of AREs (13), whereas the N-terminal domain of TTP was found to stimulate mRNA decay by recruiting components of the general mRNA decay machinery (20). To further understand the interaction of TTP with Not1 and Caf1, we expressed the N- and C-terminal domains of TTP, schematically depicted in Figure 5A, in HEK293 cells. By co-IP analysis we found that the N-terminal domain of TTP (amino acid 1–94) does not interact with either Caf1a or Flag-Not1 (Figure 5B, lane 10). The NZ fragment (amino acid 1–176) comprising the N-terminus, the zing-finger domain and a small part of the C-terminal domain showed only a very weak interaction with Flag-Not1 or Caf1a (lane 12). In contrast, the C-terminal domain (amino acid 152–319) was able to co-IP Flag-Not1 and Caf1a as efficiently as full-length TTP (compare lanes 8 and 14). Treatment of the samples during IP with RNase I, which also cleaves RNA within a poly(A) tail, did not abrogate the interaction with either Flag-Not1 or Caf1a (lanes 9 and 15). Efficient degradation of RNA is shown in the bottom panels of Figure 5B for rRNA and nucleolin as an abundant mRNA. Since TTP–Not1/Caf1a interactions are technically challenging to detect, a repeat IP is shown in Supplementary Figure S1A and B. From these results we concluded that Not1 and Caf1a primarily interact with the C-terminal domain of TTP in an RNA-independent manner. We further tested the interaction of TTP with Ccr4a and again found the C-terminal domain of TTP to associate with HA-Ccr4a (Supplementary Figure S1C).
Figure 5.
TTP C-terminal domain interacts with Not1–Caf1a and induces mRNA deadenylation. (A) Schematic representation of the murine TTP fragments used in this study. N, N-terminal domain; Z, zinc-finger domain; C, C-terminal domain; aa, amino acids. (B) HEK293 cells were transiently transfected with YFP-tagged TTP constructs as indicated together with Flag-Not1. Cytoplasmic lysates (input) were prepared after 24 h for IP with GFP-binder. Inputs were divided and one half was treated with RNase I during IP. Western blot analysis was carried out with antibodies against YFP, Flag and endogenous Caf1a. To control RNase I efficiency, RNA was isolated from unbound fractions. Ribosomal and tRNA was visualized by ethidium bromide staining, nucleolin mRNA was visualized on a northern blot. (C) HeLa cells were transiently transfected with MS2cp-TTP-C or MS2cp-TTP-N together with the Tet-Off transactivator and a pTet-Off-driven β-globin reporter gene that contains six MS2-binding sites (bs) in its 3′ UTR. In addition, empty vector or dominant negative Caf1a-AA was co-transfected. Reporter gene transcription was blocked specifically by addition of doxycycline 20 h after transfection, and total RNA was isolated after the time intervals indicated. Globin-MS2bs and nucleolin mRNA were detected by northern blot analysis. dT indicates that the RNA was treated with oligo-dT and RNase H to generate deadenylated mRNA. In the middle panel, deadenylation was visualized by quantifying the signal intensity of globin-MS2bs mRNA along the length of the signal and plotting it as a function of mRNA size. In the bottom panel, the overall signal intensity of globin-MS2bs mRNA was quantified and normalized to nucleolin mRNA. Average values ± SE were obtained from three biological repeat experiments and plotted as percentage of the initial time point.
To functionally compare the N- and C-terminal domains of TTP, we again made use of the tethering assay. TTP-N and TTP-C were fused to the MS2cp, and both fragments were found to induce very rapid deadenylation and decay of the globin-M2bs mRNA (Figure 5C). Co-expression of the dominant-negative Caf1a-AA mutant efficiently prevented deadenylation of the tethered reporter mRNA and reduced its decay rate (Figure 5C, bottom panels). Taken together, these results provide strong evidence that the interaction of the TTP C-terminal domain with Not1–Caf1a is functionally relevant for the destabilizing activity of TTP. However, it is interesting to note that the N-terminal domain can also activate deadenylation and mRNA decay, although we do not detect an interaction with Not1/Caf1a. Similarly, a previous study by Lykke-Andersen and Wagner (2005) had found that both the N- and C-terminal domains of TTP mediate mRNA degradation (20).
