Abstract
Transcription factors are involved in a number of important cellular processes. The transcription factor NF-κB has been linked with a number of cancers, autoimmune and inflammatory diseases. As a result, monitoring transcription factors potentially represents a means for the early detection and prevention of diseases. Most methods for transcription factor detection tend to be tedious and laborious and involve complicated sample preparation, and are not practical for routine detection. We describe herein the first label-free luminescence switch-on detection method for transcription factor activity using Exonuclease III and a luminescent ruthenium complex, [Ru(phen)2(dppz)]2+. As a proof of concept for this novel assay, we have designed a double-stranded DNA sequence bearing two NF-κB binding sites. The results show that the luminescence response was proportional to the concentration of the NF-κB subunit p50 present in the sample within a wide concentration range, with a nanomolar detection limit. In the presence of a known NF-κB inhibitor, oridonin, a reduction in the luminescence response of the ruthenium complex was observed. The reduced luminescence response of the ruthenium complex in the presence of small molecule inhibitors allows the assay to be applied to the high-throughput screening of chemical libraries to identify new antagonists of transcription factor DNA binding activity. This will allow the rapid and low cost identification and development of novel scaffolds for the treatment of diseases caused by the deregulation of transcription factor activity.
INTRODUCTION
Transcription factors are a class of proteins that regulate gene expression by binding to specific DNA sequences within the regulatory regions of genes (1). Due to their important role in the regulation of gene expression, transcription factors are vital for cell development, differentiation and growth in biological systems (2–4). Typically, transcription factors exist in the cell in an inactive state and become activated by the presence of a specific ligand, leading to the expression of target gene(s). As a result, the inhibition or undesired activation of transcription factors can lead to a number of diseases which include developmental disorders (5–8), abnormal hormone responses (9–11), inflammation (12,13) and cancer (14–16). Therefore, the rapid and convenient detection of transcription factor activity is important for the development of inhibitors for the treatment or prevention of these diseases.
Current methods for the detection of transcription factor activity include DNA footprinting, western blotting, the gel mobility shift assay, affinity chromatography and visual microscopy (17–19). However, the aforementioned methods are generally tedious, laborious and expensive for the routine detection of transcription factor activity in the laboratory (20). Fluorescence methodologies are an attractive alternative to the traditional methods of transcription factor activity detection due to their simplicity, low cost, high sensitivity and most importantly, amenability to high-throughput screening (21). Current fluorescence-based methods for the detection of transcription factors require labeled oligonucleotides containing the sequence recognized by the appropriate transcription factor (22–25). The basic principle behind this ‘molecular beacon’ approach for the detection of transcription factors involves monitoring the conformational change of the oligonucleotide upon binding by a transcription factor. This conformational change leads to the fluorophore and the quencher being brought closer together or further apart, leading to a ‘switch-off’ or ‘switch-on’ fluorescence effect, respectively. In 2000, Tan and co-workers (22) described a switch-on probe for the Escherichia coli single-stranded binding protein using a classical stem–loop, doubly labeled with dabcyl and tamra at the 3′- and 5′-terminus. In 2002, Heyduk and Heyduk (23) developed a switch-off detection platform that utilized two independently labeled DNA fragments each containing one-half of the transcription factor binding site. Recently, Mirkin and co-workers (25) described a fluorescence recovery assay for the detection of protein–DNA binding, utilizing a doubly labeled short DNA duplex and an exonuclease. While these fluorescence approaches to the detection of transcription factor activity are more convenient compared to the traditional methods, they are still limited by the high cost of the labeled oligonucleotides.
Luminescent transition metal complexes have received increasing attention in photochemistry, organic optoelectronics and luminescent sensors (26–33). We previously developed oligonucleotide-based, label-free detection methods for nanomolar quantities of Hg2+ and Ag+ ions by employing luminescent platinum(II) metallointercalators (34,35), as well as for assaying exonuclease activity by using crystal violet as a G-quadruplex probe (36). Consequently, we were interested in developing a label-free alternative to the molecular beacon approach through modification of the fluorescence recovery assay developed by Mirkin and coworkers by utilizing unmodified oligonucleotides and a luminescent transition metal complex as a DNA probe. Luminescent transition metal complexes typically contain a metal center bound to by organic ligands arranged in a precise 3D arrangement. The 3D nature of transition metal complexes allows selective interactions with biomolecules (36). In addition, the photophysical (i.e. emission wavelength), physical (i.e. solubility and stability) and selectivity (duplex DNA versus single-stranded DNA) of these complexes can be modulated through ligand modifications. Examples of luminescent metallointercalators used for the detection of DNA include ruthenium (37–41), osmium (42–44), iridium (45–47) and platinum complexes (48–51) that bear planar aromatic ligands suitable for intercalation. We chose the classical ‘molecular light switch’ complex [Ru(phen)2(dppz)]2+ (phen = 1,10-phenanthroline; dppz = dihydro[3,2-a:2′,3′-c]phenazine) as a probe due to its avid DNA binding affinity (>106 M−1). In addition, this complex possesses a strong luminescence response when bound to duplex DNA but is only weakly emissive when free in aqueous solution or in the presence of single-stranded DNA. The complex [Ru(phen)2(dppz)]2+ has also been employed for the detection of aptamer/protein binding using unlabeled oligonucleotides (52).
