Abstract
Here we show that c17orf42, hereafter TEFM (transcription elongation factor of mitochondria), makes a critical contribution to mitochondrial transcription. Inactivation of TEFM in cells by RNA interference results in respiratory incompetence owing to decreased levels of H- and L-strand promoter-distal mitochondrial transcripts. Affinity purification of TEFM from human mitochondria yielded a complex comprising mitochondrial transcripts, mitochondrial RNA polymerase (POLRMT), pentatricopeptide repeat domain 3 protein (PTCD3), and a putative DEAD-box RNA helicase, DHX30. After RNase treatment only POLRMT remained associated with TEFM, and in human cultured cells TEFM formed foci coincident with newly synthesized mitochondrial RNA. Based on deletion mutants, TEFM interacts with the catalytic region of POLRMT, and in vitro TEFM enhanced POLRMT processivity on ss- and dsDNA templates. TEFM contains two HhH motifs and a Ribonuclease H fold, similar to the nuclear transcription elongation regulator Spt6. These findings lead us to propose that TEFM is a mitochondrial transcription elongation factor.
INTRODUCTION
Mitochondria supply the bulk of the cell’s energy via the process of oxidative phosphorylation (OXPHOS). Although the majority of OXPHOS components are encoded in the nuclear genome, 13 subunits of the OXPHOS machinery are the products of a small, circular genome in mammalian mitochondria. Human mitochondrial DNA (mtDNA) is dependent on the nucleus for all the proteins involved in its maintenance and expression.
Recent years have witnessed significant progress towards compiling a full list of mammalian mitochondrial proteins thanks largely to advances in high-throughput methods of tandem mass spectrometry (1–3) and comparative genomics (4). However, to date only half of the estimated 1500 mitochondrial proteins have been identified and current catalogues inevitably include false positives (2,5). Moreover, functional assignment of a protein on the basis of sequence homology has limited sensitivity and reliability, as some mitochondrial enzymes differ structurally and functionally from the homologous prokaryotic or eukaryotic gene. For example, human mitochondrial RNase P (mtRNase P), a tRNA processing enzyme, consists of three subunits that do not share any significant homology with other RNase Ps. Human mtRNase P lacks a catalytic RNA component that is present in all other known RNase P enzymes and it appears to be a mélange of a tRNA methyltransferase, a short-chain dehydrogenase/reductase-family member, and a protein of unknown function and evolutionary origin (6). Two of the three subunits of mtRNase P, the tRNA methyltransferase and short-chain dehydrogenase, had been assigned to the mitochondrial proteome (2), yet this was not enough to predict their function in tRNA processing. Therefore, it is important to combine global analyses of the mitochondrial proteome with more focused studies that characterize mitochondrial proteins.
Several basic components of the machinery for the transcription of the human mitochondrial genome have been identified; they include a monomeric RNA polymerase (POLRMT), and mitochondrial transcription factors A (TFAM) and B2 (TFB2M), which collectively form an effective in vitro transcription system (7). In addition, three regulatory factors are known, mitochondrial transcription termination factor 1 (mTERF1) (8), possible transcription initiation regulator mTERF2 (9) and a negative regulator of mammalian mtDNA transcription mTERF3, (10). One of the subunits of human mitochondrial ribosomal, MRPL12, has been reported to interact directly with POLRMT and stimulate its activity in vitro (11). However, the list of mitochondrial transcription factors is unlikely to be complete, in particular, one would anticipate the existence of a mitochondrial transcription elongation factor (TEF); first because TEFs are key elements of the transcription apparatus in other systems (12) and because RNA synthesis of mammalian mitochondrial DNA yields polycistronic transcripts (13). Transcription from the heavy strand promoter in the major non-coding region (NCR) of mtDNA (HSP1) gives rise to a polycistronic transcript covering almost the entire molecule, which is subsequently processed to yield two rRNAs, 10 mRNAs and 14 tRNAs. Although the L-strand transcript can be shorter, initiation of transcription at the light strand promoter (LSP) must yield an 11 kb precursor RNA to include all the coding information of the L-strand of mtDNA.
We began to study the human gene product of c17orf42 because it shares sequence homology with known: (i) proteins involved in transcription in Pro- and Eukaryota and (ii) Holliday junction resolvases (HJRs), such as bacterial RuvC (14) and yeast mitochondrial CCE1 (15) and YDC2 (16). Recombinant c17orf42 protein proved incapable of resolving four-way junctions in vitro, and other experiments indicated a role in mitochondrial transcription elongation, hence we propose the name TEF of mitochondria, TEFM.
MATERIALS AND METHODS
Plasmid construction
In order to construct pcDNA5-TEFM.HA used for the immunofluorescence localization of the TEFM protein (Figures 1C and 6C), the cDNA encoding TEFM (c17orf42) was modified by PCR to introduce the HA epitope tag (YPYDVPDYA) to the C-terminus of the ZFP and flanked with unique KpnI (5′) and XbaI (3′) restriction sites. The resulting fragment was cloned into pcDNA5/FRT/TO (Invitrogen) using the above restriction sites.
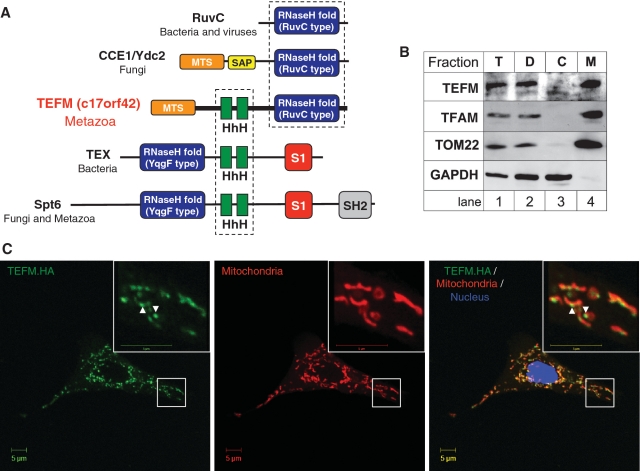
Figure 1.
In silico identification and mitochondrial localization of TEFM. (A) Domain architecture of proteins homologous to TEFM. Green blocks indicate the tandem helix-hairpin-helix domains (HhH, Pfam PF00633) found in transcription related and DNA binding proteins. The RuvC and YqgF (Pfam PF03652) motifs (blue) exhibit the typical topology and structural elements of the RNase H fold. S1 domain (red) typically binds RNA whereas the SH2 domain (grey) is found in many proteins involved in signal transduction and is responsible for protein–protein interaction. SAP motif (Pfam PF02037, yellow) represents a putative DNA binding domain found in a diverse number of proteins involved in chromosomal organization (40). The MTS present in TEFM is indicated in orange. See Supplementary Data for further details. (B) Sub-cellular location of TEFM. The HOS cells were fractionated into fraction containing unbroken cells and cell debris (‘D’, lane 2), cytosol (‘C’, lane 3) and mitochondria (‘M’, lane 4) as described ‘Materials and Methods’ section. The protein fractions were analysed by western blotting using antibodies to endogenous TEFM. The location of TEFM was compared with that of the following marker proteins: TFAM (mitochondrial matrix), TOM22 (mitochondrial outer membrane), GAPDH (cytosol). (C) The intra-cellular localization of the HA-tagged variant of TEFM (green) by immunofluorescence, in human A549 cells, as described in ‘Materials and Methods’ section. Mitochondria were stained with MitoTracker Red CMXRos (red); the nucleus was stained with DAPI (blue). Yellow signal on digitally overlaid pictures indicated that TEFM has the same cellular distribution as mitochondria.
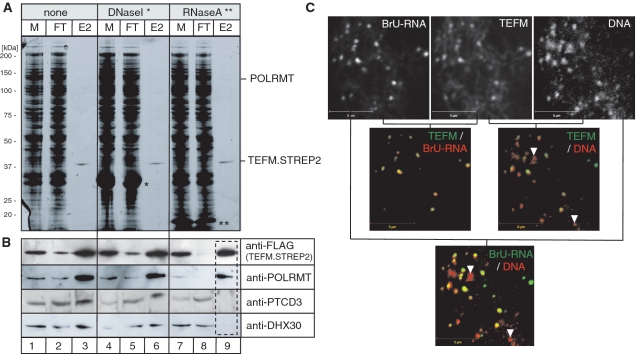
Figure 6.
Functional interaction of TEFM with RNA. (A) SDS–PAGE analysis of the affinity purified TEFM complex (lanes 1–3) treated with DNase I (lanes 4–6) lanes or RNase A (lanes 7–9) prior to the loading on the Strep-Tactin column. M, mitochondrial lysate; FT, flow through; E2, elution fraction 2; single asterisk, DNase I; two asterisks, RNase A. (B) Western blotting analysis of the SDS–PAGE gel shown in (C). The blot was incubated with the antibodies indicated to the right of the panel. (C) The co-localization of the HA-tagged version of TEFM with mtDNA and mtRNA analysed by immunofluorescence in A549 cells as described in ‘Materials and Methods’ section. Incorporation of BrU in RNA was visualized in cultured cells with BrU-specific mAbs. Black and white images shown in the top row were pseudo-coloured in red or green as indicated in the top right corner of each image and digitally overlaid; yellow staining is indicative of co-localization.