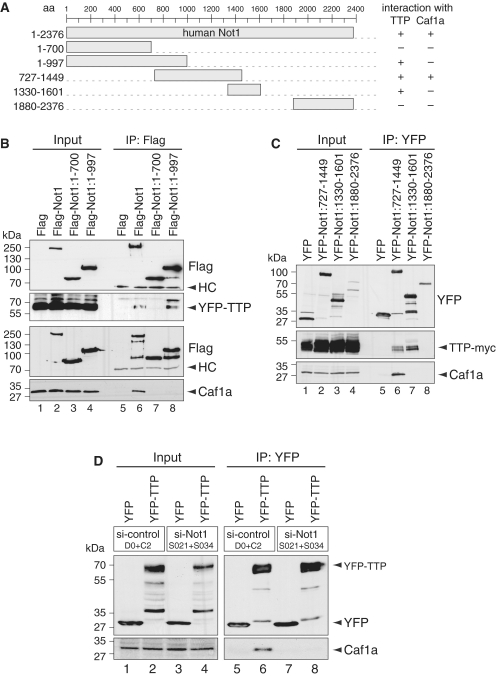
Interaction of TTP with Not1 is extensive and required for Caf1 recruitment
To further examine the interactions between TTP, Not1 and Caf1a, we divided the human Not1 cDNA, which encodes for a 2376-amino acid long protein, into several fragments (Figure 6A). The most N-terminal fragment of Not1 encompassing amino acid 1–700 did not co-IP with TTP, whereas a longer N-terminal fragment covering amino acid 1–997 did interact with TTP (Figure 6B, lanes 7 and 8). Importantly, neither of these two fragments showed an interaction with Caf1a. This result suggests that the Not1 region between amino acid 700 and 997 is important for the interaction with TTP, and that TTP can associate with Not1 independently of Caf1.
Figure 6.
Not1 associates with TTP and Caf1a differentially and is required for the interaction between TTP and Caf1a. (A) Schematic representation of the human Not1 fragments used in this study; aa, amino acids. (B) HEK293 cells were transiently transfected with Flag-tagged Not1 constructs as indicated together with YFP-TTP. Cytoplasmic lysates (input) were prepared after 24 h for IP with Flag antibody. Western blot analysis was carried out with antibodies against YFP, Flag and endogenous Caf1a. HC stands for IgG heavy chain. (C) HEK293 cells were transiently transfected with YFP-tagged Not1 constructs as indicated together with TTP-myc. Cytoplasmic lysates (input) were prepared after 24 h for IP with GFP-binder. Western blot analysis was carried out with antibodies against YFP, myc and endogenous Caf1a. (D) HeLa cells were transfected simultaneously with either two control siRNAs or two siRNAs targeting Not1. After 48 h, cells were transfected with the same siRNAs again together with YFP or YFP–TTP. Cytoplasmic lysates (input) were prepared 24 h later for IP with GFP-binder. Western blot analysis was carried out with antibodies against YFP and endogenous Caf1a.
Our previous yeast two-hybrid experiments mapping the interactions between human Ccr4-Not subunits indicated that Caf1a/CNOT7 interacts with amino acid 648–2376 of Not1 (28). We further examined the middle region of Not1 using a fragment that spans amino acid 727–1449. This fragment was able to co-IP with both TTP and Caf1a (Figure 6C, lane 6). Interestingly, a smaller Not1:1330–1601 fragment shifted slightly toward the C-terminus interacted efficiently with TTP, but not with Caf1a (lane 7). The last fragment we tested contains the C-terminus of Not1 (amino acid 1880–2373), and did not interact with either TTP or Caf1a (lane 8). Taken together, it is evident that TTP and Caf1a interact with different though overlapping regions of Not1. TTP shows a more extensive interaction with Not1 in the region between amino acid 700 and 1601, whereas the interaction with Caf1 is restricted to the region between amino acid 727–1499 of human Not1.
Finally, we wanted to test if the interaction between TTP and Caf1 is dependent on Not1. To this end, we knocked down Not1 expression in HeLa cells. Co-IP experiments clearly showed that the interaction between YFP-TTP and endogenous Caf1a is lost after knock down of Not1, whereas in a control knock down the interaction was unaffected (Figure 6D, lanes 6 and 8). Taken together, these results provide compelling evidence that Not1 provides a platform on which TTP recruits the Caf1 deadenylase to its target mRNAs.