Based on our past experience in the design of label-free oligonucleotide-based luminescent assays for metal ions (34,35), we were interested to see if we could develop a label-free detection method for the p50 subunit of the transcription factor NF-κB. The transcription factor NF-κB has been identified as an important regulator for key pro-inflammatory mediators such as TNF-α, which is involved in the immune response, apoptosis and cell cycle regulation (53). The deregulation of TNF-α has been linked with inflammatory and autoimmune diseases such as rheumatoid arthritis and osteoarthritis (54). The ability to screen a large library of compounds against an important protein target such as NF-κB using aluminescence assay amenable to high-throughput screening would be invaluable in developing new treatments and diagnostic tools for inflammation and autoimmune diseases.
MATERIALS AND METHODS
All reagents were purchased and used as received unless otherwise stated. The p50 protein was expressed and purified based on a modified procedure from Leung et al. (55). Exonuclease III (ExoIII) was purchased from New England Biolabs. [Ru(phen)2(dppz)](PF6)2 was synthesized according to literature method (38). DNA sequences were purchased from Techdragon Ltd (Hong Kong). Oridonin was purchased from China Langchem Inc (P.R. China).
Expression and purification of the p50
The pET-21b-p50 constructs were expressed in E. coli BL21(DE3) cells. The cells were grown at 37°C in a shaking incubator until the absorbance of the culture at 600 nm was 0.6. Expression of the p50 protein from the T7 promoter was induced for 5 h at 30°C by the addition of 0.1 mM isopropyl-1-thio-β-d-galactopyranoside (final concentration). The cells were then harvested in lysis buffer (25 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, β-mercaptoethanol, phenylmethylsulfonyl fluoride) and lysed by sonication. The cell debris was pelleted by ultracentrifugation (27 500 rpm, 4°C and 40 min). The supernatant was diluted with binding buffer (25 mM, Tris pH 7.4, 500 mM NaCl and 20 mM imidazole) and loaded onto a His-Bind Quick Columns (Novagen, Madison, WI, USA) and washed with washing buffer (25 mM Tris pH 7.4, 500 mM NaCl and 40 mM imidazole), then eluted with elution buffer (25 mM Tris pH 7.4, 500 mM NaCl and 200 mM imidazole). The fractions containing the p50 protein were combined and dialyzed against 10 mM Tris buffer solution (pH 7.9, 10% glycerol, 1 mM EDTA, 50 mM NaCl and β-mercaptoethanol). The purity of the expressed p50 proteins were estimated to be >90% pure using electrophoresis on SDS–PAGE gel stained with Coomassie Blue.
DNA sequences
Hairpin (HP) containing one NF-κB binding site:
5′-AGTTGAGGGGACTTTCCCAGGCCAGAAGGAGCCTGGGAAAGTCCCCTCAACT-3′
Double-strand containing one NF-κB binding site:
5′-AGTTGAGGGGACTTTCCCAGGC-3′
3′-TCAACTCCCCTGAAAGGGTCCG-5′
Double-strand containing two NF-κB binding site:
5′-TTGAGGGACTTTCCGAACATGCAGGCAAGCTGGGGACTTTCCAGG-3′
3′-AACTCCCTGAAAGGCTTGTACGTCCGTTCGACCCCTGAAAGGTCC-5′
Double-strand without NF-κB binding site:
5′-TTGTTACAACTCACTTTCCGCTGCTCACTTTCCAGGGAGGCGTGG-3′
3′-AACAATGTTGAGTGAAAGGCGACGAGTGAAAGGTCCCTCCGCACC-5′
Emission measurements
The appropriate oligonucleotide (0.02 µM) was first annealed in Tris buffer solution (10 mM, pH 7.4, 100 mM NaCl, 1 mM EDTA, final concentration) by incubating at 95°C for 5 min, followed by gradual cooling to room temperature over a period of 1 h. The p50 subunit and the annealed oligonucleotide mixture in TF buffer (10 mM Tris, pH 7.4, 50 mM KCl, 1 mM DTT, 1 mM MgCl2, 10% glycerol) were incubated for 20 min at 37°C, after which 40 units of ExoIII (NEB) were added and the mixture was incubated for an additional 50 min at 37°C. The digestion reaction was quenched by the addition of 25 mM EDTA and diluted to 1 ml with a solution of the ruthenium complex (1 µM, final concentration) and [Fe(CN)6]3− (600 µM, final concentration) in TF buffer (10 mM Tris, pH 7.4, 50 mM KCl, 1 mM DTT, 1 mM MgCl2, 10% glycerol). The solution was then allowed to stand for 10 min and the luminescence spectrum was measured using an excitation wavelength of 450 nm.