In order to construct pcDNA5-TEFM.STREP2, used to generate an inducible human embryonic kidney (HEK) cell line, two oligonucleotides encoding FLAG and STREP2 epitope tags were annealed and cloned into XbaI (5′) and ApaI (3′) restriction sites of pcDNA5/FRT/TO (Invitrogen). The resulting plasmid was named pcDNA5-FST2. Then the cDNA encoding TEFM was modified by PCR to introduce unique KpnI (5′) and XbaI (3′) and the resulting fragment was cloned into pcDNA5-FST2 using the above restriction sites.
In order to construct pcDNA5-POLRMT.STREP2, used to generate an inducible HEK cell line, the cDNA encoding POLRMT was modified by PCR to introduce flanking BamHI (5′) and XbaI (3′) sites, after digestion the resulting fragment was cloned into pcDNA5-FST2.
In order to construct pGEX-GST.TEFM the cDNA encoding TEFM without the first 32 amino acids was amplified by PCR to introduce unique EcoRI (5′) and XhoI (3′). The resulting product was cloned into the above sites into pGEX-4T1 (GE Healthcare) in-frame with the GST coding sequence.
The pcDNA3-POLRMT encoding the POLRMT of full length was constructed by introducing NheI site (5′) and the MYC-epitope tag and EcoRI (3′) into the POLRMT cDNA by PCR and cloning the resulting product into the above sites of pcDNA3.1(−) (Invitrogen).
The pcDNA3-POLTMT-MTS-786-1230 encoding the 60 amino acids-long mitochondrial targeting sequence (MTS) of POLRMT and the C-terminal portion of the protein (residues 786–1230) was cloned as follows: Firstly, the cDNA of POLRMT encoding the MTS was amplified by PCR to introduce XbaI sites on 5′- and 3′-ends and the resulting product was cloned into NheI and XbaI sites of pcDNA3.1(–) (Invitrogen). Secondly, the cDNA encoding residues 786–1230 was amplified by PCR to introduce XbaI (5′) and the MYC-epitope tag and EcoRI (3′). The resulting product was fused to the MTS harbouring pcDNA3.1(–) using XbaI and EcoRI.
The pcDNA-POLRMT-1-801 encoding the first 801 amino acids of POLRMT was constructed by introducing NheI site (5′) and the MYC-epitope tag and EcoRI (3′) into the POLRMT cDNA encoding resides 1–801 by PCR and cloning the resulting product into the above sites of pcDNA3.1(−) (Invitrogen).
Maintenance and transfection of mammalian cell lines
Human A549 adenocarcinoma and 143B osteosarcoma (HOS) cells that are routinely used by us for immunofluorescence localization of mitochondrial proteins and/or siRNA gene silencing were cultured in DMEM containing 2 mM l-glutamine (Invitrogen) with 10% FBS. For immunofluorescence experiments A549 cells were grown in 6-well plates to 90–95% confluence and transfected with 0.5 µg of plasmid DNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Twelve hours after transfection the cells were trypsinized and transferred onto coverslips placed in 60-mm culture dishes.
For the siRNA experiments electroporations of 143B cells were performed using Cell Line Nucleofector (Lonza), buffer kit V (Lonza), dsRNA (100 pmol) and applying programme I-13. For 6-day siRNA experiments the cells were split 2 days after the first electroporation and electroporated as above on day three.
Flp-In T-Rex™ HEK 293T cells (Invitrogen) that allows for generating stable, tetracycline inducible expression of transgenes by the FLP recombinase-mediated integration was used to express of TEFM.STREP2. HEK cells were grown in DMEM containing 2 mM l-glutamine (Invirtogen), 10% tetracycline free FCS (Autogen Bioclear) supplemented with 100 µg/ml Zeocin (Invivogen) and 15 µg/ml Blasticidin (Invitrogen). Twenty-four hours prior to, transfection cells were split to 10-cm plates and grown to 80–90% confluence. HEK cells were transfected according to the manufacturer’s instruction using Cell Line Nucleofector (Lonza), buffer kit V (Lonza) applying programme A-23. The pcDNA5/FRT/TO plasmid DNA encoding appriopriate TEFM.STREP2 was purified by Qiafilter MidiPrep Kit (Qiagen). Twenty-four hours after the transfection the selective antibiotics hygromycin (100 µg/ml) and blasticidin (15 µg/ml) were added and the selective medium was replaced every 3–4 days.
Immunofluorescence
A549 cells were cultured in DMEM with 10% FBS as above. For RNA labelling with BrU the medium was supplemented with 2.5 mM bromouridine (BrU, Sigma) for 30 min prior to fixation. To visualize mitochondria, 30 min prior to fixation the culture medium was replaced with one containing 100 nM MitoTracker Red CMXRos (Invitrogen). Cells were then washed 3 times with PBS and fixed for 15 min at room temperature using 3.7% formaldehyde 5% sucrose (w/v) solution in PBS. Fixed cells were permeabilized with 1% Triton X-100 10% FBS solution in PBS for 5 min. They were then blocked for 1 h with 10% FBS solution in PBS. Cells were incubated with primary antibodies for 1 h in 10% FBS solution and subsequently for 1 h in the dark with secondary antibodies in 10% FBS solution. Finally, cells were stained with DAPI by incubation in 50 nM DAPI solution in PBS for 1 min and washed. The whole procedure was performed at room temperature and the cells were washed 3 times with PBS between each step. After the final washing the coverslips were mounted on Superfrost slides in Mowiol medium and left in the dark for 4 h at room temperature and then overnight at 4°C; immunofluorescence was viewed using a Zeiss LSM 510 META confocal microscope.
The following primary antibodies were used (dilution in brackets): rat anti-HA IgG (Roche, 1:200); mouse anti-DNA IgM (Progen, 1:100); mouse anti-BrdU IgG (Roche, 1:50). Secondary antibodies: Cy5-conjugated anti-rat IgG (Abcam, 1:200); FITC-conjugated anti-rat IgG (Abcam, 1:200); FITC-conjugated anti-mouse IgM (Sigma, 1:200); TRITC-conjugated anti-mouse IgG (Sigma, 1:200).
Western blotting
For immunoblot analysis equal amounts of proteins corresponding to total cell lysates or protein fractions were subjected to SDS–PAGE or blue native/SDS–PAGE 2D electrophoresis, semi-dry transferred to nitrocellulose membranes, blocked in 5% non-fat milk (Marvel) in PBS for 1 h and incubated with specific primary antibodies in 5% non-fat milk in PBS for 1 h or overnight. The blots were further incubated with HRP-conjugated secondary antibodies in 5% non-fat milk in PBS for 1 h and visualized using ECL (Amersham).
The primary antibodies used were: mouse anti-FLAG IgG (Sigma, 1:5000), rabbit anti-Myc (Abcam, 1:1000), rabbit anti-POLRMT IgG (1:1000, Abcam), rabbit anti-PTCD3 IgG (1:1000, Santa Cruz Biotechnology), rabbit anti-DHX30 IgG (1:1000, Abcam), rabbit anti-VDAC-1 IgG (1:1000, Abcam), mouse anti-Cox2 IgG (1:5000, Abcam), rabbit anti-TFAM IgG (kindly provided by Dr D. Kang, Kyushu University, Japan, 1:4000), goat anti-TFB2M (1:1000, Abcam), mouse anti-TOM22 IgG (Abcam, 1:5000), mouse anti-GAPDH IgG (Abcam, 1:10000), rabbit anti-c17orf42 (Sigma, 1:1000), mouse anti-Complex I subunit NDUFB8 (MitoSciences, 1:2000), mouse anti-Complex II subunit 30 kDa (MitoSciences, 1:1000), mouse anti-Complex III subunit Core 2 (MitoSciences, 1:1000), rabbit anti-Complex V subunit G (prepared by the ATP synthase group in the MRC Mitochondrial Biology Unit, 1:1000).
Secondary antibodies were: anti-rat IgG-HRP (Sant Cruz Biotechnology, 1:2000), anti-rabbit IgG-HRP (Promega, 1:2000), anti-mouse IgG-HRP (Promega, 1:2000), anti-goat IgG-HRP (Sigma, 1:1000).
Oligonucleotides for TEFM siRNA
dsRNAs (iGene Therapeutics) for the TEFM siRNA experiments were:
Oligo 1 forward: UCCAAAGACUGGACGGGAAAAAAGA
Oligo 1 reverse: AGGUUUCUGACCUGCCCUUUUUUCU
Oligo 2 forward: GGACUAGUGGAAAAGAGCUAGUGAA
Oligo 2 reverse: CCUGAUCACCUUUUCUCGAUCACUU
The control RNA oligonucleotides to GFPmax were from the siRNA Test Kit (Lonza)
Measurement of mitochondrial respiration
Human 143B cells were seeded at 1.5–2 × 104 cells/well in 200 µl growth medium in XF 24-well cell culture microplates (Seahorse Bioscience, Billerica, MA, USA) and incubated at 37°C in 5% CO2 for 36–40 h. One hour before the assay growth medium was removed and replaced with assay medium (low buffered DMEM, 10 mM l-glutamine, 1 mM sodium pyruvate, 2 mM glucose), with one rinse with assay medium, and left to stabilize for 1 h in a 37°C non-CO2 incubator. Analysis was performed in quadruplicates using XF24 Extracellular Flux Analyzer (Seahorse Bioscience, Billerica, MA, USA). The wells containing cells were sequentially injected with 20 mM 2-deoxyglucose (2-DG) to inhibit glycolysis, 100 nM oligomycin to inhibit ATP-synthase, 500–1000 nM carbonylcyanide-4-trifluorometho-xyphenylhydrazone (FCCP) to uncouple the respiratory chain, and 200 nM rotenone to inhibit complex I. Oxygen consumption rate (OCR) was measured for each well every 5 min before and after each injection. Respiratory control ratio (RCR = OCRFCCP/OCRoligomycin) was calculated for each well each time using average of three OCR measurements. Test compounds: 2-DG, oligomycin, FCCP and rotenone were obtained from Sigma (St Louis, MO, USA).
