Abstract
The dynamics of histone methylation have emerged as an important issue since the identification of histone demethylases. We studied the regulatory function of Rph1/KDM4 (lysine demethylase), a histone H3K36 demethylase, on transcription in Saccharomyces cerevisiae. Overexpression of Rph1 reduced the expression of PHR1 and increased UV sensitivity. The catalytically deficient mutant (H235A) of Rph1 diminished the repressive transcriptional effect on PHR1 expression, which indicates that histone demethylase activity contributes to transcriptional repression. Chromatin immunoprecipitation analysis demonstrated that Rph1 was associated at the upstream repression sequence of PHR1 through zinc-finger domains and was dissociated after UV irradiation. Notably, overexpression of Rph1 and H3K36A mutant reduced histone acetylation at the URS, which implies a crosstalk between histone demethylation and acetylation at the PHR1 promoter. In addition, the crucial checkpoint protein Rad53 acted as an upstream regulator of Rph1 and dominated the phosphorylation of Rph1 that was required for efficient PHR1 expression and the dissociation of Rph1. The release of Rph1 from chromatin also required the phosphorylation at S652. Our study demonstrates that the histone demethylase Rph1 is associated with a specific chromatin locus and modulates histone modifications to repress a DNA damage responsive gene under control of damage checkpoint signaling.
INTRODUCTION
The eukaryotic genome is assembled with histones into a highly ordered chromatin structure. The basic unit of chromatin is the nucleosome, which consists of 146 bp of DNA wrapped around the histone octamer (1). Histones are subjected to various post-translational modifications (PTM), including phosphorylation, acetylation, ubiquitination and methylation (2,3). These covalent modifications alter chromatin dynamics to regulate DNA processes (4,5).
For a long time, histone lysine methylation was considered to be irreversible until the identification of the first histone demethylase, lysine-specific demethylase 1 (LSD1) (6). However, the LSD1/KDM1 (lysine demethylase) type of histone demethylase can only demethylate mono- and di-methylated lysine because of its catalytic characteristics. A distinct family of histone demethylases containing Jumonji C (JmjC) domains (JHDMs) is highly conserved from budding yeast to humans. The catalytic JmjC domain is required for oxidative demethylation; it requires Fe (II) and α-ketoglutarate as cofactors and can demethylate mono-, di- and tri-methylated substrates (7,8).
Dozens of JHDMs have been discovered to have various functions in regulation of gene expression, cell growth and development (9,10). The role of JHDMs in transcriptional regulation has been reported in mammals. Mammalian JHDM1 and JHDM2 subfamilies reverse mono- and di-methylated H3K36 and H3K9, respectively, and the JHDM3 subfamily preferentially antagonizes di- and tri-methylation of H3K36 and H3K9 (8,11,12). JHDM2A acts on mono-/di-methyl H3K9 and plays important roles in nuclear hormone receptor-mediated gene activation, male germ-cell development, obesity control and metabolic gene expression (12–14). JHDM3A, the tri-methyl-specific demethylase for H3K36 and H3K9, negatively regulates ASCL2 transcription (11).
Previously, we characterized four JmjC-containing proteins (Rph1, Jhd1, Gis1 and Jhd2) in the budding yeast Saccharomyces cerevisiae and demonstrated that Rph1, Jhd1 and Gis1 are specific to H3K36, whereas Jhd2 is H3K4 specific (15). Notably, Rph1 is the only demethylase targeting H3K36 tri-methylation; Jhd1 and Gis1 specifically demethylate mono-/di-methylated H3K36. In general, histone methylation on H3K36 is thought to be involved in transcriptional elongation (16). In S. cerevisiae, H3K36 is methylated by the histone methyltransferase Set2 (17). Set2 interacts with the phosphorylated C-terminal domain of RNA polymerase II to methylate H3K36 during transcriptional elongation within the body of actively transcribed genes (18). This methylation is recognized by Eaf3, a subunit of the small Rpd3 (Rpd3S) histone deacetylase complex (HDAC). The recruitment of HDAC further removes the acetyl group from hyper-acetylated histones of transcribed genes to re-build a compact chromatin structure. During transcriptional elongation, H3K36 methylation prevents aberrant intragenic transcription (19). Recently, two JmjC-domain-containing proteins, Rph1 and Jhd1, were shown to bypass the lethality of deletion of BUR1, a cyclin-dependent kinase of RNA polymerase II. Therefore, the demethylase activity toward H3K36 methylation could be related to transcriptional elongation (20). In addition, H3K36 methylation was reported to be involved in regulating the transcriptional initiation at the MET16 promoter region (21). Even though H3K36 methylation is known to be involved in the regulation of transcription, the detailed functions of reversible H3K36 demethylation remain unclear.
The H3K36 demethylase Rph1, also known as KDM4 (22), was originally defined as a repressor of the PHR1 gene (23). PHR1 encodes the apoenzyme for the DNA repair enzyme photolyase, which catalyzes the repair of pyrimidine dimers in the presence of visible light (24–27). Previous in vitro footprinting studies showed that Rph1 represses the expression of PHR1 by associating with the upstream repression sequence (URS) of the PHR1 promoter (23). Although Rph1 is now known to be a histone demethylase, whether its H3K36 demethylase activity plays a role in the transcriptional regulation of PHR1 remains to be elucidated.
Here, we investigated the role of Rph1 in repressing the transcription of PHR1 in a histone demethylase-dependent manner. We revealed Rph1 associated with the URS region of the PHR1 promoter via its zinc-finger (ZF) domains and was dissociated after UV irradiation. Rph1-mediated histone demethylation influenced the dynamic crosstalk between histone methylation and acetylation with Rpd3 at the URS region of PHR1 promoter. Furthermore, we revealed that Rad53 functions as an upstream activator of PHR1 by phosphorylation of Rph1 in a Rad53 kinase-dependent manner. Phosphorylation at S652 of Rph1 potentially contributes to its dissociation from chromatin and modulates the transcriptional de-repression of PHR1 in response to DNA damage. Our study demonstrates that the H3K36 demethylase Rph1 regulates PHR1 expression by association with the promoter and by altering chromatin modifications under the control of DNA damage checkpoint signaling.