RESULTS AND DISCUSSION
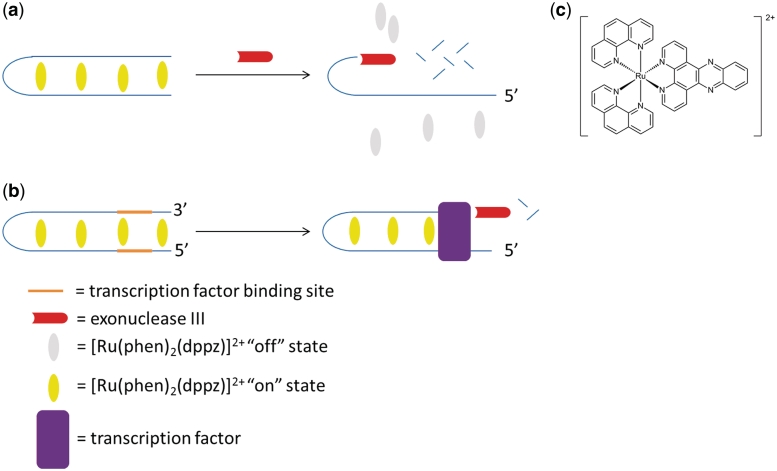
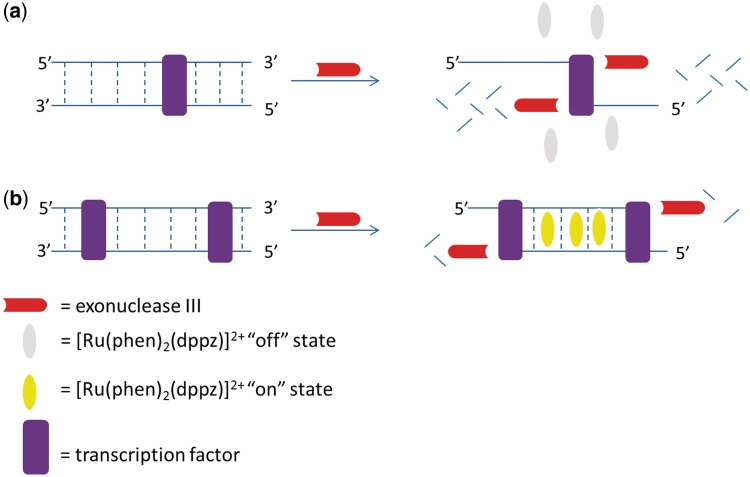
The principle behind our assay for the detection of transcription factor activity is based on the 3′→5′ activity of ExoIII and a luminescent transition metal complex which is ‘switched-on’ in the presence of double-stranded DNA (Scheme 1). In the presence of double-stranded DNA, the ruthenium complex [Ru(phen)2(dppz)]2+ (Scheme 1) intercalates into the double-stranded DNA and is emissive, presumably through suppression of non-radiative decay by solvent interactions. A 3′→5′ ExoIII is added to the reaction mixture and digests the double-stranded oligonucleotide from the 3′-end, leading to the formation of single-stranded fragments. Due to the weak binding affinity of the ruthenium complex to single-stranded DNA, the luminescence response of the complex is reduced (Scheme 1a). In the presence of a transcription factor that binds to the double-stranded substrate with the cognate binding site, the digestion of the oligonucleotide is blocked, allowing the oligonucleotide to retain its double-stranded structure in the presence of ExoIII (Scheme 1b). The intercalation of the ruthenium complex into the double-stranded DNA substrate leads to a strong luminescence response.
Scheme 1.
The principle of the label-free detection of transcription factor activity using a combination of a luminescent ruthenium-based metallointercalator [Ru(phen)2(dppz)]2+ and 3′→5′ ExoIII.
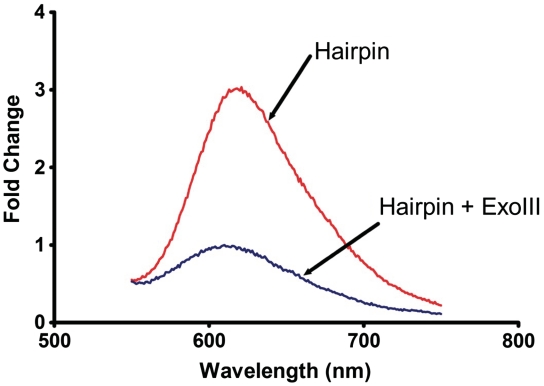
To validate our label-free detection assay for transcription factors, we designed a hairpin oligonucleotide that contained the NF-κB binding site [-GGGACTTTC-] (56). The hairpin substrate was incubated at 95°C for 5 min, followed by gradual cooling to room temperature to ensure the formation of the double-stranded structure. The luminescence response of the ruthenium complex in the presence of the hairpin substrate was enhanced by 4.6-fold due to intercalation of the ruthenium complex into the DNA (Figure 1). The addition of ExoIII leads to the digestion of the oligonucleotide, converting the hairpin structure into short single-stranded DNA fragments. Due to the weak binding of the ruthenium complex with the single-stranded DNA, emission intensity was decreased by 3.0-fold. Potassium ferrocyanide K3[Fe(CN)6] was used to quench the background emission of the ruthenium complex in aqueous solution or when bound to single-stranded DNA. A ferrocyanide concentration of 600 μM was found to give the highest degree of discrimination between double-stranded and single-stranded DNA.
Figure 1.
The luminescence response of [Ru] (1 µM) in TF buffer solution containing K3[Fe(CN)6] (600 µM) in the presence of (a) the hairpin DNA (0.02 µM); and (b) the hairpin DNA (0.02 µM) and ExoIII (40 U).
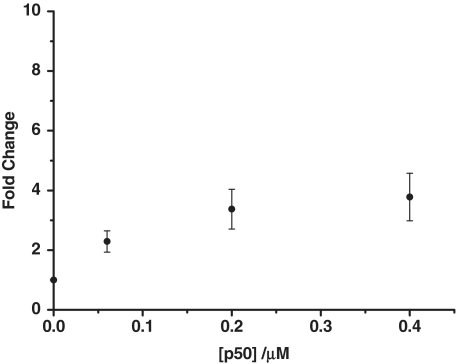
After confirming the activity of the exonuclease, we next investigated the effect of adding the transcription factor. When the hairpin substrate was incubated with the NF-κB subunit p50 before the addition of ExoIII, a luminescence enhancement of 3.6 was observed relative to the control (no transcription factor added) (Figure 2). Presumably, the p50 subunit bound the hairpin substrate and inhibited the digestion of the oligonucleotide, allowing the ruthenium metallointercalator to bind to the intact double-stranded DNA.
Figure 2.
The fold change luminescence response of [Ru] (1 µM) in TF buffer solution containing K3[Fe(CN)6] (600 µM) in the presence of the digestion mixture containing the hairpin DNA substrate (0.02 µM) and 40 U of ExoIII as a function of the concentration of the p50 subunit (0, 0.06, 0.20 and 0.40 µM).