RNA isolation and northern blotting
Total RNA from HOS cells was isolated using Trizol (Invitrogen) according to the manufacturer’s instructions. In order to analyse mitochondrial mRNAs and rRNAs in northern blots, RNA was resolved on 1% agarose gels containing 0.7 M formaldehyde in 1× MOPS buffer, transferred to a nylon membrane in 2× SSC and hybridized with radioactively labelled PCR fragments corresponding to appropriate regions of mtDNA. In order to analyse mitochondrial tRNAs in northern blots, RNA was resolved on 5% UREA polyacrylamide gel in 1× GTE buffer containing 90 mM Tris base, 30 mM Taurine, 0.5 mM EDTA and electrotransferred to a nylon membrane in 0.5× TBE buffer. The blots were hybridized with appropriate antisense mitochondrial tRNAs produced by in vitro transcription using MAXIscript® T7 Kit (Ambion) in the presence of α-32P UTP (3000 Ci/mmol, Perkin Elmer) according to the manufacturer’s instructions.
Isolation of mitochondria
HEK cells were cultured in DMEM with 10% tetracycline-free FBS (Autogen Bioclear). Expression of TEFM.STREP2 was induced 24 h after the last plating by adding doxycycline to a final concentration of 20 ng/ml. Twenty-four hours after induction the cells were harvested and mitochondria were prepared based on a modified procedure adopted from (17) as follows. The whole procedure was performed on ice or at 4°C. Cells were detached by pipetting and pelleted by centrifugation for 10 min at 300 gmax, then washed twice with PBS and the final pellet was weighed to determine its volume (assuming a density of 1.25 g/ml). Cells were then resuspended in nine volumes of hypotonic buffer containing 20 mM HEPES (pH 8.0), 5 mM KCl, 1.5 mM MgCl2 with 1 mg/ml BSA, 2 mM DTT, 0.2 PMSF and protease inhibitors (Roche), incubated on ice for 10 min and homogenized with 10 strokes of a tight-fitting 15 ml glass Dounce homogenizer (cell disruption was monitored by microscopic examination). Next, two-third of total volume of 2.5× MSH buffer [525 mM mannitol, 175 mM sucrose, 20 mM HEPES (pH 8.0), 5 mM EDTA with 1 mg/ml BSA, 2 mM DTT, 0.2 PMSF and protease inhibitor cocktail (Roche)] was immediately added and the homogenate was centrifuged for 10 min at 1600gmax. The pellet was discarded and the supernatant was centrifuged again. The pellet containing the cell debris and nuclei was resuspended in 0.5 ml of 1× MSH buffer [210 mM mannitol, 70 mM sucrose, 20 mM HEPES (pH 8.0), 2 mM EDTA with 1 mg/ml BSA, 2 mM DTT, 0.2 PMSF and protease inhibitors (Roche)] and stored in −20°C. The cytoplasmic fraction was centrifuged for 10 min at 8900gmax and a 0.5 ml sample the supernatant containing the cytosolic fraction was stored in −20°C. The pellet containing the mitochondrial fraction was resuspended in 1× MSH buffer supplemented with 10 mM MgCl2 to a final protein concentration of 2 mg/ml (assuming 10 mg of mitochondrial protein per 1 g of cell pellet) and DNase I (Sigma) was added to a final concentration of 0.2 mg/ml (DNase I treatment removes contamination by residual nuclear DNA and chromatin proteins). The sample was rotated for 30 min and the reaction was terminated by adding EDTA to a final concentration of 15 mM. The sample was washed three times with 5 ml of MSH buffer without BSA by centrifugation for 10 min at 8 900 gmax. After washing, the pellet was suspended in 0.1–0.2 of volume of MSH buffer without BSA, loaded onto a sucrose gradient (see below) and centrifuged for 40 min at 117 000gmax (33 000 rpm in a MLS50 rotor, Beckman Coulter). The gradient was prepared by layering 1 M and 1.5 M sucrose solutions in fresh gradient buffer [10 mM HEPES (pH 7.8), 5 mM EDTA, 2 mM EDTA, 2 mM DTT] in polyallomer tubes (Beckman Coulter). After the centrifugation, the interface was collected and four volumes of gradient buffer were slowly added with gentle vortexing. The sample was then centrifuged for 10 min at 8 900gmax, the supernatant discarded and the mitochondria resuspended in MSH buffer without BSA.
Isolation of STREP2-tagged complexes from HEK cells
Mitochondria derived from HEK cells expressing the TEFM.STREP2 transgene were isolated as described above and resuspended to 8 mg protein/ml in 40 mM HEPES (pH 7.6), 10 mM EDTA, 4 mM DTT, 0.4 mM PMSF, 300 mM NaCl and protease inhibitors cocktail (Roche). Next, the mitochondria were lysed by mixing with an equal volume of 0.8% dodecylmaltoside (DDM) in water, on a roller at 4°C for 30 min. The lysate was centrifuged at 1600gmax for 10 min to remove insoluble debris and the supernatant was load onto a gravity flow Strep-Tactin column (IBA) and the flow-through re-loaded twice. The column was washed sequentially with five column volumes (CV) of washing buffer containing 20 mM HEPES (pH 7.6), 1 mM EDTA, 2 mM DTT, 0.2 mM PMSF, 150 mM NaCl, 0.05% DDM, protease inhibitors (Roche) and eluted with six lots of 0.5 CV elution buffer (washing buffer plus 5 mM dethiobiotin). Strep-Tactin purified proteins were separated by SDS–PAGE, Coomassie-stained protein bands were excised from gels and identified by mass spectrometry.
Mass spectrometry
Strep-Tactin purified proteins were separated by 4–12% gradient SDS–PAGE (NuPage, Invitrogen). Coomassie-stained protein bands were excised from gels and digested by ‘in-gel’ cleavage (18) at 37°C overnight with trypsin (Roche Diagnostic GmbH). Peptide products were analysed in an ABI plus MALDI-TOF-TOF Mass Spectrometer using α-cyano-4–hydroxy-tans-cinnamic acid as the matrix. The mass spectral data were analysed by Peaks to MASCOT® Tool and proteins were identified using MASCOT® database searching. The criteria for identification of proteins were a significant PMF score, as defined by MASCOT (>70 ppm), and at least two MS/MS peptide matches.
Production of recombinant TEFM in E. coli
The BL21(DE3) Escherichia coli strain harbouring the pGEX-GST.TEFM plasmid was grown at 37°C until OD600 = 0.6 and induced with 50 µM IPTG for 16 h at 25°C. The pellet was resuspended in a buffer containing 20 mM Tris–Cl (pH 8.0), 150 mM NaCl and 1 mM DTT and sonicated for 3 min (10 s with 60 s intervals, Misonix 3000 sonicator, output 6). Cell lysate was filtered through a 0.45 µm syringe filter and run twice through a column with 1 ml Glutathione-Sepharose 4B (GE Healthcare). The column-bound protein was eluted with five column volumes of buffer containing 40 mM Glutathione (Sigma), 35 mM NaOH, 20 mM Tris–Cl (pH 8.0), 150 mM NaCl and 1 mM DTT. Peak fractions were subjected to FPLC on a Supedex G200 column. The fractions from the Supedex G200 column were assessed by SDS–PAGE and the peak fractions as indicated on Supplementary Figure S5 were concentrated using Vivaspin 2 Centrifugal Concentrators (Sartorius stedim) and stored at −80°C in a buffer containing 20 mM Tris–Cl (pH 8.0), 150 mM NaCl, 1 mM DTT and 20% glycerol.
In vitro RNA synthesis on ssDNA and dsDNA
The reactions were performed essentially as described by (19) with the following modifications. The reaction mixture of 20 µl containing 200 ng of single stranded M13mp18 DNA (Amersham) or 1 pmol of 3′-tailed dsDNA of various length, 10 mM Tris–Cl, (pH 8.0), 20 mM MgCl2, 1 mM DTT, 100 µg/ml BSA, 400 µM ATP, 150 µM CTP, 150 µM GTP, 10 µM UTP (all ribonucleotides were from Ambion), 0.084 µM α-32P UTP (3000 Ci/mmol, Perkin Elmer), 4 U of RNasin (Promega), 0.35 pmol of POLRMT (kindly provided by Drs Gustafsson and Falkenberg, Karolinska Institutet, Stockholm, Sweden) and indicated concentrations of recombinant TEFM. The mixture was incubated for 1 h for the ssDNA template or indicated times for the 3′-tailed templates at 32°C. The samples were analysed on a 5% UREA polyacrylamide gel in 1× GTE buffer containing 90 mM Tris base, 30 mM Taurine and 0.5 mM EDTA.