DISCUSSION
Initially, the Not proteins were identified by genetic studies in S. cerevisiae as general repressors of transcription that preferentially act on promoters lacking a canonical TATA box (Negative on TATA, Not). This function is mediated through regulation of the TFIID transcription complex and histone modification by the Not proteins (48,52). In parallel, catabolite repressor protein 4 (Ccr4) was identified as a factor required for the expression of genes that are induced under non-fermentative conditions. Biochemical studies then revealed that Ccr4 and Not proteins are actually part of the same complex (53). In addition to its functions in the nucleus, the yeast Ccr4–Not complex was found to play an important role in the cytoplasm where Ccr4 and Caf1 represent the two major exoribonucleases responsible for cytoplasmic mRNA deadenylation (34,35,37).
In metazoa, the Ccr4–Not complex shares many of its characteristics with the yeast complex. In the nucleus, the human complex serves as a negative regulator of transcription through its subunits Not1, Not2 and Caf40 (30,31). Moreover, Not1 interacts with the ligand-binding domain of estrogen receptor alpha and represses estrogen receptor-mediated transcription (31). The general role of the cytoplasmic Ccr4–Not complex in mRNA deadenylation is also conserved in metazoa (22,49). In contrast to yeast, however, Caf1 was found to be the major deadenylase in trypanosomes as well as in Drosophila and human cells, whereas Ccr4 is either lacking (in trypanosomes) or of minor importance for deadenylation (25,27,49). Aside from Caf1 that carries the exoribonucleolytic activity, a detailed analysis in Drosophila cells showed that Not1, Not2 and Not3 are also essential for deadenylation of both bulk and Hsp70 mRNA (38). The exact role of these proteins in mRNA deadenylation, however, is not known.
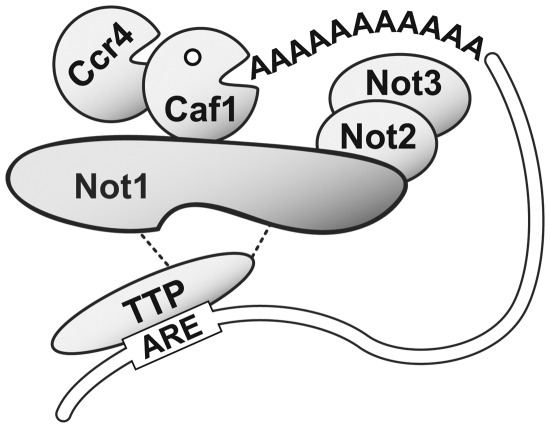
In this study, we provide evidence through biochemical purification and co-IP experiments that TTP associates with Not1 (Figure 1). TTP binds to mRNAs containing AREs and mediates their rapid degradation, and indeed we found that Not1 is required for efficient AMD (Figure 2). Using a tethering assay whereby TTP is bound to a reporter mRNA through a heterologous interaction, we could further show that mRNA decay that is strictly dependent on TTP also requires Not1 (Figure 3). Moreover, we found that TTP co-IPs with Caf1a, and that TTP-induced mRNA deadenylation is inhibited by a dominant-negative mutant of Caf1a (Figure 4). Thus, both Caf1 and Not1 are essential for the ability of TTP to deadenylate and destabilize its target mRNAs. A key finding was that TTP interacts with Caf1a only when Not1 is present (Figure 6D). Taken together, these results provide strong evidence that TTP interacts with Not1 in order to recruit the Caf1a deadenylase, which in turn induces rapid deadenylation of the mRNA bound to TTP. Our view of the functional relationship between TTP, Not1 and Caf1 is schematically depicted in Figure 7. In this model, TTP plays a central role in selective deadenylation of specific mRNAs. Recent experiments in mouse germ cells indicate that the Nanos2 protein performs an analogous role in recruiting the Ccr4–Not complex to meiosis-specific mRNAs for degradation in cytoplasmic P-bodies (54).
Figure 7.