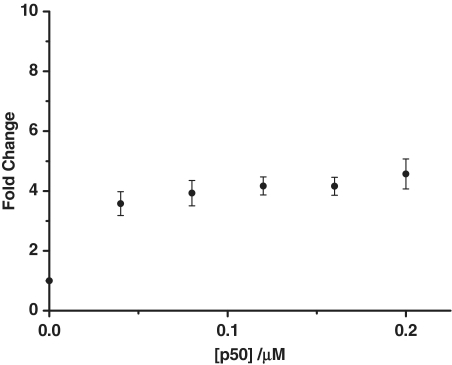
It has been previously shown that the structure of the oligonucleotide substrate can influence the digestion rate of ExoIII (57). To examine the effect of a double-stranded DNA substrate on the performance of this label-free assay, we annealed a duplex DNA sequence containing the NF-κB binding site. In the absence of the p50 subunit, a weak luminescence response was observed due to the exonuclease digestion of the double-stranded DNA. As the concentration of the p50 subunit was increased, there was a corresponding enhancement in the luminescence response of the ruthenium complex, with saturation occurring at about 160 nM (Figure 3). A maximum fold change of 4.5 was observed.
Figure 3.
The fold change luminescence response of [Ru] (1 µM) in TF buffer solution containing K3[Fe(CN)6] (600 µM) in the presence of the digestion mixture containing the double-stranded DNA substrate (0.02 µM) and 40 U of ExoIII as a function of the concentration of the p50 subunit (0, 0.04, 0.08, 0.12, 0.16, 0.20 µM).
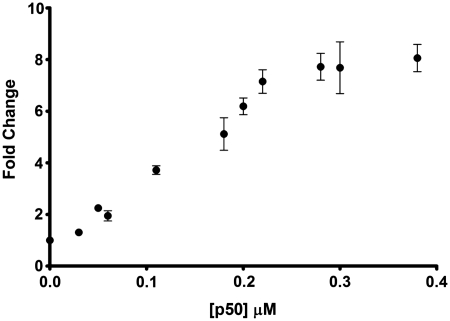
We next investigated the effect of introducing two binding sites on the double-stranded oligonucleotide substrate. The luminescence spectrum of the ruthenium complex in the presence of the double-stranded substrate containing two binding sites after digestion is shown in Figure 4. When this oligonucleotide was incubated in the presence of the p50 subunit and subjected to Exo III digestion, a maximal 8-fold increase in the luminescence response was observed, compared to only 4.5-fold for the oligonucleotide containing one binding site. We postulate that in the case of the double-stranded substrate containing one binding site, complete digestion by Exo III from the 3′ would be expected to generate long 5′-overhangs with limited duplex regions (Figure 5a). Thus, even though digestion was inhibited relative to the control, the luminescence response of the ruthenium complex would still be reduced. However, when two p50 subunit binding sites are present, the complete digestion of the double-stranded substrate from the two 3′-termini does not occur (Figure 5b), preserving the duplex structure of the substrate and allowing the intercalation of the ruthenium complex. Using the double-stranded substrate with two p50 subunit binding sites, we observed a linear luminescence response (up to 8-fold intensity enhancement) to changes in the concentration of the p50 subunit in the concentration range of 30–220 nM with a detection limit of 30 nM.
Figure 4.
The fold change luminescence response of [Ru] (1 µM) in TF buffer solution containing K3[Fe(CN)6] (600 µM) in the presence of the digestion mixture containing the double-stranded DNA substrate with two NF-κB binding sites (0.02 µM) and 40 U of ExoIII as a function of the concentration of the p50 subunit (0, 0.03, 0.05, 0.06, 0.11, 0.18, 0.20, 0.22, 0.28, 0.30 and 0.38 µM).
Figure 5.
A schematic representation of the digestion products for oligonucleotides containing one (a) and two (b) NF-κB subunit binding sites.
In the original report by Heyduk and Heyduk (23), the molecular beacon approach could effectively sense down to 10 nM of catabolite activator protein, a bacterial transcription factor. The fluorescence recovery assay developed by Mirkin and co-workers (25) gave a 32% decrease in the fluorescence intensity upon addition of 130 nM of estrogen receptor-α. Thus, the sensitivity of our assay for transcription factor activity is at least comparable to that of previously reported methods. Furthermore, a significant advantage of the method presented here is its label-free nature, which obviates the requirement for the expensive labeling of oligonucleotides, contrasting favorably with previous methods. Reducing the cost of the assay is important for potential adaptation into a high-throughput format. Finally, both literature methods were ‘switch-off’ with respect to transcription factors, which may suffer from false positives due to non-specific quenching by environmental samples. The ‘switch-on’ detection mode reported here is advantageous and is generally preferable for analytical purposes.
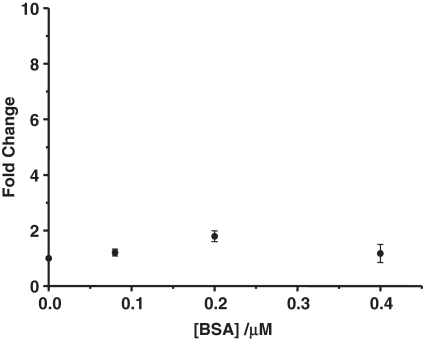
This label-free assay is based on the inhibition of ExoIII catalyzed digestion of the oligonucleotide by the binding of the p50 subunit. To validate the mechanism of this method, we replaced the p50 subunit with the non-DNA binding protein bovine serum albumin (BSA). The luminescence response of the ruthenium complex in the presence of the oligonucleotide containing the double p50 binding site and BSA after ExoIII digestion is shown in Figure 6.