The T7 RNA polymerase (T7RNAP) is used in the bacterial expression system based on the BL21 E. coli strain that was employed to over express TEFM and hence might have contaminated the recombinant TEFM preparations. In order to exclude a potential effect of the presence of T7RNAP on the in vitro RNA synthesis on ssDNA by POLRMT, we performed the reaction as described above together with various concentrations of T7RNAP (Fermentas) (Supplementary Figure S6).
The long 3′-tailed dsDNA templates were constructed according to our new procedure as follows: a 400 bp fragment containing the NCR region of human mtDNA downstream from the LSP promoter lacking the conserved sequence block II was amplified by PCR. The forward primer used in the reaction contained an overhang of 15 uracils. After the PCR reaction the DNA product was digested with the mixture of Uracil DNA glycosylase (UDG) and the DNA glycosylase-lyase Endonuclease VIII (USER Enzyme, New England Biolabs) and agarose gel purified. The short 3′-tailed dsDNA templates of 20 or 100 bp were constructed by annealing two complementary oligonucleotides that had the same sequence as the first 20 or 100 bp from the 5′ part of the long 400 bp 3′-tailed template, respectively.
Other procedures
Other procedures including: identification of RNAs co-purified with the TEFM complex, assessing steady-state levels of mtRNAs and mtDNA copy number by quantitative PCR and measuring of the stability of mitochondrial transcripts are described in Supplementary data.
RESULTS
In silico identification and domains of TEFM
We initially identified the c17orf42 gene, hereafter TEFM, as a putative human mitochondrial Holliday Junction Resolvase (HJR), as it exhibits significant sequence homology with the well-characterized bacterial HJR RuvC (14) (Supplementary text and Supplementary Figure S1). However, this hypothesis was not supported by experiment, as recombinant TEFM isolated from human mitochondria or purified from E. coli lacked any detectable HJR activity (data not shown). In addition to the RuvC-like RNase H fold TEFM contains two tandemly repeated helix-hairpin-helix (HhH) motifs that share sequence homology with the HhH motifs of bacterial TEX protein, which functions in a variety of transcriptional processes, and nuclear transcription elongation regulator Spt6 (Figure 1A) (20). Like TEFM, both Spt6 and TEX contain an RNase H fold, although its position in the protein differs in relation to the HhH motif (Figure 1A, blue boxes). The RNase H fold of Spt6 and TEX belongs to the YqgF family (21) and preserves the overall topology and the structural elements of the same fold of RuvC HJRs; however there are no reports of YqgF functioning as a HJR (22,23).
TEFM is a mitochondrial protein
We analysed the sequence of the human TEFM protein with several computer programmes that scan the N-terminal region of proteins for the presence of a putative MTS. Most of the algorithms used returned a high probability of a MTS in TEFM (MultiLoc, 95.0%; MitoProt II, 92.4%; TargetP, 78.2%; PSORT II, 51.2%).
In order to determine by experiment the cellular location of TEFM, HOS cells were disrupted and fractionated. The endogenous TEFM protein was concentrated in the mitochondrial fraction similarly to well-characterized mitochondrial matrix protein TFAM and the mitochondrial outer membrane TOM22 (Figure 1B). In a parallel experiment, a haemagglutinin (HA) tagged version of the protein (TEFM.HA) was transiently expressed in human A549 adenocarcinoma cells; immunocytochemistry using an anti-HA antibody and staining with the mitochondrial probe (Mitotracker Red CMXRos) indicated that TEFM was targeted to mitochondria, as the HA tagged protein was distributed in a punctate pattern within mitochondria (Figure 1C, arrow heads). Therefore, it is concluded that TEFM protein is present inside the mitochondria of human cells.
TEFM is necessary for OXPHOS function
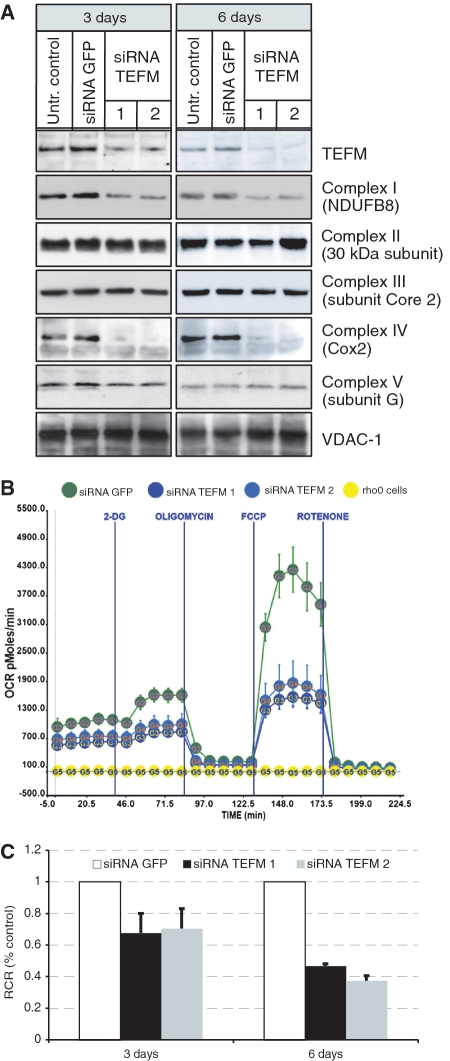
In order to test experimentally the role of TEFM in OXPHOS function, the expression of the gene was inactivated by RNA interference. We identified two siRNAs that efficiently depleted the target mRNA (Supplementary Figure S2A) and protein (Figure 2A). We inactivated TEFM in HOS cells and analysed steady-state levels of respiratory chain subunits, as OXPHOS dysfunction is often associated with aberrant assembly or instability of mitochondrial respiratory complexes. TEFM RNAi markedly reduced the abundance of the complex IV subunit, COX2, and to a lesser extent, NDUFB8, a component of complex I (Figure 2A). The decreases in respiratory chain components were accompanied by reduced cellular OCR by ∼50% (Figure 2B) and uncoupling of the respiratory chain and ATP synthase based on the respiratory control ratio (Figure 2C). These results indicated that TEFM plays a key role in mitochondrial energy production and given its homology to nucleic acids modifying proteins (Figure 1A) this was most likely to occur via a contribution to mtDNA maintenance or expression.
Figure 2.
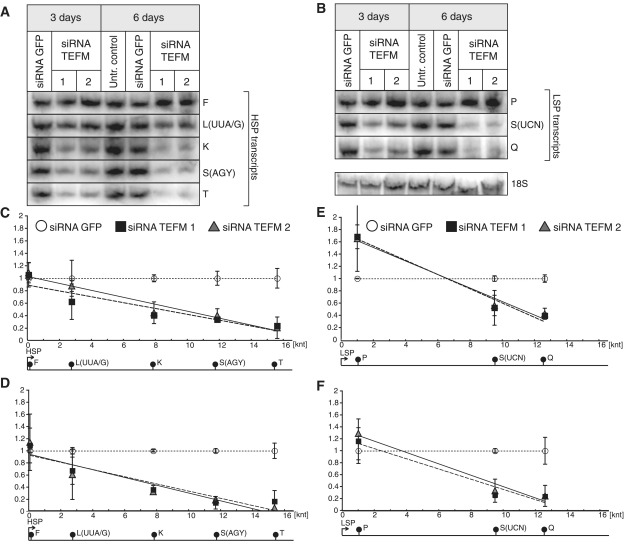
Defects in respiratory chain function upon TEFM gene silencing. (A) Western blot analyses of steady-state protein level of TEFM and subunits of respiratory chain complexes in control cells (untransfected cells and siRNA GFP) and cells treated with siRNA TEFM for 3 and 6 days. (B) Oxygen consumption rate (OCR) measured in quadruplicate population of control cells transfected siRNA GFP (green), cells treated with siRNA TEFM for 3 days (siRNA 1 in dark blue and siRNA 2 in light blue) and cells lacking mtDNA (Rho0, yellow). (C) Respiratory control ratio (RCR) in cells treated with siRNA GFP or siRNA TEFM for 3 or 6 days.