MATERIALS AND METHODS
Plasmids and yeast strains
All recombinant plasmids were constructed by use of the Gateway system [(28), Invitrogen]. The coding region of RPH1 was synthesized by PCR and recombined to the pDONR2.1 to generate BP-RPH1. The mutants of Rph1, including the catalytic-deficient mutant H235A (15), phosphorylation sites and ZF deletion mutants were generated by site-directed mutagenesis and verified by DNA sequencing. For inducible expression of Rph1, BP-RPH1 was cloned into the BG1805 vector (Open Biosystems) by the Gateway system (29). For constitutive expression of Rph1 in yeast, we first generated a set of yeast destination vectors by modification of pRS vectors (pRS415 and pRS425) with a GPD1 promoter or a RPH1 promoter (800 bp upstream of ATG), a CYC1 terminator and the chloramphenicol/ccdB cassette for recombinational cloning. The pET21 vector (Merck) was used for expression of recombinant proteins in the bacterial system. All recombinant plasmids identified from individual Escherichia coli colonies were verified by sequencing. The yeast strains used in this study are described in Supplementary Table S1. Saccharomyces cerevisiae BY4742 and BY4743 were used as the wild-type (WT) yeast strain. Homozygous knockout strains of RPH1 in the BY4742 or BY4743 background were obtained from the Saccharomyces Genome Deletion Project (30).
UV irradiation treatment
Yeast cultures were grown in synthetic-complete selection medium until the A600 reached 0.6–0.8, then transferred to YP medium containing 2% galactose at 30°C to induce gene expression. After 4 h induction, cell pellets were collected and spread on YP galactose (YPGal) plates. The plates were UV irradiated (Stratalinker, Stratagene) with the indicated doses (0–30 mJ/cm2). After 30 min of recovery, the cells were harvested for further analysis. The procedure for UV-sensitivity testing was as described (15). The induced cells were spotted on the indicated plates by serial dilutions. After UV irradiation, plates were grown for 2–3 days at 30°C before data collection.
Reverse transcription
Total RNA was extracted from yeast cells by use of a total RNA mini kit (Geneaid). RNA (1 μg) was treated with DNAse I (Promega) followed by reverse transcription with use of Moloney Murine Leukemia Virus High Performance Reverse Transcriptase (MMLV HP RT) (Epicentre) and oligo-dT primer (Protech) according to the manufacturer’s instructions. Reverse transcription of RNA samples from each biological experiment was performed and the resulting cDNA was used for PCR analysis.
Real-time quantitative PCR
Real-time quantitative PCR (qPCR) with SYBR green detection was performed as described (31) by use of an ABI Prism 7000 thermocycler with fluorescence detection (Applied Biosystems). All primers used are indicated in Supplementary Table S2. Appropriate non-template controls were included in each PCR reaction, and dissociation analysis was performed at the end of each run to confirm the specificity of the reaction.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) was performed as described (32,33). To maintain proper size of fragmented chromatin, we optimized the sonication and sheared DNA with an average size of 100–200 bp (Supplementary Figure S1) corresponding to 1 or 2 nucleosomes. For Rpd3 ChIP, samples were fixed with dimethyl adipimidate (Sigma) and formaldehyde (34). of chromatin solution (0.75–1 mg) was immunoprecipitated (IP) with indicated antibodies [HA-tag/Myc-tag (Roche), H3K36me3 (Abcam), acetylated histone H3(K9/14)/H4(K5/8/12/16) (Upstate) and Rpb3 (Neoclone)] and purified with protein A or G sepharose (Upstate). The precipitated DNA was analyzed by semi-quantitative PCR or real-time qPCR. For semi-quantification, PCR products were separated by electrophoresis and followed by quantification (Image Quant software, GE). Real-time qPCR analysis was performed as described above. For each ChIP, the signal for each gene primer pair in the immunoprecipitation was normalized to that of the input and then divided by the control vector to determine the fold change. Quantification of data was based on the number of independent biological and experimental replicates indicated in each figure.
In vitro kinase assay
Rad53 kinase assay was performed as described (35,36). Briefly, yeast carrying WT or kinase-dead Rad53 fused with V5 and 6 copies of His tags was treated with 0.05% MMS for 1 h, and total proteins were extracted for immunoprecipitation with anti-V5 antibody. Protein G-bound Rad53 was incubated with 1 μg recombinant Rph1 or BSA (New England Biolabs) and 5 μCi of γ32P-ATP (3000 Ci/mmol, PerkinElmer) in 20 μl kinase buffer (40 mM Hepes-NaOH pH 8.0, 1 mM DTT, 20 mM MgCl2, 20 mM MnCl2, 100 μM sodium orthovanadate and 0.02 mM ATP) at 30°C for 30 min. The reaction was stopped by addition of SDS-loading buffer and followed by SDS–PAGE electrophoresis. The autoradiography signal was captured and quantitated by phospho-imager (Typhoon 9200 Scanner, GE Healthcare).
Statistical analysis
Comparison of 2 groups involved Student’s t-test. A P < 0.05 was considered statistically significant. A regression procedure was used to explore the correlation between the experimental condition and PHR1 expression and to identify potentially important predictors. Statistical analyses involved use of Microsoft Excel and SPSS (SPSS Inc., Chicago, IL, USA) (37).