Model of the cytoplasmic Ccr4–Not complex in association with TTP. TTP interacts through its C-terminal domain with the central region of Not1, whereas Caf1a binds to a more restricted though partially overlapping region of Not1 (this study). As shown previously, Not2 binds to the C-terminal region of human Not1, whereas Not3 is connected to the complex via Not2 (28). In accordance with the biochemical analysis of the Ccr–Not complex in yeast, human Ccr4a/b are connected to the complex via their interaction with Caf1a (28,55). TTP recruits the Caf1a deadenylase to its target mRNA containing AREs via its interaction with Not1.
Interaction studies in S. cerevisiae suggested that Ccr4/Caf1 bind to a middle fragment of yeast Not1 comprising amino acid 667–1152 (55). This region corresponds to amino acid 918–1453 in human Not1, which is in good agreement with our observation that the human Not1:727–1449 fragment interacts with Caf1a (Figure 6C). In contrast to Caf1a, we found that TTP associates with a larger region of Not1 that comprises amino acid 700–1601. Importantly, TTP interacts not only with the Not1:727–1449 fragment that also co-IPs with Caf1a, but in addition with the two non-overlapping fragments Not1:1–997 and Not1: 1330–1601. From this we concluded that TTP engages in an extensive interaction with Not1 that involves at least two separate interaction domains. However, because co-IP experiments cannot prove that two proteins are in direct contact, future experiments using recombinant proteins will have to address whether TTP and Not1 directly bind to each other.
Our analysis of TTP subfragments showed that the C-terminal domain of TTP interacts with both Not1 and Caf1a (Figure 5B). Interestingly, both the N- and C-terminal domains were able to cause mRNA deadenylation of a tethered reporter mRNA in a Caf1a-dependent manner (Figure 5C). Indeed, a previous study by Lykke-Andersen and Wagner (2005) had shown that the N-terminal domain of TTP interacts with various factors of the RNA decay machinery including Ccr4, the decapping enzyme Dcp2, the 5′-3′ exonuclease Xrn1 and Rrp4, a component of the exosome that has 3′-5′ exonuclease activity. This analysis could not assign any interactions to the C-terminal domain of TTP although the C-terminal domain was found to cause mRNA decay more efficiently in a tethering assay than the N-terminal domain (20). Our finding that the C-terminal domain efficiently associates with Not1–Caf1 provides an explanation for the high activity of the C-terminal domain. It remains unclear why Ccr4a interacts with the C-terminal domain of TTP in our hands (Supplementary Figure S1C), whereas it was immunoprecipitated with the N-terminal domain of TTP in the study of Lykke-Andersen and Wagner (2005).
TTP is known to be regulated in its activity through phosphorylation at two specific serine residues (S52 and S178) by the MK2 kinase (40,56). Phosphorylation at these two sites causes binding of 14-3-3 adaptor proteins and inhibition of TTP, which allows for a transient stabilization of TTP-target mRNAs (40). Very recently, phosphorylation of TTP was shown to reduce its interaction with Ccr4 and Caf1 (25,57). It will now be interesting to determine whether the TTP–Not1 interaction we describe here is also affected by phosphorylation, or whether the Not1–Caf1 interaction might be subject to regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Young investigator grant HZ-NG-210 from the Helmholtz Gemeinschaft (to G.S.); Netherlands Organization for Scientific Research grant NWO-TOP #700.57.302 (to H.Th.M.T.). Funding for open access charge: Helmholtz Gemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are very grateful to Tilmann Bürckstümmer (Center for Molecular Medicine of the Austrian Academy of Sciences, Vienna, Austria) for the GS- and SG-tag vectors; Jens Lykke-Andersen (University of California at San Diego, USA) for plasmids b-6bs and pcNMS2; Keith Blackwell (Joslin Diabetes Center, Harvard Medical School, Boston, USA) for the TTP-M1,2 mutant; Tobias Dick (German Cancer Research Center, Heidelberg) for the T-REx-HeLa cells and the pcDNA4/TO vector and Ann-Bin Shyu (University of Texas, Houston, USA) for the Caf1a antibody. We also thank Julia Luther, Sahil Sharma, Oksana Seibert, Frederic Bethke, Nicole Sakellari (all at the German Cancer Research Center, Heidelberg) and Nga-Chi Lau (University Medical Center Utrecht) for contributions to cloning plasmids.