TEFM gene silencing lowers the abundance of promoter-distal mitochondrial transcripts
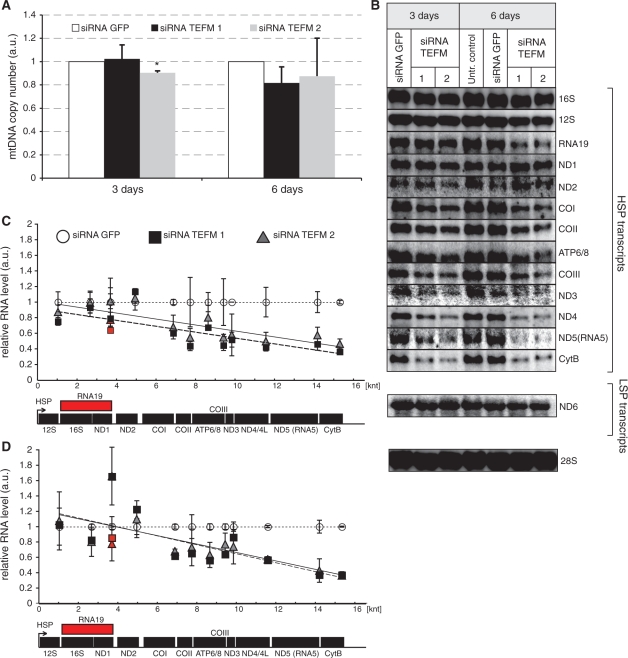
Inactivation of TEFM had little effect on mtDNA copy number (Figure 3A) or mitochondrial replication intermediates (data not shown) in HOS cells. Next, mitochondrial transcripts were extracted from HOS cells after TEFM RNAi. Quantification of the steady-state levels of mRNAs and rRNAs generated by transcription from HSP and LSP of mtDNA revealed marked reductions in many mitochondrial transcripts of cells subjected to TEFM RNAi (Figure 3B–E). There was a clear pattern to the changes in transcript levels: transcripts at promoter-distal locations on the H-strand polycistronic transcript were much less abundant than transcripts mapping closer to the promoter, with the exception of ND1 and ND2 mRNAs (Figure 3C–D). The exceptions could be explained by changes in transcript stability, as the half-life of ND1 was markedly increased in the TEFM silenced cells (Supplementary Figure S2B). Up-regulation of the steady-state level of ND1 mRNA has been observed previously in cells with inactivated genes that play a role in mitochondrial transcription and/or processing (24). RNA19, which contains 16S rRNA, tRNA-LeuUUR and ND1, followed the general trend of the other transcripts (Figure 3C and D, red). There was no detectable increase in any precursor RNA in northern blots (Supplementary Figure S3) and so the changes in mitochondrial mRNAs levels were not attributable to perturbed RNA processing.
Figure 3.
Steady-state levels of mtDNA and mitochondrial transcripts in cells with inactivated TEFM. (A) mtDNA copy number as measured by comparative qPCR of the mitochondrial Cox2 gene and single copy nuclear gene (APP) in controls (Untransfected and siRNA GFP) and cells treated with TEFM siRNA (siRNA TEFM 1 and 2). *P < 0.05, n = 3, error bars indicate 1 SD. (B) Northern blot analyses of mitochondrial transcripts transcribed from the HSP1 or LSP promoter in control cells (untransfected and treated with GFP siRNA) and cells treated with TEFM siRNA for 3 or 6 days. Nuclear 28S rRNA was used as a loading control. (C and D) Quantification of steady-state levels of the H-strand mitochondrial transcripts in cells treated with TEFM siRNA for 3 days (C) and 6 days (D) analysed by northern blots. The values of the relative RNA level (mtRNA/28S rRNA) were obtained by quantifying PhosphoImager scans of blots in the ImageQuant software and normalized for the values obtained for control cells transfected with siRNA GFP. The relative RNA level of each transcript for siRNA TEFM 1 (square) and 2 (triangle) was plotted in the function of the distance of its 3′ end from HSP. Dotted line, trend for siRNA GFP control; solid line, trend for siRNA TEFM 1; dashed line, trend for siRNA TEFM 2. Red symbols indicate the RNA19 transcript. n = 3, error bars = 1 SD. The P-values (two-tailed Student’s t-test) for each transcript calculated for combined values for both TEFM siRNAs for 3 days: 12S = 0.103, 16S = 0.719, RNA19 = 0.124, ND1 = 0.492, ND2 = 0.234, COI = 0.009, COII = 0.031, ATP6/8 = 0.064, COIII = 0.023, ND3 = 0.890, ND4/4L = 0.006, ND5 = 0.007, CytB < 0.001, ND6 = 0.502; and for 6 days: 12S = 0.813, 16S = 0.092, RNA19 = 0.187, ND1 = 0.026, ND2 = 0.285, COI = 0.003, COII = 0.129, ATP6/8 = 0.025, COIII = 0.027, ND3 = 0.169, ND4/4L < 0.001, ND5 = 0.002, CytB < 0.001, ND6 = 0.137. The quantification of the steady-state level of the ND6 transcript that is transcribed from LSP is shown in Supplementary Figure S2.
In addition, we measured the abundance of several mitochondrial tRNAs (mt-tRNA) encoded on the H- and L-strands, from cells treated with TEFM-targeted dsRNAs (Figure 4A–B). As with mitochondrial mRNAs and rRNAs, TEFM gene-silencing decreased the steady-state level of promoter-distal tRNAs encoded both on H- and L-strand to a greater extent than promoter-proximal tRNAs (Figure 4C–F). The steady-state levels of mt-tRNAs that map in the last third of the mitochondrial genome (with respect to the promoter) were decreased by ∼90% (e.g. tRNA-SerAGY or tRNA-Thr) in cells treated with TEFM siRNA for 6 days. The effective loss of 90% tRNAs due to pathological mutation has a substantial effect on complex I activity and mitochondrial translation (Dunbar et al., 1996), and so the decrease in mt-tRNAs caused by TEFM siRNA can explain the associated severe decreases in OCR and mitochondrially encoded respiratory chain components (Figure 2A). In vertebrates mitochondria, transcription from the HSP and LSP promoters produces polycistronic precursor RNAs that are processed to yield the individual mRNAs, tRNAs and rRNAs, and so reduced processivity of POLRMT is the most straightforward explanation for the larger decreases in the levels of promoter-distal RNAs than promoter-proximal RNAs. Thus, the data are consistent with the hypothesis of TEFM enhancing transcription processivity of both stands of mtDNA.
Figure 4.
Steady-state levels of mitochondrial tRNAs in TEFM-depleted cells. (A)–(B) Northern blot analyses of mitochondrial tRNAs transcribed from the HSP (A) or LSP (B) promoter in control cells (untransfected and treated with GFP siRNA) and cells treated with TEFM siRNA for 3 or 6 days. Nuclear 18S rRNA was used as a loading control. (C)–(F) Quantification of steady-state levels of the H-strand (C and D) or L-strand (E and F) mitochondrial tRNAs in cells treated with TEFM siRNA for 3 days (C and E) and 6 days (D and F) analysed by Northern blots. The values of the relative RNA level (tRNA/28S rRNA) were obtained by quantifying PhosphoImager scans of blots in the ImageQuant software and normalized for the values obtained for control cells transfected with siRNA GFP. The relative RNA level of each tRNA for siRNA TEFM 1 (square) and 2 (triangle) was plotted in the function of the distance of its 3′-end from the promoters. Dotted line, trend for siRNA GFP control; solid line, trend for siRNA TEFM 1; dashed line, trend for siRNA TEFM 2. n = 3, error bars = 1 SD. The P-values (two-tailed Student’s t-test) for each tRNA calculated for combined values for both TEFM siRNAs for 3 days: F = 0.553, L(UUA/G) = 0.303, K = 0.002, S(AGY) = 0.001, T = 0.002, P = 0.103, S(UCN) = 0.004, Q < 0.001; and for 6 days: F = 0.656, L(UUA/G) = 0.154, K < 0.001, S(AGY) < 0.001, T = 0.002, P = 0.297, S(UCN) = 0.002, Q = 0.004.
TEFM interacts with the mitochondrial RNA polymerase and other mitochondrial proteins with presumed roles in RNA metabolism
In order to identify mitochondrial proteins that interact with TEFM a FLAG- and STREP2-tagged version of TEFM (TEFM.STREP2) was introduced into Flp-In T-Rex™ HEK 293T cells. The STREP2 binding group was used in order to purify TEFM and potential interacting proteins via a streptavidin-coated matrix after induction of the transgene for 24 h with 20 ng/ml of doxycycline (Figure 5A). Streptavidin immobilized proteins from mitochondrial lysates of HEK cells expressing TEFM.STREP2 or control cells were analysed by mass spectrometry. Three mitochondrial proteins were identified consistently in TEFM.STREP2 expressing mitochondrial lysates: mitochondrial RNA polymerase—POLRMT (25), Pentatricopeptide repeat domain 3—PTCD3 (26) and a putative DEAH-box RNA helicase—DHX30 (27) (Figure 5B). The mass spectrometry analysis also revealed a variable set of mitochondrial ribosomal proteins co-purifying with TEFM (Supplementary Table S2).
Figure 5.