RESULTS
The H3K36 demethylase Rph1 regulates PHR1 transcription
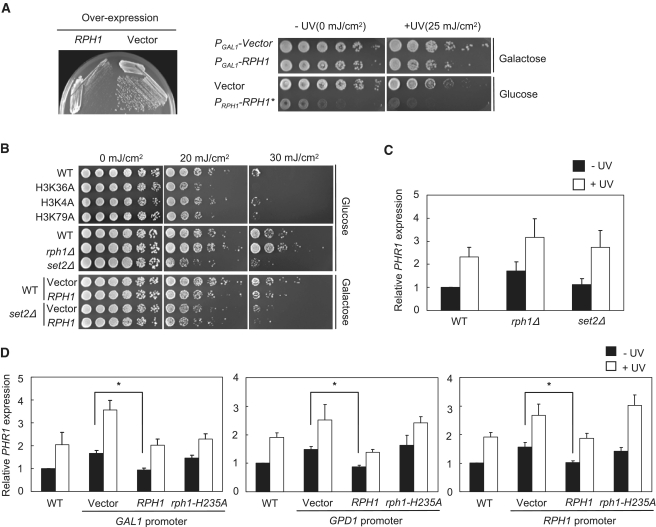
To characterize the biological function of histone demethylases, including Rph1, Jhd1 and Jhd2, we first performed phenotypic analyses and observed that deletion of any histone demethylase caused no overt growth phenotype (15), which may be due to overlapping activities of the multiple histone lysine demethylases in yeast. In an independent approach, we used overexpression to probe the function of individual histone demethylases. Of these, only Rph1 overexpression showed elevated sensitivity to UV-induced damage (15). However, the constitutive overexpression of Rph1 with its own promoter in a multi-copy (2μ) plasmid caused a severe defect in cell growth (Figure 1A, left panel and 2 lower rows in right panel). Thus, we used an inducible expression strategy to study the biological phenotype resulting from temporally increased Rph1 levels. Cells with galactose-inducible overexpression of RPH1 grew well without UV treatment yet displayed hypersensitivity to UV irradiation at 25 mj/cm2 (Figure 1A, 2 upper rows in right panel). Because previous experiments suggested that Rph1 is a histone demethylase specific to tri-methylated-H3K36 (15,38), we surmised that the demethylation at H3K36 may be linked to UV sensitivity. To test the possibility, we used alanine-substituted mutants blocking methylation at histone H3 K4, K36 and K79 in UV-sensitivity assays. Interestingly, only histone H3K36A and H3K79A mutants showed increased UV sensitivity (Figure 1B, upper panel). Dot1-mediated H3K79 methylation is linked with H2B ubiquitination and is involved in the DNA damage response (39). However, H3K36 methylation in the UV damage response has not been well-established. Furthermore, we found that the deletion of the H3K36 methyl-transferase Set2 (set2Δ) also enhanced the UV sensitivity (Figure 1B, middle panel). Moreover, overexpression of Rph1 in the WT conferred high sensitivity to UV irradiation, whereas overexpression of Rph1 combined with set2Δ caused an additive effect (Figure 1B, lower panel), which was more significant at a higher dosage. These observations suggest that Set2 and Rph1 likely work in parallel pathways to control UV sensitivity.
Figure 1.
Histone H3K36 demethylase Rph1 regulates the transcription of PHR1. (A) Left: The rph1Δ strains carrying constitutively overexpressed RPH1 or control 2-µ vector were streaked on selective plate. Right: The UV-sensitivity test was performed with indicated strains. Cells were spotted on plates containing galactose or glucose with 5-fold serial dilution and subjected to UV irradiation. Asterisks denotes the constitutive expression of RPH1 with its own promoter in 2-µ vector. (B) UV sensitivity was tested with indicated strains. Yeast strains (WT, histone mutants, rph1Δ and set2Δ) grown in glucose in log-phase or induced with galactose for 4 h were spotted on selective plates. (C) PHR1 expression in WT, rph1Δ and set2Δ strains. The cells were cultured to early log phase and subjected to 20 mj/cm2 UV irradiation. RT–qPCR was performed and transcription of PHR1 of each strain was normalized to ACT1. Error bars indicate the SD from three biological repeats. (D) PHR1 level in WT or rph1Δ yeast containing control vector (Vector), WT RPH1 or H235A-mutated RPH1 under different promoters before or after 20 mJ/cm2 UV irradiation (right). Error bars are the SD of five biological replicates. *P < 0.05.
Because Rph1 was originally identified as a repressor of the DNA repair gene PHR1, we sought to elucidate whether the demethylase activity is linked to the transcriptional regulation of PHR1. RT-qPCR was used to measure the levels of PHR1 expression in the WT and rph1-deletion (rph1Δ) and set2-deletion (set2Δ) strains treated with UV irradiation. Deletion of rph1 led to approximately 2-fold enhancement of PHR1 (Figure 1C) under normal conditions (−UV). These results agree with a previous report that Rph1 is a repressor of PHR1 expression (23). Surprisingly, lack of Set2 did not interfere with the expression of PHR1 (Figure 1C), which suggests that Set2 methyl-transferase plays a minor role in PHR1 expression. Although set2Δ increased the UV sensitivity (Figure 1B, middle panel), our results suggest that Set2 may regulate factors other than PHR1 that are involved in UV response such as the DNA repair gene RNR3 (40,41).
To investigate the role of Rph1 in PHR1 expression, we used three different promoters, representing an inducible (GAL1), a constitutive (CEN-GPD1), or a native promoter (CEN-RPH1), to express RPH1 in the rph1Δ background. In addition to WT RPH1, a catalytic-deficient rph1 (rph1-H235A) was expressed by the aforementioned promoters to determine the involvement of demethylase activity (Figure 1D). In the absence of UV treatment (−UV), the expression of RPH1 by any of the three promoters was sufficient to suppress PHR1 expression, which is higher in rph1Δ (vector) than in the WT (Figure 1D). In contrast, PHR1 expression in the rph1-H235A mutant did not significantly differ from that in rph1Δ (Figure 1D), which indicates that the histone H3K36 demethylase activity of Rph1 is required to suppress PHR1 expression.
In response to UV irradiation, PHR1 expression was induced ranging from 2.5- to 3.4-fold in rph1Δ (Figure 1D, ‘Vector+UV’) as compared to that in WT (Figure 1D, ‘WT−UV’, defined as 1), which indicates that Rph1 represses PHR1 expression in the absence of DNA damage. However, the levels of PHR1 expression with UV irradiation were induced in both RPH1 and rph1-H235A under control of the three promoters (Figure 1D), which suggests that histone H3K36 demethylation-independent pathways are involved in the transcriptional activation of PHR1 responding to UV damage signal. Taken together, these data suggest that the demethylase activity of Rph1 plays an important role in the repression of PHR1 expression, rather than the UV-inducible transcriptional activation.