Figure 6.
The fold change luminescence response of [Ru] (1 µM) in TF buffer solution containing K3[Fe(CN)6] (600 µM) in the presence of the digestion mixture containing the double-stranded DNA substrate with two NF-κB binding sites (0.02 µM) and 40 U of ExoIII as a function of the concentration of BSA (0, 0.08, 0.20 and 0.40 µM).
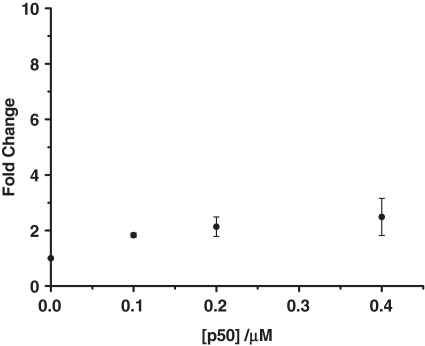
The emission spectrum in Figure 6 shows that incubation of the oligonucleotide substrate with BSA did not produce the same emission enhancement (fold change of 1.1) as observed for the p50 subunit. To further provide evidence that the inhibition of ExoIII digestion was due to the selective binding of the p50 subunit, we replaced the oligonucleotide substrate with a DNA sequence that cannot bind to the p50 subunit. The non-NF-κB-binding substrate was incubated in the presence of the p50 subunit and the luminescence spectrum of the ruthenium complex in the presence of the digestion mixture was measured (Figure 7). In the presence of 0.4 µM p50 subunit, a fold change of 2.4 was observed, which was significantly lower than the 8-fold enhancement observed with the wild-type sequence.
Figure 7.
The fold change luminescence response of [Ru] (1 µM) in TF buffer solution containing K3[Fe(CN)6] (600 µM) in the presence of the digestion mixture containing double-stranded non-NF-κB-binding substrate (0.02 µM) and 40 U of ExoIII as a function of the concentration of the p50 subunit (0, 0.10, 0.20 and 0.40 µM).
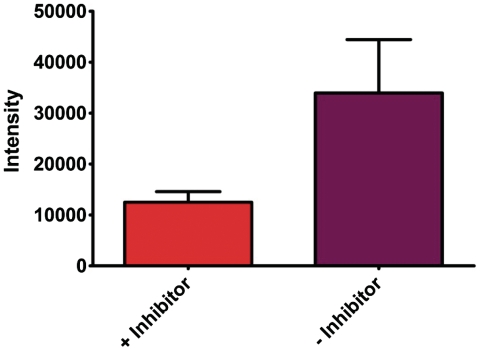
To provide additional evidence that the selective binding of the p50 subunit is responsible for the inhibition of ExoIII catalyzed digestion of the double-stranded substrate, we repeated the luminescence measurements in the presence of oridonin, a known inhibitor of NF-κB DNA binding activity (58). In the presence of the NF-κB inhibitor (Figure 8, orange), a significant reduction in the luminescence response of the ruthenium complex was observed compared to the sample that does not contain the inhibitor (Figure 8, purple). Oridonin is presumed to inhibit binding of p50 to the double-stranded substrate and ExoIII is able to digest the DNA into short single-stranded fragments resulting in a reduced luminescence response. Taken together, these series of negative control experiments demonstrate that the luminescence enhancement observed in the presence of the p50 subunit is probably due to the binding of the transcription factor to the oligonucleotide, inhibiting the ExoIII catalyzed digestion of the double-stranded substrate.
Figure 8.
The luminescence response of [Ru] (1 µM) in TF buffer solution containing K3[Fe(CN)6] (600 µM) in the presence of the digestion mixture with the double-stranded DNA substrate (0.02 µM) containing two NF-κB binding sites incubated with p50 (0.12 µM) with or without oridonin (20 µM).
The above results also highlight the amenability of this assay to the high-throughput screening of small molecules as inhibitors of the p50 subunit of NF-κB. NF-κB is found in the cytoplasm bound to the inhibitory protein IκB (59). In the presence of activators, such as ultraviolet irradiation, cytokines, bacterial and viral products, NF-κB is released from IκB and becomes activated (59). The overactivation of NF-κB has been associated number of autoimmune and inflammatory diseases and it is thus considered an important drug target (53,54). The label-free assay described herein can be readily applied to a high-throughput format using 96-well plates. Wells showing a reduction in luminescence intensity of the ruthenium complex contain a potential inhibitor of the p50 subunit. Due to the low cost of the label-free oligonucleotides and the ruthenium metallointercalator, large chemical libraries can be screened in an inexpensive and high-throughput manner, allowing the identification of small molecule NF-κB inhibitors for treating autoimmune and inflammatory diseases.
CONCLUSION
In conclusion, we have described the first label-free luminescence detection method for transcription factor activity. Our method is based on the principle that the binding of the transcription factor prevents the ExoIII catalyzed digestion of a double-stranded substrate. A luminescent ruthenium metallointercalator is used to probe the double-stranded substrate leading to a ‘switch-on’ effect in the presence of the transcription factor. The luminescence enhancement was shown to be proportional to the concentration of the transcription factor NF-κB subunit p50. This method allows the detection of transcription factor activity without the need for time-consuming experiments such as gel mobility shift assays or DNA footprinting. We have also demonstrated that in the presence of a known NF-κB inhibitor oridonin, the luminescence response of the ruthenium complex was decreased. Therefore, this assay can be used to identify modulators that can activate or inhibit transcription factor–DNA binding, for the diagnosis and treatment of diseases linked with irregular transcription factor activity. Furthermore, this technique is readily amenable to high-throughput screening, allowing rapid and economical identification of the target compounds. We anticipate that this assay can be adapted to selectively detect any transcription factor simply by changing the binding site sequences. Due to the modular synthesis of the transition metal complexes, we envisage that there is considerable scope to adjust the selectivity of the complexes toward particular DNA structures which would further improve this assay for transcription factor detection.