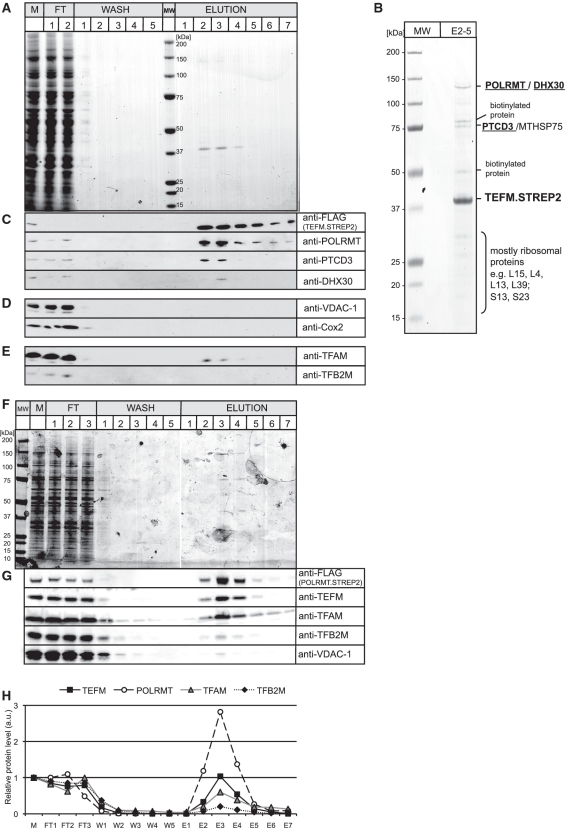
Mitochondrial RNA polymerase co-purifies with TEFM. (A) A SDS–PAGE gel stained with Coomassie Brilliant Blue showing the protein profile of the affinity purification of the TEFM.STREP2 from the mitochondria of HEK cells. The most intense protein band of ∼40 kDa seen in the elution fractions 2–4 corresponds to the purified TEFM.STREP2 protein. M, total mitochondrial lysate; FT, flow-through; MW, Molecular weight marker. (B) A SDS–PAGE gel stained with Coomassie Brilliant Blue with concentrated fractions from 2 to 5 (E2–5). Protein bands were cut from the gel and analysed by mass spectrometry. The identities of the protein are shown on the left-hand side. Some endogenous mitochondrial biotinylated proteins (e.g. 3-hydroxyacyl-CoA dehydrogenase α-subunit or hydroxyacyl dehydrogenase, subunit B) were detected in the analysis as affinity purification system used here is based on the interaction between the STREP2 tag and engineered streptavidin (Strep-Tactin), which also binds biotin. Only biotinylated proteins were detected in mock affinity capture experiments performed on parental HEK cells (data not shown). The presence of the mitochondrial chaperone protein, MTHSP75, in our preparation could well result from a mitochondrial stress response caused by overexpression of TEFM (41). (C)–(E) Western blots confirming the identity of the proteins that co-purify with TEFM that were detected by mass spectrometry (C) or documenting that there was no enrichment of highly abundant mitochondrial proteins (D) or mitochondrial proteins involved in the initiation of mtDNA transcription (E). (F) A SDS–PAGE gel stained with Coomassie Brilliant Blue showing the protein profile of the affinity purification of the POLRMT.STREP2 from the mitochondria of HEK cells. The most intense protein band of ∼140 kDa seen in the elution fractions 3–4 corresponds to the purified POLRMT.STREP2 protein. M, total mitochondrial lysate; FT, flow-through; MW, Molecular weight marker. (G) Western blots illustrating the protein profile of the affinity purification of the POLRMT.STREP2 from the mitochondria of HEK cells. (H) Relative abundance of proteins that co-purify with POLRMT.STREP2. The values were obtained by quantifying PhosphoImager scans of western blots from (G) in the ImageQuant software and normalized for the values obtained for the total mitochondrial lysate.
The co-purification of POLRMT, DHX30 and PTCD3 with TEFM was confirmed by immunoblotting (Figure 5B). In contrast to these three proteins, highly abundant mitochondrial proteins such as voltage-dependent anion channel 1, VDAC-1 and cytochrome c oxidase subunit II, Cox2 were depleted during the purification procedure (Figure 5D). Nor was there any appreciable enrichment of known proteins of the mitochondrial transcription initiation machinery (TFAM or TFB2M) in our preparations of tagged TEFM (Figure 5E). Thus, a substantial enrichment of a specific subset of mitochondrial proteins had been achieved by the affinity purification procedure.
In the reciprocal experiment, a tagged version of POLRMT that carried a FLAG and a STREP2 binding group (POLRMT.STREP2) was expressed in HEK cells. POLRMT.STREP2 and interacting proteins were purified on a streptavidin column as described above for TEFM.STREP2 (Figure 5F). The TEFM protein co-purified with POLRMT.STREP2 as confirmed by western blot (Figure 5G). Importantly, there was twice and five times as much of TEFM present in the POLRMT.STREP2 peak elution fractions compared to the known transcription initiation factors: TFAM and TFB2M, respectively (Figure 5H). These results suggest a tight association between TEFM and POLRMT in human mitochondria.
TEFM is found in complexes containing RNA
Fractionation of affinity-purified TEFM on blue native gels revealed several complexes, the largest of which had a molecular mass in excess of one mega-dalton (Supplementary Figure S4A). This complex contained a fraction of POLRMT and most of the PTCD3 co-purifying with TEFM (Supplementary Figure S4A). Treatment of the mitochondrial lysate with RNase A, but not DNase I, disrupted the complex suggesting that RNA forms part of the complex (Supplementary Figure S4B).
In order to determine which mitochondrial transcripts were present in the TEFM complex, RNA was extracted from the complex, radioactively labelled and hybridized with immobilized fragments of human mtDNA (Supplementary Figure S4C). All mitochondrial transcripts co-purified with TEFM, with rRNAs being the most abundant (Supplementary Figure S4C–D). The enrichment of rRNAs associated with TEFM reflects their natural overabundance (28).
The contribution of RNA to the interaction between TEFM and POLRMT, DHX30 and PTCD3 was investigated further, by screening mitochondrial lysates of TEFM.STREP2 expressing cells that had, or had not, been treated with nucleases, prior to loading the lysate on a STREP2-affinity column (Figure 6A). After RNase A treatment neither PTCD3 nor DHX30 co-purified with TEFM, whereas the association with POLRMT persisted. DNase I had no effect on the proteins co-purifying with TEFM (Figure 6B), which suggests PTCD3 and DHX30 are held in a complex with TEFM by RNA.
In our immunofluorescence studies TEFM was distributed in a punctate pattern within mitochondria. Such a pattern is reminiscent of proteins found in the mitochondrial nucleoid or newly synthesized mtRNA (29). Because RNA is present in a complex containing TEFM, and it interacts with POLRMT, we examined the distribution of newly synthesized RNA, DNA and a HA tagged version of TEFM in human cultured cells. Mitochondrial RNA was visualized by growing A549 adenocarcinoma cells in the presence of bromouridine (BrU) for 30 min and applying a specific monoclonal antibody to BrU-containing RNA. Some nucleoids lacked both RNA and TEFM, whereas nucleoids with newly synthesized RNA generally had TEFM.HA present (Figure 6C), suggesting that TEFM is a component of transcriptionally active nucleoids.
TEFM interacts with the catalytic domain of POLRMT
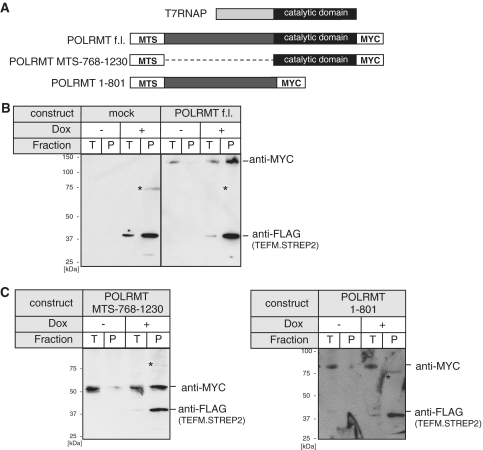
Human POLRMT displays significant homology to the RNA polymerase of T-odd bacteriophages, such as T7 (25,30). However, mitochondrial RNA polymerases contain N-terminal extensions not present in the T7 polymerase (T7RNAP) (Figure 7A) and so this was considered to be a potential binding region for TEFM. In order to map the region of human mitochondrial RNA polymerase that binds to TEFM, pull-down experiments with truncated variants of POLRMT were performed. Myc-tagged truncated versions of POLRMT (Figure 7A) were transiently expressed in HEK cells that simultaneously expressed TEFM, under the control of a doxycycline inducible promoter. Control experiments showed that full-length, Myc-tagged POLRMT could be pulled down only if TEFM was induced (Figure 7B). The POLRMT variant lacking residues 61–767 interacted with TEFM (Figure 7C, left), whereas the C-terminally truncated variant of POLRMT (lacking residues 802–1230) was no longer able to form a complex with TEFM (Figure 7C, right). Therefore, we concluded that TEFM interacts with the catalytic region of POLRMT, suggesting that it might be involved directly in the regulation of polymerization.
Figure 7.
TEFM interacts with C-terminal, catalytic part of POLRMT. (A) Schematic representation of the myc-tagged POLRMT constructs used to map the interaction with TEFM in comparison with the T7 phage RNA polymerase (T7RNAP). The N-terminal extension present in the POLRMT (dark gray) is missing in the T7RNAP. f.l., full-length. (B) Western blot of the control pull-down experiment with mock (left) and the full-length POLRMT (right) transfected HEK cells that inducibly express TEFM.STREP2. The blot was incubated with the antibodies indicated to the right. T, total cell lysate; P, pulled-down material. The asterisk indicates a non-specific band. (C) Western blot of the pull-down experiment with the POLRMT lacking the N-terminal extension (POLRMT MTS-768-1230, left) or the C-terminal catalytic part (POLRMT 1-801, right) with TEFM.STREP2. The asterisk indicates a non-specific band.