Rph1 binds to the URS of PHR1 through ZF domains and modulates chromatin modifications in specific regions of the PHR1 promoter
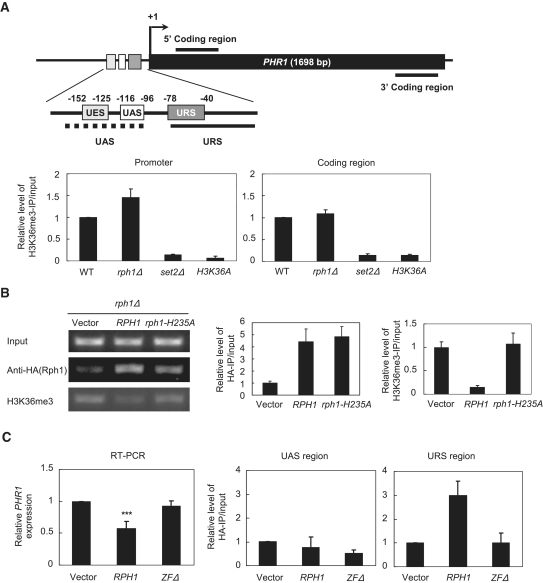
We have previously demonstrated that Rph1 plays a major role in transcriptional repression of PHR1 (Figure 1). We next used ChIP assays to evaluate whether the Rph1-mediated repression of PHR1 affects the chromatin structure. The primary protein-coding gene structure of PHR1 is illustrated in Figure 2A (upper panel). We first determined whether deletion of RPH1 changed the H3K36 tri-methylation at the PHR1 gene region (Figure 2A, lower panel). To confirm the specificity of the H3K36me3 signal, we used set2Δ and H3K36A mutants for H3K36me3-ChIP and found extremely low levels of H3K36me3 in set2Δ and H3K36A mutants as compared with that in WT and rph1Δ. Interestingly, rph1Δ showed an increased level at the promoter of PHR1 but not the 3′ coding region of PHR1, suggesting that Rph1-mediated demethylation participates in the regulation of PHR1 promoter activity. We next investigated whether Rph1 is physically associated with chromatin in vivo. To determine the temporal regulation of PHR1 expression by Rph1, we used the inducible GAL1 promoter to analyze the immediate effect of overexpressed Rph1 on the transcriptional regulation of PHR1. ChIP assays were performed to detect the relative abundance of HA-tagged Rph1 and H3K36me3 at the PHR1 promoter region containing a 300 bp 5′-upstream sequence (Figure 2B). In agreement with in vitro electrophoretic mobility shift assay (EMSA) and footprinting analyses, which demonstrated that Rph1 binds to a specific PHR1 promoter sequence (23), our ChIP analysis also revealed that Rph1 binding was enriched at the PHR1 promoter by at least 4-folds in both the WT (RPH1) and mutant rph1-H235A as when compared with that ine rph1Δ mutant (vector alone) (Figure 2B, HA-IP). However, only the WT Rph1 but not the rph1-H235A reduced H3K36 tri-methylation (Figure 2B, H3K36me3-IP), which indicates that the enzymatic activity of Rph1 is required for H3K36 demethylation at the promoter of PHR1. This result was not merely due to the induced overexpression of Rph1 by the GAL1 promoter because we also found similar results by using the GPD1 promoter to drive a constitute expression of Rph1 in a low-copy (CEN) plasmid (Supplementary Figure S2). Therefore, Rph1 is associated with the promoter of PHR1 resulting in a decreased H3K36 methylation to influence transcriptional repression.
Figure 2.
Rph1 binds to the upstream repression sequence (URS) of PHR1 through ZF domains. (A) Top panel: The schematic representation of primers specific to different regions on PHR1 for PCR. +1 indicates the transcription start site of PHR1. The primer sequences are in Supplementary Table S2. Lower panel: The specificity of H3K36me3 at the PHR1 promoter and coding region were detected by ChIP in WT, rph1Δ, set2Δ and H3K36A mutants. Bar graph represents the quantified results from three biological repeats. (B) ChIP with anti-HA and anti-H3K36me3 antibodies were performed with the indicated strains. The right panels show the fold change relative to the control (vector), which was normalized by input. (C) The ZF domains are required for transcriptional repression on PHR1 and for specific association with URSPHR1. Left panel: PHR1 expression in rph1Δ (vector), induced WT RPH1 or ZF-deleted RPH1 (ZFΔ). ***P < 0.001 compared with vector. Right panel: ChIP with anti-HA antibody at the UAS or URS regions. Data are from three different biological samples.
Because histone H3K36 demethylation is involved in transcriptional repression at the PHR1 promoter, we next identified the specific region of the PHR1 promoter associated with Rph1 and the specific domain of Rph1 required for chromatin association. Three putative cis-elements of PHR1 promoter were characterized previously by β-galactosidase assays: an upstream activation sequence (UASPHR1), a novel essential sequence (UESPHR1) and an upstream repression sequence (URSPHR1) (25). To define the Rph1-controlled transcriptional events, we used a series of ChIP experiments with primers to amplify specific regions of the PHR1 promoter (URSPHR1, 78∼−40; and UAS + UESPHR1, 152∼−96). Results from ChIP assays with anti-HA (Rph1) uncovered that Rph1 was specifically associated with the URS (URSPHR1) but not the UAS (UES + UASPHR1) region (Figure 2C).
A C-terminal domain of Rph1 is required for DNA binding in vitro (23). We attempted to dissect the essential motif of Rph1 required for the DNA binding affinity in vivo. Domain analysis by SMART (http://smart.embl-heidelberg.de/) predicted that Rph1 contains two ZFs at the C-terminus that may contribute to DNA binding. We introduced a ZF-domain deleted Rph1 construct (ZFΔ) into the rph1Δ strain and found a higher expression level of PHR1 than that in WT (RPH1) (Figure 2C, left panel). ChIP assays with anti-HA (Rph1) further demonstrated that ZF deletion reduced the binding of Rph1 with the URSPHR1 region, which was negatively correlated to the expression level of PHR1 (Figure 2C, right panel). Therefore, the ZF domains of Rph1 directed its DNA binding and transcriptional repression in vivo.