FUNDING
Funding for open access charge: The Hong Kong Baptist University (FRG2/09-10/070).
Conflict of interest statement. None declared.
REFERENCES
- 1.Papavassiliou AG. Molecular medicine. Transcription factors. N. Engl. J. Med. 1995;332:45–47. doi: 10.1056/NEJM199501053320108. [DOI] [PubMed] [Google Scholar]
- 2.Pabo CO, Sauer RT. Transcriptional factors: structural families and principles of DNA recognition. Annu. Rev. Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 3.Latchman DS. Transcription-factor mutations and disease. N. Engl. J. Med. 1996;334:28–33. doi: 10.1056/NEJM199601043340108. [DOI] [PubMed] [Google Scholar]
- 4.Wang K, Wan Y-JY. Nuclear receptors and inflammatory diseases. Exp. Biol. Med. 2008;233:496–506. doi: 10.3181/0708-MR-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfaeffle RW, DiMattia GE, Parks JS, Brown MR, Wit JM, Jansen M, van der Nat H, van der Brande JL, Rosenfeld MG, Ingraham HA. Mutation of the POU-specific domain of Pit-1 and hypopituitarism without pituitary hypoplasia. Science. 1992;257:1118–1121. doi: 10.1126/science.257.5073.1118. [DOI] [PubMed] [Google Scholar]
- 6.Radovick S, Nations M, Du Y, Berg LA, Weintraub BD, Wondisford FE. A mutation in the POU-homeodomain of Pit-1 responsible for combined pituitary hormone deficiency. Science. 1992;257:1115–1118. doi: 10.1126/science.257.5073.1115. [DOI] [PubMed] [Google Scholar]
- 7.Hanson IM, Fletcher JM, Jordan T, Brown A, Taylor D, Adams RJ, Punnett HH, van Heyningen V. Mutation at the PAX6 locus are found in heterogeneous anterior segment malformations including Peters' anomaly. Nat. Genet. 1994;6:168–173. doi: 10.1038/ng0294-168. [DOI] [PubMed] [Google Scholar]
- 8.Sanyanusin P, Schimmenti LA, McNoe LA, Ward TA, Pierpont MEM, Sullivan MJ, Dobyns WB, Eccles MR. Mutation of the PAX2 gene in a family with optic nerve colobomas, renal anomalies and vesicoureteral reflux. Nat. Genet. 1995;9:358–364. doi: 10.1038/ng0495-358. [DOI] [PubMed] [Google Scholar]
- 9.Baniahmad A, Koehne AC, Renkawitz R. A transferable silencing domain is present in the thyroid hormone receptor, in the v-erbA oncogene product and in the retinoic acid receptor. EMBO J. 1992;11:1015–1023. doi: 10.1002/j.1460-2075.1992.tb05140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baniahmad A, Tsai SY, O'Malley BW, Tsai MJ. Kindred S thyroid hormone receptor is an active and constitutive silencer and a repressor for thyroid hormone and retinoic acid responses. Proc. Natl Acad. Sci. USA. 1992;89:10633–10637. doi: 10.1073/pnas.89.22.10633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zanaria E, Muscatelli F, Bardoni B, Strom TM, Guioli S, Guo W, Lalli E, Moser C, Walker AP, Mc Cabe ER, et al. An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature. 1994;372:635–641. doi: 10.1038/372635a0. [DOI] [PubMed] [Google Scholar]
- 12.Chang H-C, Sehra S, Goswami R, Yao W, Yu Q, Stritesky GL, Jabeen R, McKinley C, Ahyi A-N, Han L, et al. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat. Immunol. 2010;11:527–534. doi: 10.1038/ni.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat. Rev. Immunol. 2010;10:365–376. doi: 10.1038/nri2748. [DOI] [PubMed] [Google Scholar]
- 14.Spencer CA, Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv. Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- 15.Forrest D, Curran T. Crossed signals: oncogenic transcription factors. Curr. Opin. Genet. Dev. 1992;2:19–27. doi: 10.1016/s0959-437x(05)80316-8. [DOI] [PubMed] [Google Scholar]
- 16.Dyck JA, Maul GG, Miller WH, Jr, Chen JD, Kazizuka A, Evans RM. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 17.Galas DJ, Schmitz A. DNasefootprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fried M, Crothers DM. Equilibriums and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavski V, Chris LX. Ultrasensitive protein-DNA binding assays. Curr. Opin. Biotechnol. 2003;14:65–73. doi: 10.1016/s0958-1669(02)00016-2. [DOI] [PubMed] [Google Scholar]
- 20.Lymperopoulos K, Crawford R, Torella JP, Heilemann M, Hwang LC, Holden SJ, Kapanidis AN. Single-molecule DNA biosensors for protein and ligand detection. Angew. Chem. Int. Ed. 2010;49:1316–1320. doi: 10.1002/anie.200904597. [DOI] [PubMed] [Google Scholar]
- 21.Hill JJ, Royer CA. Fluorescence approaches to study of protein-nucleic acid complexation. Methods Enzymol. 1997;278:390–416. doi: 10.1016/s0076-6879(97)78021-2. [DOI] [PubMed] [Google Scholar]