Recombinant TEFM enhances POLRMT processivity
Despite significant sequence similarity between the T-odd phage RNA polymerases and mitochondrial RNA polymerases, there are important functional differences between the two types of enzyme. For example, unlike the single-subunit T7 RNA polymerase, POLRMT requires auxiliary factors to initiate transcription at promoter sequences (7). Previous reports have also shown that recombinant POLRMT is non-processive on ssDNA templates, synthesizing only relatively short RNA species of 25–75 nt compared to the >500 nt achieved by T7 RNA polymerase (19). By analogy with transcription initiation, we hypothesized that POLRMT might require additional factors to regulate its processivity. TEFs that increase the processivity of RNA polymerases are known to operate in bacteria and the nucleus [reviewed in (12)]. Moreover, animal mitochondria require a highly processive RNA polymerase (complex) to produce polycistronic transcripts of 15, 11 and 2.7 knt, from which all mtDNA products derive. TEFM was adjudged a plausible candidate for enhancing processivity, as it interacts with the catalytic region of POLRMT, but does not co-purify with the initiation factors TFAM or TFB2M (see above).
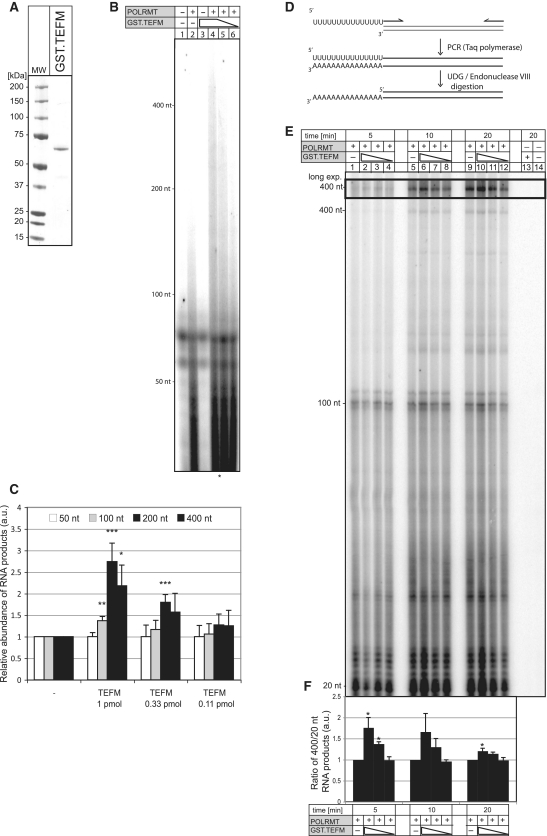
To test the above hypothesis, recombinant GST.TEFM protein was purified to homogeneity (Figure 8A, Supplementary Figure S5 and ‘Materials and Methods’ section) and the polymerase activity of recombinant POLRMT on ssDNA assayed in vitro, with or without recombinant GST.TEFM. POLRMT incubated with ssDNA in the presence of radiolabelled UTP yielded short RNA fragments (∼25–75 nt), and longer RNAs when recombinant GST.TEFM was included in the reaction mixture (Figure 8B and C). The highest concentration of GST.TEFM tested (50 pM), revealed a >2-fold increase in RNA products of 200 and 400 nt in length, compared to POLRMT transcripts synthesized without TEFM (Figure 8C). In a further test, the promoter independent activity of POLRMT was assayed for 5–20 min on short or long 3′-tailed dsDNA of 20, 100 or 400 bp, with or without recombinant GST.TEFM (Figure 8D–F). In 5 min reactions containing the highest concentration of GST.TEFM the ratio of 400:20 nt product was 75% higher than that of reactions lacking TEFM (Figure 8F). Therefore, POLRMT in concert with TEFM needs less time to make transcripts 400 nt in length than POLRMT alone; i.e. POLRMT processivity is enhanced by TEFM in vitro.
Figure 8.
Stimulation of the POLRMT activity by TEFM. (A) Coomassie Brilliant Blue stained SDS–PAGE gel showing E. coli purified the GST.TEFM protein fusion. MW, molecular weight marker. (B) The synthesis of 32P-labelled RNA by POLRMT (0.35 pmol) on M13mp18(+) ssDNA in the absence (lane2) and the presence of 1.0 pmol (lane 4), 0.33 pmol (lane 5) and 0.11 pmol (lane 6) of GST.TEFM was performed as described in ‘Materials and Methods’ section. The products were separated on a 5% UREA polyacrylamide gel and subjected to autoradiography. (C) Relative abundance of the RNA products of indicated lengths synthesized by POLRMT on the ssDNA template in the presence of different concentrations of GST.TEFM. The values were obtained by quantifying PhosphoImager scans of dried UREA gels in the ImageQuant software and normalized for the values obtained from the reaction with POLRMT only. *P < 0.05, **P < 0.01, ***P < 0.001; two-tailed Student’s t-test; n = 4, Error bars = 1 SD. (D) Schematic representation of the construction of long 3′-tailed dsDNA templates. (E) The synthesis of 32P-labelled RNA by POLRMT (0.35 pmol) on 3′-tailed dsDNA of different length (20, 100 and 400 bp) in the absence (lanes 1, 5 and 9) and the presence of 1.0 pmol (lanes 2, 6 and 10), 0.33 pmol (lanes 3, 7 and 11) and 0.11 pmol (lanes 4, 8 and 12) of GST.TEFM for the indicated time. (F) The ratio between the 400 and 20 nt RNA products synthesized by POLRMT on the 3′-tailed dsDNA template in the presence of different concentrations of TEFM. The values were normalized with respect to the reaction with POLRMT alone. *P < 0.05; two-tailed Student’s t- test; n = 3, Error bars = 1 SD.
DISCUSSION
We show here that the gene product of c17orf42, named TEFM, is a human mitochondrial protein, which is important for the synthesis of polycistronic RNAs, from which the full panoply of mature mitochondrial transcripts is generated. Transcription of DNA is divided into an initiation stage, entailing recruitment of transcription factors, including an RNA polymerase, to a promoter and the commencement of RNA synthesis; and an elongation step, during which the RNA polymerase travels along the DNA template synthesizing an RNA transcript. Several nuclear accessory factors have been identified that facilitate transcription elongation. The mechanism may be direct, modulating RNA polymerase activity; or indirect, modifying chromatin structure (12).
TEFM is an accessory factor of mitochondrial RNA polymerase that enhances its processivity
TEFM can be used to affinity purify mitochondrial RNA polymerase (POLRMT) (Figure 5) indicating a tight association between the two proteins. TEFM is deemed likely to facilitate the elongation step of mitochondrial transcription because: (i) TEFM gene-silencing lowers the abundance of promoter-distal mitochondrial transcripts; (ii) the protein co-localizes with newly synthesized mtRNA; (iii) TEFM interacts with the catalytic domain of POLRMT, but not the mitochondrial transcription initiation factors TFAM and TFB2M; and (iv) it stimulates POLRMT processivity in vitro.
TEFM’s similarities with known TEFs also support its assignment as a mitochondrial TEF (Figure 1A). In addition to sequence homology, TEFM shares several functional resemblances with Spt6 and TEX. Both Spt6 and TEX interact directly with RNA polymerse II (22,31); and Spt6 stimulates transcription elongation by RNA polymerase II in vitro (32). Taken together, the presence of the characteristic HhH motif and the Ribonuclease H fold in Spt6, Tex and TEFM and the functional parallels between the three proteins point to a similar role in transcription.
Interdependence of transcription and translation in mitochondria
The putative TEFM complex, in addition to POLRMT, contains all mitochondrial mRNAs and rRNAs, mitochondrial ribosomal proteins and two other proteins: PTCD3 (pentatricopeptide repeat domain 3) and DHX30 (a putative DEAD-box RNA helicase). The presence of RNA is integral to the complex, as RNase A treatment abolished TEFM association with DHX30 and PTCD3; it remains to be determined whether the three proteins bind RNA independently, or have direct contacts with each other that are stabilized by RNA. PTCD3 contains a pentatricopeptide repeat (PPR) and was first identified as an RNA-binding factor associated with the mitochondrial ribosome (26) and a recent report has suggested that PTCD3 is involved in mitochondrial translation in human cells (33). The PPR proteins constitute one of the largest gene families in plants and the majority of them are, or are at least predicted to be, localized in mitochondria and chloroplasts where they play a variety of roles in RNA metabolism, including splicing, editing, regulation of transcript stability and translation (34). The mitochondrial isoform of the putative RNA helicase DHX30 was identified as a candidate nucleoid protein (27). Of note, DHX30 forms foci within mitochondria that are juxtaposed rather than coincident with mtDNA (27). It is therefore probable that DHX30, like TEFM, co-localizes with mtRNA, especially as they are both found in the same RNA-containing complex (Figure 5). Moreover, DHX30 is a DEAH-box helicase, and so was always likely to participate in RNA metabolism; its purpose may be to prevent secondary structure formation of RNA and thereby facilitate ribosome loading in mitochondria.