Crosstalk between H3K36 tri-methylation and H3 acetylation occurs at the PHR1 promoter
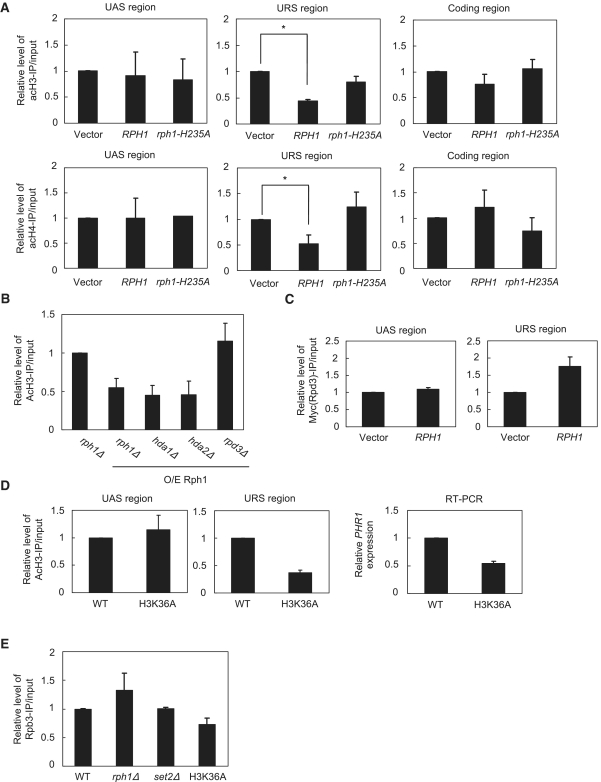
The synergistic crosstalk of histone modifications has been suggested as an important regulatory mechanism in gene expression (33,42,43). We hypothesized that gene-specific synergistic histone modifications also take place on the PHR1 promoter by interacting with DNA-bound transcription factors. Because Rph1 bound to the URSPHR1 but not UES + UASPHR1 region (Figure 2C) and consequently decreased H3K36 methylation in the absence of UV irradiation, we examined whether histone H3K36 demethylation affected histone acetylations. Results from ChIP assays with anti-acetylated histone H3K9/14 (acH3) and H4K5/8/12/16 (acH4) revealed that Rph1 altered histone acetylation in different regions (Figure 3A). The levels of histone H3/H4 acetylations were reduced by about 50% only at the URSPHR1 but not the UASPHR1 or coding region (Figure 3A). Thus, the association of WT Rph1 at URSPHR1 reduced the histone acetylations in this region. Furthermore, the catalytic-deficient rph1-H235A mutant displayed patterns similar to those of the rph1Δ (vector) in histone acetylations, which indicates that Rph1 demethylase activity is critical for the crosstalk of histone modifications at the PHR1 promoter. The reduced histone acetylation implies an involvement of histone deacetylase(s). To examine this possibility, we performed acH3-ChIP using various histone deacetylase (HDAC)-deleted strains, including HDA1, HDA2 (Type II) and RPD3 (Type I), in the presence of overexpressed RPH1. Lack of RPD3 restored the H3 acetylation level at the URSPHR1 in the presence of Rph1 (Figure 3B). Moreover, results from ChIP experiments revealed the significant association of Rpd3 at the URSPHR1 but not the UASPHR1 region in the presence of Rph1 (Figure 3C), which indicates that Rpd3 specifically deacetylated histones at the URSPHR1 region responsible for the transcriptional repression of PHR1.
Figure 3.
Demethylation at H3K36 coexists with a reduction of histone acetylations specifically at the URS of PHR1. (A) Histone H3/H4 acetylation was altered at the URS and coding region of PHR1. ChIP-qPCR from the indicated strains was performed with anti-AcH3 and anti-AcH4. *P < 0.05. Data are from three biological repeats. (B) RPD3 deletion restored the reduction of H3 acetylation at the URS. Indicated HDAC deletion strains with overexpressed Rph1 were harvested for acH3-ChIP. (C) Rpd3-Myc is associated at URS region in the presence of Rph1. The rph1Δ cells containing control vector or overexpressed Rph1 with Myc-tagged Rpd3 were harvested for Myc-ChIP and qPCR. Error bars represents the SD from two biological repeats. (D) Left: H3K36A showed the reduction of H3 acetylation at the URS of PHR1. Cells carrying WT or H3K36 mutated (H3K36A) histones were subject to acH3-ChIP followed by qPCR. Right: PHR1 expression in WT or H3K36A mutant. (E) ChIP with anti-Rpb3 at URS region of PHR1 in WT, rph1Δ, set2Δ and H3K36A mutants. Error bars shows the SD from two biological samples.
To confirm the crosstalk between histone methylation and acetylation, we performed acH3-ChIP experiments in WT yeast and the H3K36A mutant. Reduced acetylation was revealed exclusively at the URSPHR1 region in the H3K36A mutant (Figure 3D, left panel); therefore, Rph1 suppresses PHR1 expression by modulating the chromatin structure in a demethylase-dependent manner. Our current findings support that the Rph1-mediated H3K36 demethylation and crosstalk with histone acetylation also take place at the PHR1 promoter to regulate gene transcription. To determine whether the promoter association of Rph1 and the crosstalk of histone modifications are specific to the PHR1 gene, we performed ChIP assay with anti-acetylated histone H3 (acH3) and Rph1-HA at the ADH1 promoter. However, we did not find a significant difference in H3 acetylation or Rph1 association on the ADH1 promoter and coding region (Supplementary Figure S3).
The recruitment of RNA polymerase II (Pol II) to promoters to initiate transcription has been the central dogma in regulation of active gene expression. However, recent studies from genome-wide analysis revealed that Pol II is associated with the promoters of many non-actively transcribed genes in murine embryonic stem cells and Drosophila (44,45). To determine the preoccupancy of Pol II at the repressed PHR1 promoter, we performed ChIP experiments with Rpb3 (a subunit of RNA Pol II core complex). Binding of Pol II at the URSPHR1 region was reduced in the H3K36A mutant but remained unchanged in the set2Δ mutant as compared with the WT (Figure 3E). The binding was only slightly increased in the rph1Δ mutant. Thus, Rph1 and modification of H3K36 but not Set2 regulate the pre-occupancy of Pol II at the PHR1 promoter. Rph1 likely specifically binds and modulates histone methylation and acetylation and consequently influences Pol II recruitment at the URSPHR1 region.