- 22.Li JJ, Fang X, Schuster SM, Tan W. Molecular beacons: a novel approach to defect protein-DNA interactions. Angew. Chem., Int. Ed. 2000;39:1049–1052. doi: 10.1002/(sici)1521-3773(20000317)39:6<1049::aid-anie1049>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Heyduk T, Heyduk E. Molecular beacons for detecting DNA binding proteins. Nat. Biotechnol. 2002;20:171–176. doi: 10.1038/nbt0202-171. [DOI] [PubMed] [Google Scholar]
- 24.Heyduk E, Knoll E, Heyduk T. Molecular beacons for detecting DNA binding proteins: mechanism of action. Anal. Biochem. 2003;316:1–10. doi: 10.1016/s0003-2697(03)00004-6. [DOI] [PubMed] [Google Scholar]
- 25.Xu X, Zhao Z, Qin L, Wei W, Levine JE, Mirkin CA. Fluorescence recovery assay for the detection of protein-DNA binding. Anal. Chem. 2008;80:5616–5621. doi: 10.1021/ac8007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalyanasundaram K, Gratzel M. Applications of functionalized transition metal complexes in photonic and optoelectronic devices. Coord. Chem. Rev. 1998;177:347–414. [Google Scholar]
- 27.Tamayo AB, Alleyne BD, Djurovich PI, Lamansky S, Tsyba I, Ho NN, Bau R, Thompson ME. Synthesis and characterization of facial and meridional tris-cyclometalated Iridium(III) complexes. J. Am. Chem. Soc. 2003;125:7377–7387. doi: 10.1021/ja034537z. [DOI] [PubMed] [Google Scholar]
- 28.Coppo P, Plummer EA, De CL. Tuning iridium(III) phenylpyridine complexes in the “almost blue” region. Chem. Commun. 2004:1774–1775. doi: 10.1039/b406851c. [DOI] [PubMed] [Google Scholar]
- 29.Sandee AJ, Williams CK, Evans NR, Davies JE, Boothby CE, Koehler A, Friend RH, Holmes AB. Solution-processible conjugated electrophosphorescent polymers. J. Am. Chem. Soc. 2004;126:7041–7048. doi: 10.1021/ja039445o. [DOI] [PubMed] [Google Scholar]
- 30.Jung S, Kang Y, Kim H-S, Kim Y-H, Lee C-L, Kim J-J, Lee S-K, Kwon S-K. Effect of substitution of methyl groups on the luminescence performance of Ir(III) complexes: preparation, structures, electrochemistry, photophysical properties and their applications in organic light-emitting diodes (OLEDs) Eur. J. Inorg. Chem. 2004:3415–3423. [Google Scholar]
- 31.Ma D-L, Wong W-L, Chung W-H, Chan F-Y, So P-K, Lai T-S, Zhou Z-Y, Leung Y-C, Wong K-Y. A highly selective luminescence switch-on sensor for histidine/histidine-rich proteins and its application in protein staining. Angewandte Chemie. 2008;47:3735–3739. doi: 10.1002/anie.200705319. [DOI] [PubMed] [Google Scholar]
- 32.Lo KK-W. Luminescent transition metal complexes as biological labels and probes. Struct. Bonding. 2007;123:205–245. [Google Scholar]
- 33.Ma D-L, Chan DS-H, Man BY-W, Leung C-H. Oligonucleotide-based luminescent detection of metal ions. Chem. -Asian. J. 2011 doi: 10.1002/asia.201000870. doi:10.1002/asia.201000870. [DOI] [PubMed] [Google Scholar]
- 34.Chan DS-H, Lee H-M, Che C-M, Leung C-H, Ma D-L. A selective oligonucleotide-based luminescent switch-on probe for the detection of nanomolar mercury(II) ion in aqueous solution. Chem. Commun. 2009:7479–7481. doi: 10.1039/b913995h. [DOI] [PubMed] [Google Scholar]
- 35.Man BY-W, Chan DS-H, Yang H, Ang S-W, Yang F, Yan S-C, Ho C-H, Wu P, Che C-M, Leung C-H, et al. A selective G-quadruplex-based luminescent switch-on probe for the detection of nanomolar silver(I) ions in aqueous solution. Chem. Commun. 2010;46:8534–8536. doi: 10.1039/c0cc01201g. [DOI] [PubMed] [Google Scholar]
- 36.Leung C-H, Chan DS-H, Man BY-W, Wang C-J, Lam W, Cheng Y-C, Fong W-F, Hsiao W-LW, Ma D-L. Simple and convenient G-quadruplex-based turn-on fluorescence assay for 3′->5′ exonuclease activity. Anal. Chem. 2011;83:463–466. doi: 10.1021/ac1025896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedman AE, Chambron JC, Sauvage JP, Turro NJ, Barton JK. A molecular light switch for DNA: [Ru(bpy)2(dppz)]2+ J. Am. Chem. Soc. 1990;112:4960–4962. [Google Scholar]
- 38.Franklin SJ, Treadway CR, Barton JK. A reinvestigation by circular dichroism and NMR: ruthenium(II) and rhodium(III) metallointercalators do not bind cooperatively to DNA. Inorg. Chem. 1998;37:5198–5210. [Google Scholar]
- 39.Rueba E, Hart JR, Barton JK. [Ru(bpy)2(L)]Cl2: luminescent metal complexes that bind DNA base mismatches. Inorg. Chem. 2004;43:4570–4578. doi: 10.1021/ic0499291. [DOI] [PubMed] [Google Scholar]
- 40.Ma D-L, Che C-M, Siu F-M, Yang M, Wong K-Y. DNA binding and cytotoxicity of ruthenium(II) and rhenium(I) complexes of 2-amino-4-phenylamino-6-(2-pyridyl)-1,3,5-triazine. Inorg. Chem. 2007;46:740–749. doi: 10.1021/ic061518s. [DOI] [PubMed] [Google Scholar]