REFERENCES
- 1.Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat. Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raghavan A, Ogilvie RL, Reilly C, Abelson ML, Raghavan S, Vasdewani J, Krathwohl M, Bohjanen PR. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002;30:5529–5538. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Natl Acad. Sci. USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw G, Kamen R. A conserved AU sequence from the 3′untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 5.Stumpo DJ, Lai WS, Blackshear PJ. Inflammation: cytokines and RNA-based regulation. Wiley Interdiscipl. Rev. RNA. 2010;1:60–80. doi: 10.1002/wrna.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 7.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 8.Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- 9.Ogilvie RL, Abelson M, Hau HH, Vlasova I, Blackshear PJ, Bohjanen PR. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J. Immunol. 2005;174:953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- 10.Stoecklin G, Ming XF, Looser R, Moroni C. Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Mol. Cell. Biol. 2000;20:3753–3763. doi: 10.1128/mcb.20.11.3753-3763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, Blackshear PJ, Anderson P. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J. Biol. Chem. 2008;283:11689–11699. doi: 10.1074/jbc.M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai WS, Parker JS, Grissom SF, Stumpo DJ, Blackshear PJ. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol. Cell. Biol. 2006;26:9196–9208. doi: 10.1128/MCB.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to AU-rich elements and destabilization of mRNA. J. Biol. Chem. 2000;275:17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 14.Stoecklin G, Colombi M, Raineri I, Leuenberger S, Mallaun M, Schmidlin M, Gross B, Lu M, Kitamura T, Moroni C. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 2002;21:4709–4718. doi: 10.1093/emboj/cdf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shyu A-B, Belasco JG, Greenberg ME. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 16.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 17.Anderson JR, Mukherjee D, Muthukumaraswamy K, Moraes KC, Wilusz CJ, Wilusz J. Sequence-specific RNA binding mediated by the RNase PH domain of components of the exosome. RNA. 2006;12:1810–1816. doi: 10.1261/rna.144606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee D, Gao M, O′Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao M, Wilusz CJ, Peltz SW, Wilusz J. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 2001;20:1134–1143. doi: 10.1093/emboj/20.5.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 23.Stoecklin G, Gross B, Ming XF, Moroni C. A novel mechanism of tumor suppression by destabilizing AU-rich growth factor mRNA. Oncogene. 2003;22:3554–3561. doi: 10.1038/sj.onc.1206418. [DOI] [PubMed] [Google Scholar]
- 24.Lai WS, Kennington EA, Blackshear PJ. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol. Cell. Biol. 2003;23:3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchese FP, Aubareda A, Tudor C, Saklatvala J, Clark AR, Dean JL. MAPKAP kinase 2 blocks tristetraprolin-directed mRNA decay by inhibiting CAF1 deadenylase recruitment. J. Biol. Chem. 2010;285:27590–27600. doi: 10.1074/jbc.M110.136473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mauxion F, Faux C, Seraphin B. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J. 2008;27:1039–1048. doi: 10.1038/emboj.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwede A, Ellis L, Luther J, Carrington M, Stoecklin G, Clayton C. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res. 2008;36:3374–3388. doi: 10.1093/nar/gkn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau NC, Kolkman A, van Schaik FM, Mulder KW, Pijnappel WW, Heck AJ, Timmers HT. Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem. J. 2009;422:443–453. doi: 10.1042/BJ20090500. [DOI] [PubMed] [Google Scholar]
- 29.Albert TK, Lemaire M, van Berkum NL, Gentz R, Collart MA, Timmers HT. Isolation and characterization of human orthologs of yeast CCR4-NOT complex subunits. Nucleic Acids Res. 2000;28:809–817. doi: 10.1093/nar/28.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zwartjes CG, Jayne S, van den Berg DL, Timmers HT. Repression of promoter activity by CNOT2, a subunit of the transcription regulatory Ccr4-not complex. J. Biol. Chem. 2004;279:10848–10854. doi: 10.1074/jbc.M311747200. [DOI] [PubMed] [Google Scholar]