Rph1 is dissociated from the PHR1 promoter in response to DNA damage
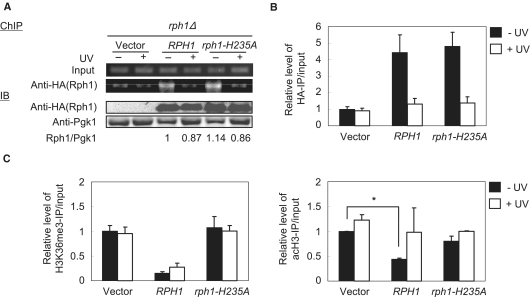
In response to DNA damage, transcriptional induction of PHR1 should require a priori de-repression. Previously, Rph1 was named photolyase regulatory protein (PRP), which bound to the PHR1 URS and regulated the induction of PHR1 transcription after DNA damage (24). We performed ChIP assays to study the dynamics of Rph1 in response to DNA damage in vivo. After UV irradiation, the association of both Rph1 and the rph1-H235A mutant was significantly decreased, which suggests that Rph1 was released from the promoter of PHR1 after DNA damage (Figure 4A). To determine whether the protein level of Rph1 was affected by DNA damage, Rph1-HA expression was analyzed by immunoblotting (IB; Figure 4A). Rph1 levels were only slightly decreased after UV irradiation, which suggests that the dissociation from the promoter cannot simply be attributed to protein expression levels of Rph1 and rph1-H235A. The quantitative-ChIP results also confirmed the association and dissociation of Rph1 at the PHR1 promoter in vivo (Figure 4B).
Figure 4.
Rph1 is dissociated from PHR1 promoter in response to UV irradiation. (A) The indicated strains were irradiated (UV: 20 mJ/cm2) and harvested for ChIP with anti-HA antibody. IB with anti-HA antibody showed the expression of Rph1. Anti-Pgk1 was the loading control and the ratio of Rph1/Pgk1 is indicated. (B) The quantitative result is shown from comparable samples in (A). (C) ChIP assay from samples in (A) with anti-H3K36me3 and anti-acH3 antibodies was followed by qPCR to monitor the URS region signals. Data are from three biological repeats. *P < 0.05.
We further examined the synergistic crosstalk of chromatin modifications in the repressed PHR1 promoter in response to UV irradiation. We monitored histone acetylation and methylation levels at the URSPHR1 before and after UV irradiation by ChIP with antibodies against H3K36me3 and acH3. The level of H3K36me3 was elevated slightly in the presence of RPH1 but not in rph1Δ (vector) or in the rph1-H235A mutant by UV treatment (Figure 4C, left panel). However, in the presence of UV irradiation (+UV), the levels of histone H3 acetylation were comparable among RPH1, rph1-H235A and rph1Δ (vector) at the URSPHR1 region (Figure 4C, right panel). These results suggest that dissociation of Rph1 responding to UV irradiation results in an elevated level of histone acetylation. Furthermore, our observation demonstrated that dynamic interaction of Rph1 on the promoter of PHR1 plays a role in H3K36 demethylation and histone acetylation to regulate PHR1 expression in response to DNA damage.
Methylation at H3K36 mediated by Set2 plays a major role in transcriptional elongation (18,46,47), which was suggested to involve the H3K36 demethylase Rph1 (15,20). To investigate the role of H3K36 demethylation in transcriptional elongation, we used ChIP of the transcribed region with primers specific to 5′ (11–349) and 3′ (907–1434) coding regions of PHR1. Surprisingly, in the coding regions, the relative HA-IP signals were similar to those for rph1Δ (vector), which indicates no enhancement of Rph1 binding, yet H3K36 methylation was still decreased (Supplementary Figure S4). It is possible that the histone H3K36 methylation is transient or too weak to be detected by ChIP in the coding region. Our observations are similar to results of study of ADH1, PMA1 and YEF3 genes (20). However, the detailed mechanism for the demethylation in the coding regions remains to be established.
Rad53 regulates the expression of PHR1 and dissociation of Rph1 in response to DNA damage
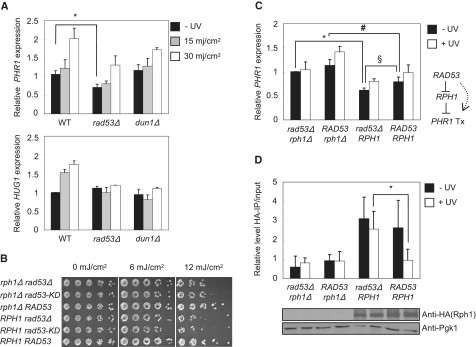
It has been suggested that Rph1 phosphorylation is under the control of the Mec1-Rad53 damage checkpoint pathway but distinct from the Dun1-Crt1 pathway (48). In budding yeast, Rad53, the ortholog of mammalian Chk2, is a crucial checkpoint protein, and Dun1 is considered the main downstream kinase of Rad53 responding to DNA damage (49,50). However, the connection between the expression of PHR1, a DNA repair gene and Rad53 is still unknown. To examine whether Rad53 plays a regulatory role in PHR1 expression in DNA damage signaling, we analyzed the PHR1 transcription level in a rad53Δ strain. Deletion of RAD53 is lethal. Therefore, we evaluated the role of RAD53 by using a rad53Δ sml1Δ strain, which loss of SML1 suppresses the lethality of RAD53 deletion (51). By using RT–qPCR analysis, we found the expression level of PHR1 in the absence of UV irradiation decreased by 30% in the rad53Δ mutant but not in the dun1Δ mutant (Figure 5A). With UV irradiation (30 mj/cm2), the induction of PHR1 was reduced by 35% in the rad53Δ mutant but only slightly in the dun1Δ strain. The Rad53-Dun1-regulated gene HUG1 was not induced in either the rad53Δ or dun1Δ mutant (Figure 5A, right panel). Therefore, our data demonstrate that an efficient PHR1 expression requires RAD53 but is less dependent on DUN1.
Figure 5.