- 41.Lim MH, Song H, Olmon ED, Dervan EE, Barton JK. Sensitivity of [Ru(bpy)2dppz]2+ luminescence to DNA defects. Inorg. Chem. 2009;48:5392–5397. doi: 10.1021/ic900407n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holmlin RE, Barton JK. [Os(phen)2(dppz)]2+: a red-emitting DNA probe. Inorg. Chem. 1995;34:7–8. [Google Scholar]
- 43.Holmlin RE, Stemp EDA, Barton JK. [Os(phen)2dppz]2+ in photoinduced DNA-mediated electron transfer reactions. J. Am. Chem. Soc. 1996;118:5236–5244. [Google Scholar]
- 44.Holmlin RE, Yao JA, Barton JK. Dipyridophenazine complexes of Os(II) as red-emitting DNA probes: synthesis, characterization, and photophysical properties. Inorg. Chem. 1999;38:174–189. [Google Scholar]
- 45.Stinner C, Wightman MD, Kelley SO, Hill MG, Barton JK. Synthesis and spectroelectrochemistry of [Ir(bpy)(phen)(phi)]3+, a tris(heteroleptic) metallointercalator. Inorg. Chem. 2001;40:5245–5250. doi: 10.1021/ic010376t. [DOI] [PubMed] [Google Scholar]
- 46.Shao F, Barton JK. Long-range electron and hole transport through DNA with tethered cyclometalatediridium(III) complexes. J. Am. Chem. Soc. 2007;129:14733–14738. doi: 10.1021/ja0752437. [DOI] [PubMed] [Google Scholar]
- 47.Lo KK-W. Exploitation of luminescent organometallic rhenium(I) and iridium(III) complexes in biological studies. Top. Organomet. Chem. 2010;29:115–158. [Google Scholar]
- 48.Ma D-L, Che C-M. A bifunctionalplatinum(II) complex capable of intercalation and hydrogen-bonding interactions with DNA: binding studies and cytotoxicity. Chem. Eur. J. 2003;9:6133–6144. doi: 10.1002/chem.200304964. [DOI] [PubMed] [Google Scholar]
- 49.Chan H-L, Ma D-L, Yang M, Che C-M. Synthesis and biological activity of a platinum(II) 6-phenyl-2,2'-bipyridine complex and its dimeric analogue. ChemBioChem. 2003;4:62–68. doi: 10.1002/cbic.200390015. [DOI] [PubMed] [Google Scholar]
- 50.Eryazici I, Moorefield CN, Newkome GR. Square-Planar Pd(II), Pt(II), and Au(III) terpyridine complexes: their syntheses, physical properties, supramolecular constructs, and biomedical activities. Chem. Rev. 2008;108:1834–1895. doi: 10.1021/cr0781059. [DOI] [PubMed] [Google Scholar]
- 51.Ma D-L, Che C-M, Yan S-C. Platinum(II) complexes with dipyridophenazine ligands as human telomerase inhibitors and luminescent probes for G-quadruplex DNA. J. Am. Chem. Soc. 2009;131:1835–1846. doi: 10.1021/ja806045x. [DOI] [PubMed] [Google Scholar]
- 52.Jiang Y, Fang X, Bai C. Signaling aptamer/protein binding by a molecular light switch complex. Anal. Chem. 2004;76:5230–5235. doi: 10.1021/ac049565u. [DOI] [PubMed] [Google Scholar]
- 53.Fujisawa K, Aono H, Hasunuma T, Yamamoto K, Mita S, Nishioka K. Activation of transcription factor NF-κB in human synovial cells in response to tumor necrosis factor α. Arthritis Rheum. 1996;39:197–203. doi: 10.1002/art.1780390205. [DOI] [PubMed] [Google Scholar]
- 54.Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat. Rev. Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 55.Leung C-H, Grill SP, Lam W, Gao W, Sun H-D, Cheng Y-C. Eriocalyxin B inhibits nuclear factor-κB activation by interfering with the binding of both p65 and p50 to the response element in a noncompetitive manner. Mol. Pharmacol. 2006;70:1946–1955. doi: 10.1124/mol.106.028480. [DOI] [PubMed] [Google Scholar]
- 56.Chaturvedi MM, Kumar A, Darnay BG, Chainy GB, Agarwal S, Aggarwal BB. Sanguinarine (pseudochelerythrine) is a potent inhibitor of NF-κB activation, IκBα phosphorylation, and degradation. J. Biol. Chem. 1997;272:30129–30134. doi: 10.1074/jbc.272.48.30129. [DOI] [PubMed] [Google Scholar]
- 57.Xodo LE, Manzini G, Quadrifoglio F, van der Marel G, van Boom J. DNA hairpin loops in solution. Correlation between primary structure, thermostability and reactivity with single-strand-specific nuclease from mung bean. Nucleic Acids Res. 1991;19:1505–1511. doi: 10.1093/nar/19.7.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leung C-H, Grill SP, Lam W, Han Q-B, Sun H-D, Cheng Y-C. Novel mechanism of inhibition of nuclear factor-κB DNA-binding activity by diterpenoids isolated from Isodon rubescens. Mol. Pharmacol. 2005;68:286–297. doi: 10.1124/mol.105.012765. [DOI] [PubMed] [Google Scholar]
- 59.Chen F, Castranova V, Shi X, Demers LM. New insights into the role of nuclear factor-κB, a ubiquitous transcription factor in the initiation of diseases. Clin. Chem. 1999;45:7–17. [PubMed] [Google Scholar]