- 31.Winkler GS, Mulder KW, Bardwell VJ, Kalkhoven E, Timmers HT. Human Ccr4-Not complex is a ligand-dependent repressor of nuclear receptor-mediated transcription. EMBO J. 2006;25:3089–3099. doi: 10.1038/sj.emboj.7601194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulder KW, Brenkman AB, Inagaki A, van den Broek NJ, Timmers HT. Regulation of histone H3K4 tri-methylation and PAF complex recruitment by the Ccr4-Not complex. Nucleic Acids Res. 2007;35:2428–2439. doi: 10.1093/nar/gkm175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laribee RN, Shibata Y, Mersman DP, Collins SR, Kemmeren P, Roguev A, Weissman JS, Briggs SD, Krogan NJ, Strahl BD. CCR4/NOT complex associates with the proteasome and regulates histone methylation. Proc. Natl Acad. Sci. USA. 2007;104:5836–5841. doi: 10.1073/pnas.0607996104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daugeron MC, Mauxion F, Seraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 36.Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. Embo J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J, Chiang YC, Denis CL. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. Embo J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, Simonelig M, Wahle E. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA. 2010;16:1356–1370. doi: 10.1261/rna.2145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J. Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 42.Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods. 2006;3:1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- 43.Ozgur S, Chekulaeva M, Stoecklin G. Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing-bodies. Mol. Cell. Biol. 2010;30:4308–4323. doi: 10.1128/MCB.00429-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Catrein I, Herrmann R, Bosserhoff A, Ruppert T. Experimental proof for a signal peptidase I like activity in Mycoplasma pneumoniae, but absence of a gene encoding a conserved bacterial type I SPase. FEBS J. 2005;272:2892–2900. doi: 10.1111/j.1742-4658.2005.04710.x. [DOI] [PubMed] [Google Scholar]
- 45.Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Sandler H, Stoecklin G. Control of mRNA decay by phosphorylation of tristetraprolin. Biochem. Soc. Trans. 2008;36:491–496. doi: 10.1042/BST0360491. [DOI] [PubMed] [Google Scholar]
- 47.Johnson BA, Stehn JR, Yaffe MB, Blackwell TK. Cytoplasmic localization of Tristetraprolin involves 14-3-3-dependent and -independent mechanisms. J. Biol. Chem. 2002;277:18029–18036. doi: 10.1074/jbc.M110465200. [DOI] [PubMed] [Google Scholar]
- 48.Collart MA, Timmers HT. The eukaryotic Ccr4-not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog. Nucleic Acid Res. Mol. Biol. 2004;77:289–322. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- 49.Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23:2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson BA, Geha M, Blackwell TK. Similar but distinct effects of the tristetraprolin/TIS11 immediate-early proteins on cell survival. Oncogene. 2000;19:1657–1664. doi: 10.1038/sj.onc.1203474. [DOI] [PubMed] [Google Scholar]
- 51.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 52.Denis CL, Chen J. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 2003;73:221–250. doi: 10.1016/s0079-6603(03)01007-9. [DOI] [PubMed] [Google Scholar]
- 53.Liu HY, Badarinarayana V, Audino DC, Rappsilber J, Mann M, Denis CL. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998;17:1096–1106. doi: 10.1093/emboj/17.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki A, Igarashi K, Aisaki K, Kanno J, Saga Y. NANOS2 interacts with the CCR4-NOT deadenylation complex and leads to suppression of specific RNAs. Proc. Natl Acad. Sci. USA. 2010;107:3594–3599. doi: 10.1073/pnas.0908664107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bai Y, Salvadore C, Chiang YC, Collart MA, Liu HY, Denis CL. The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol. Cell. Biol. 1999;19:6642–6651. doi: 10.1128/mcb.19.10.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chrestensen CA, Schroeder MJ, Shabanowitz J, Hunt DF, Pelo JW, Worthington MT, Sturgill TW. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding. J. Biol. Chem. 2004;279:10176–10184. doi: 10.1074/jbc.M310486200. [DOI] [PubMed] [Google Scholar]
- 57.Clement SL, Scheckel C, Stoecklin G, Lykke-Andersen J. Phosphorylation of TTP by MK2 impairs ARE-mRNA decay by preventing deadenylase recruitment. Mol. Cell. Biol. 2011;31:256–266. doi: 10.1128/MCB.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.