Rph1 regulates transcription of PHR1 mediated by Rad53 in response to DNA damage. (A) The expression of PHR1 and HUG1 in sml1Δ (WT), sml1Δ rad53Δ (rad53Δ) and sml1Δ dun1Δ (dun1Δ) mutants responding to different doses of UV irradiation. HUG1 was used as an indicator of intact DNA damage signaling. *P < 0.05. (B) UV-sensitivity test of indicated strains in the rph1Δ rad53Δ background with different combinations of control vector, overexpressed RAD53 or kinase-dead (KD) and WT RPH1. (C) PHR1 expression of strains as in (B). Cells harvested from early-log phase underwent 30 mJ/cm2 irradiation. *, #, §P < 0.05. (D) ChIP with anti-Rph1 (HA) at URS from comparable samples as in Figure 4C. Bar graphs show qPCR results in URS of PHR1 promoter. IB indicated the protein expression of Rph1, and Pgk1 is a loading control. All RT-qPCR data are at least from three different biological samples. Results of ChIP are from 2 biological samples. *P < 0.05.
To further address the interplay between Rad53 and Rph1 for PHR1 expression, we performed genetic epistasis analysis. We generated the triple deletion strain (rad53Δ rph1Δ sml1Δ) as the genetic background for overexpression of Rad53, Rph1 or both in yeast. UV sensitivity assays and RT–qPCR analysis were used to evaluate the functional relationship between Rad53 and Rph1. In the absence of Rph1, overexpression of the WT Rad53, but neither kinase-dead (rad53-KD) nor control vector (rad53Δ), restored the growth of rad53Δ in response to UV irradiation (Figure 5B). Furthermore, overexpression of WT Rph1 enhanced the UV sensitivity, whereas expression of WT Rad53, but not kinase-dead (rad53-KD) or control vector (rad53Δ), compensated the defective growth phenotype (Figure 5B). Therefore, Rad53 may be involved in the regulation of PHR1 expression in response to UV irradiation, possibly through modulating Rph1 function. To understand the effect of Rad53 on Rph1 in regulating gene transcription, we analyzed the expression of PHR1 in the absence or presence of Rad53 and Rph1 (Figure 5C). The basal transcription of PHR1 remained high in rph1Δ, regardless of the presence of Rad53 (asterisks and hash symbols in Figure 5C). Overexpression of WT Rph1 reduced the basal transcription of PHR1 to 68% (asterisks in Figure 5C). In the presence of overexpressed Rph1, Rad53 could enhance the basal expression of PHR1 (section symbol in Figure 5C), whereas UV-induced damage slightly increased the transcription of PHR1. To further elucidate the relation among Rph1, Rad53 and UV irradiation in regulating PHR1 expression, the data were subjected to regression analysis (Table 1). Rph1 is the most effective factor suppressing the transcription of PHR1 (standardized coefficient of Rph1: −0.738), whereas Rad53 and UV irradiation play a moderate but positive role in regulating PHR1 (standardized coefficient of Rad53 and UV: 0.453 and 0.375, respectively). These observations (Figure 5 and Table 1) indicate that Rph1 is the major regulator of PHR1 under experimental conditions and emphasize the role of the checkpoint protein Rad53 in the modulation of Rph1 during the regulation of PHR1.
Table 1.
Regression analysis to evaluate the effective strength to PHR1 expression
| Model | Unstandardized coefficients | Standard. error | Standardized coefficients | t | Significance |
|---|---|---|---|---|---|
| B | β | ||||
| (Constant) | 0.969 | 0.041 | 23.83 | 3.71e-01 | |
| RPH1 | −0.405 | 0.041 | −0.738 | −9.946 | 3.47e-00 |
| RAD53 | 0.248 | 0.041 | 0.453 | 6.108 | 5.72e-00 |
| UV | 0.206 | 0.041 | 0.375 | 5.054 | 6.07e-00 |
Dependent variable: PHR1 expression.
R2 = 0.943.
To determine the role of Rad53 in regulating PHR1 expression, we used ChIP analysis to examine the effect of Rad53 on the recruitment of Rph1 and the levels of H3K36me3 at the URSPHR1. In the absence of UV irradiation, Rph1 was associated with URSPHR1, as shown above, regardless of the presence of Rad53 (Figure 5D, black bars). Distinct from our previous observation that Rph1 dissociated from the URSPHR1 after UV irradiation (Figure 3A), Rph1 remained bound to URSPHR1 in the absence of Rad53 (asterisksin Figure 5D), which strongly indicates that Rad53 functions as a crucial regulator for Rph1 to dissociate from PHR1 promoter upon UV irradiation (Figure 5D). Furthermore, the levels of H3K36me3 and H3 acetylation are correlated to the presence of Rph1 at the URSPHR1 region (Supplementary Figure S5). Chromatin-bound Rph1 retained low levels of histone H3K36 tri-methylation in the rad53Δ strain after DNA damage. ChIP analysis indicated that Rad53 was required for the dissociation of Rph1 from URSPHR1. Our results suggest a potentially novel damage checkpoint pathway that is directed by a Rad53-Rph1 cascade of regulatory events.