The human mitochondrial ribosome recycling factor (mtRRF) co-immunoprecipitates mitochondrial ribosomes and a large number of proteins involved in mitochondrial RNA and DNA metabolism, including POLRMT, PTCD3 and DHX30 (35). In another report, the mitoribosomal protein, MRPL12, was shown to interact directly with POLRMT and stimulates its activity in vitro (11). TEFM also interacts with the transcription machinery (POLRMT) and protein synthesis apparatus (mitochondrial ribosomal RNAs, MRPs and PTCD3) of mitochondria and so provides further evidence of a physical coupling of transcription, and translation in mammals, as previously proposed (11,36–39).
In summary, we propose the TEFM protein identified here is a critical component of the transcription apparatus of human mitochondria. Further studies of this protein should provide new insights into regulatory mechanisms of mitochondrial transcription. There are also other areas of research that could benefit from additional knowledge of TEFM as its coding gene, c17orf42, is located within the region of 17q11 microdeletions that encompass the NF1 gene. Deletions of this region cause 5–10% of cases of neurofibromatosis type 1 (OMIM 162200); the remainder are associated with intergenic mutations in the NF1 gene. The phenotype of neurofibromatosis type 1 generated through 17q11 microdeletions differs from that associated with intragenic NF1 mutations and might be related to haplo-insufficiency of specific genes within the deleted interval, such as TEFM.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
Medical Research Council; Biotechnology and Biological Sciences Research Council; Federation of the Societies of Biochemistry and Molecular Biology (short-term fellowship to A.C.); the Netherlands Genomics Initiative (Horizon Programme, project 050-71-555). Funding for open access charge: MRC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank John Berrisford and Rouslan Efremov for their help in purifying recombinant TEFM, and Hiroshi Sembongi for help in designing siRNA oligos and qPCR primers. We also would like to thank Massimo Zeviani and Erika Fernandez-Vizarra for the gift of POLRMT cDNA and Claes Gustafsson and Maria Falkenberg for providing the POLRMT enzyme. We are indebted to Michael Harbor and Ian Fearnley for their help in protein identification by M.S.
REFERENCES
- 1.Forner F, Foster LJ, Campanaro S, Valle G, Mann M. Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol. Cell Proteomics. 2006;5:608–619. doi: 10.1074/mcp.M500298-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, et al. Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 2003;21:281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- 4.Calvo S, Jain M, Xie X, Sheth SA, Chang B, Goldberger OA, Spinazzola A, Zeviani M, Carr SA, Mootha VK. Systematic identification of human mitochondrial disease genes through integrative genomics. Nat. Genet. 2006;38:576–582. doi: 10.1038/ng1776. [DOI] [PubMed] [Google Scholar]
- 5.Lopez MF, Kristal BS, Chernokalskaya E, Lazarev A, Shestopalov AI, Bogdanova A, Robinson M. High-throughput profiling of the mitochondrial proteome using affinity fractionation and automation. Electrophoresis. 2000;21:3427–3440. doi: 10.1002/1522-2683(20001001)21:16<3427::AID-ELPS3427>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 6.Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 7.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 8.Kruse B, Narasimhan N, Attardi G. Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell. 1989;58:391–397. doi: 10.1016/0092-8674(89)90853-2. [DOI] [PubMed] [Google Scholar]
- 9.Wenz T, Luca C, Torraco A, Moraes CT. mTERF2 regulates oxidative phosphorylation by modulating mtDNA transcription. Cell Metab. 2009;9:499–511. doi: 10.1016/j.cmet.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Park CB, Asin-Cayuela J, Camara Y, Shi Y, Pellegrini M, Gaspari M, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, et al. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Cotney J, Shadel GS. Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J. Biol. Chem. 2007;282:12610–12618. doi: 10.1074/jbc.M700461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 13.Montoya J, Christianson T, Levens D, Rabinowitz M, Attardi G. Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA. Proc. Natl Acad. Sci. USA. 1982;79:7195–7199. doi: 10.1073/pnas.79.23.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connolly B, Parsons CA, Benson FE, Dunderdale HJ, Sharples GJ, Lloyd RG, West SC. Resolution of Holliday junctions in vitro requires the Escherichia coli ruvC gene product. Proc. Natl Acad. Sci. USA. 1991;88:6063–6067. doi: 10.1073/pnas.88.14.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleff S, Kemper B, Sternglanz R. Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J. 1992;11:699–704. doi: 10.1002/j.1460-2075.1992.tb05102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White MF, Lilley DM. Characterization of a Holliday junction-resolving enzyme from Schizosaccharomyces pombe. Mol. Cell. Biol. 1997;17:6465–6471. doi: 10.1128/mcb.17.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higuchi Y, Linn S. Purification of all forms of HeLa cell mitochondrial DNA and assessment of damage to it caused by hydrogen peroxide treatment of mitochondria or cells. J. Biol. Chem. 1995;270:7950–7956. doi: 10.1074/jbc.270.14.7950. [DOI] [PubMed] [Google Scholar]
- 18.Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 19.Wanrooij S, Fuste JM, Farge G, Shi Y, Gustafsson CM, Falkenberg M. Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc. Natl Acad. Sci. USA. 2008;105:11122–11127. doi: 10.1073/pnas.0805399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ponting CP. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 2002;30:3643–3652. doi: 10.1093/nar/gkf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aravind L, Makarova KS, Koonin EV. SURVEY AND SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson SJ, Close D, Robinson H, Vallet-Gely I, Dove SL, Hill CP. Crystal structure and RNA binding of the Tex protein from Pseudomonas aeruginosa. J. Mol. Biol. 2008;377:1460–1473. doi: 10.1016/j.jmb.2008.01.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu D, Wang YS, Wyss DF. Solution structure of the hypothetical protein YqgF from Escherichia coli reveals an RNAse H fold. J. Biomol. NMR. 2003;27:389–392. doi: 10.1023/a:1025840121177. [DOI] [PubMed] [Google Scholar]
- 24.Metodiev MD, Lesko N, Park CB, Camara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson NG. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Tiranti V, Savoia A, Forti F, D’Apolito MF, Centra M, Rocchi M, Zeviani M. Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the Expressed Sequence Tags database. Hum. Mol. Genet. 1997;6:615–625. doi: 10.1093/hmg/6.4.615. [DOI] [PubMed] [Google Scholar]
- 26.Koc EC, Spremulli LL. RNA-binding proteins of mammalian mitochondria. Mitochondrion. 2003;2:277–291. doi: 10.1016/S1567-7249(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Bogenhagen DF. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- 28.Gelfand R, Attardi G. Synthesis and turnover of mitochondrial ribonucleic acid in HeLa cells: the mature ribosomal and messenger ribonucleic acid species are metabolically unstable. Mol. Cell. Biol. 1981;1:497–511. doi: 10.1128/mcb.1.6.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iborra FJ, Kimura H, Cook PR. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004;2:9. doi: 10.1186/1741-7007-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaspari M, Falkenberg M, Larsson NG, Gustafsson CM. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. EMBO J. 2004;23:4606–4614. doi: 10.1038/sj.emboj.7600465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 2007;21:160–174. doi: 10.1101/gad.1503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Endoh M, Zhu W, Hasegawa J, Watanabe H, Kim DK, Aida M, Inukai N, Narita T, Yamada T, Furuya A, et al. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol. Cell. Biol. 2004;24:3324–3336. doi: 10.1128/MCB.24.8.3324-3336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies SM, Rackham O, Shearwood AM, Hamilton KL, Narsai R, Whelan J, Filipovska A. Pentatricopeptide repeat domain protein 3 associates with the mitochondrial small ribosomal subunit and regulates translation. FEBS Lett. 2009;583:1853–1858. doi: 10.1016/j.febslet.2009.04.048. [DOI] [PubMed] [Google Scholar]
- 34.Delannoy E, Stanley WA, Bond CS, Small ID. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochem. Soc. Trans. 2007;35:1643–1647. doi: 10.1042/BST0351643. [DOI] [PubMed] [Google Scholar]
- 35.Rorbach J, Richter R, Wessels HJ, Wydro M, Pekalski M, Farhoud M, Kuhl I, Gaisne M, Bonnefoy N, Smeitink JA, et al. The human mitochondrial ribosome recycling factor is essential for cell viability. Nucleic Acids Res. 2008;36:5787–5799. doi: 10.1093/nar/gkn576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol. Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 37.Krause K, Lopes de Souza R, Roberts DG, Dieckmann CL. The mitochondrial message-specific mRNA protectors Cbp1 and Pet309 are associated in a high-molecular weight complex. Mol. Biol. Cell. 2004;15:2674–2683. doi: 10.1091/mbc.E04-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markov DA, Savkina M, Anikin M, Del Campo M, Ecker K, Lambowitz AM, De Gnore JP, McAllister WT. Identification of proteins associated with the yeast mitochondrial RNA polymerase by tandem affinity purification. Yeast. 2009;26:423–440. doi: 10.1002/yea.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodeheffer MS, Shadel GS. Multiple interactions involving the amino-terminal domain of yeast mtRNA polymerase determine the efficiency of mitochondrial protein synthesis. J. Biol. Chem. 2003;278:18695–18701. doi: 10.1074/jbc.M301399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 41.Broadley SA, Hartl FU. Mitochondrial stress signaling: a pathway unfolds. Trends Cell Biol. 2008;18:1–4. doi: 10.1016/j.tcb.2007.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.