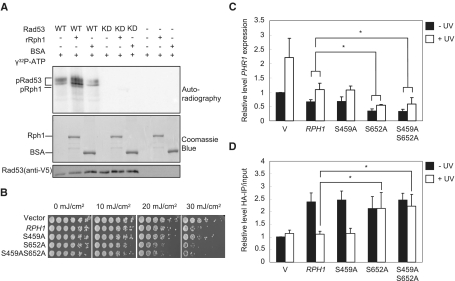
Activated Rad53 complex phosphorylates Rph1 and S652A-mutated Rph1 impairs the dissociation in response to DNA damage
To test whether Rph1 is a substrate of Rad53, we performed an in vitro kinase assay by incubating IP-activated Rad53 (WT or KD) with recombinant Rph1. By autoradiography, we detected a specific signal of Rph1 phosphorylation in the presence of WT Rad53 but not rad53-KD, which established that Rad53 kinase dominated the phosphorylation of Rph1 (Figure 6A). Previous proteomic studies had revealed that Rph1 was phosphorylated at multiple serine residues induced by DNA damage or cell cycle arrest (52–55). To determine the functional role of phosphorylation, we generated a series of alanine-substituted mutations on putative phosphorylated serine residues (S412, S459, S557, S561, S652 and S689) to analyze the UV sensitivity of these mutants. Because the Rph1 phosphorylation triggered by, UV irradiation may reflect a transient response, we attempted to examine the immediate response by use of the GAL inducible expression system. However, we found severe growth defects in the WT and phospho-mutants of Rph1 (Figure 1A and Supplementary Figure S6) cultured on synthetic complete selective medium in the presence of galactose (SCM-URA + galactose) but not glucose (SCM-URA + glucose), presumably because of constitutively overexpressed Rph1. To avoid this potential issue to obscure the growth phenotype, we modified our experimental protocol to use the GAL1 promoter to induce the overexpression of the WT and phospho-mutants of Rph1 4 h before UV irradiation, then scored the phenotype 2 days later (see ‘Methods and Materials’ section). When the UV dose was increased to 30 mJ/cm2, we found that the rph1-S652A mutant began to show a hypersensitivity to UV irradiation, even greater than that of Rph1 overexpression (Figure 6B, Supplementary Figure S7). From bioinformatics studies (Scansite, http://scansite.mit.edu/and GPS2.1, http://gps.biocuckoo.org/), we selected S459 (embedded within the putative bipartite nuclear localization region) and S652 (a consensus phosphorylated site detected in genome-wide LC/MS analysis) (52–55) to further characterize the functional role of Rph1 phosphorylation in PHR1 expression. The rph1-S459A mutant displayed a similar UV-sensitivity phenotype to WT Rph1, whereas rph1-S652A and rph1-S459AS652A mutants were hypersensitive to UV irradiation (Figure 6B), which indicates that phosphorylation at S652 may play a critical role in Rph1 function responding to DNA damage. We subsequently measured the expression of PHR1 and association of Rph1 phospho-mutants with URSPHR1. The expression levels of PHR1 were comparable in rph1-S459A and WT Rph1 but were reduced to 30% (P < 0.05) in rph1-S652A and rph1-S459AS652A mutants, regardless of UV treatment (Figure 6C). Remarkably, results from HA (Rph1)-ChIP assays showed that S652A mutation did not affect the Rph1 association with URSPHR1 but greatly impaired the dissociation from URSPHR1 on UV treatment (Figure 6D), which indicates that phosphorylation at S652 is important for Rph1 to dissociate from URSPHR1 in the presence of UV irradiation. These data support that chromatin association and dissociation of Rph1 on the PHR1 promoter mediated by protein phosphorylation is the major regulatory mechanism for PHR1 expression responding to DNA damage.
Figure 6.
The phospho-mutant at S652 of Rph1 increases UV sensitivity and impairs the dissociation after UV irradiation. (A) In vitro kinase assay was performed by recombinant Rph1 or BSA incubated with or without V5-IP WT or KD Rad53 supplied by γ32P-ATP. The signal was detected by autoradiography. pRad53 indicated the autophosphorylation of Rad53. pRph1 indicated the phosphorylation of Rph1. Coomassie Blue and immnoblotting (anti-V5) showed the loading controls. (B) UV sensitivity of rph1Δ cells containing control vector, WT Rph1 (RPH1) or phospho-defective Rph1 mutants. (C and D) The indicated strains as in (B) were harvested for RT-qPCR to detect PHR1 expression in response to UV or not (C) and for HA-ChIP to measure the association of Rph1 at URS of PHR1 (D). Error bars show the SD of three biological repeats. *P < 0.05.
DISCUSSION
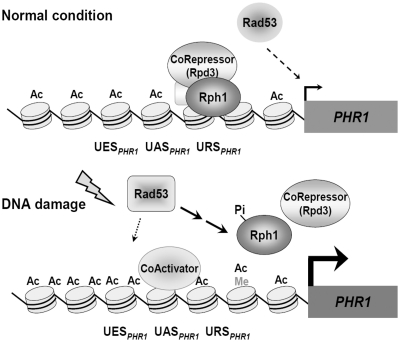
Collectively, we hypothesize a model to describe the regulatory event modulated by the H3K36 demethylase Rph1 at the PHR1 promoter in response to DNA damage signals in Figure 7. Roeder proposed a model of a ‘two-step process’ of transcriptional activation in eukaryotes: (i) The overall level of induction in response to activating signal involving first a ‘de-repression step’ that restores activity to the basal level, and followed by (ii) A ‘net-activation’ step that leads to the higher induction expression level (56). Here, we show that Rph1-mediated H3K36 demethylase activity is required to repress PHR1 expression and is involved in regulating the early step of transcriptional activation. Rph1 is specifically associated with URSPHR1 to generate a repressed or ground state of chromatin structure in the absence of UV irradiation. The physiological repressed chromatin structure at the URSPHR1 subsequently leads to decreased histone acetylation by cooperatively associating with the Rpd3 co-repressor complex. The checkpoint kinase Rad53 is required for the basal and inducible expression of PHR1. Upon UV-induced DNA damage, the fully activated Rad53 modulates the phosphorylation of Rph1, which subsequently dissociates from URSPHR1 to relieve the suppressed expression of PHR1. In addition, Rad53 may mediate the recruitment of other co-activators to increase the histone acetylations at the promoter in response to UV irradiation, which induces PHR1 expression for efficient DNA repair. This study highlights a distinct mechanism of the histone demethylase in transcriptional regulation at the promoter region instead of coding sequence. Thus, we reveal that the key regulatory step of Rph1 is to maintain a low level of H3K36 methylation at the PHR1 promoter in the basal state. Dismissal of Rph1 from the URSPHR1 region is mediated by a DNA damage signal to allow immediate histone acetylations as well as transcriptional initiation by recruiting RNA pol II.
Figure 7.
A model for Rph1-regulated PHR1 expression in response to DNA damage. Under normal conditions, Rph1 associates with URSPHR1, and PHR1 transcription is repressed (upper panel). Under DNA damage signaling, Rph1 dissociates from the PHR1 promoter to induce the expression of PHR1. Ac, histone acetylation; Me, H3K36 tri-methylation; Pi, phosphorylation; See ‘Discussion’ section for details.
Histone H3K36 demethylase activity cooperates with DNA-binding affinity of Rph1 in the repression of PHR1 transcription
Given the genome-wide distribution of histone modifications, H3K36me3 is enriched in the transcribed region of generally active genes (57). A major question concerns the mechanism of transition from methylation–demethylation involving in the transcriptional repression to activation. The transitions could be a simple matter of equilibrium enzyme reactions, the histone methyltransferases (HMTs) and histone demethylases (JHDMs). Kim and Buratowski (20) reported that JHDMs (Jhd1 and Rph1) antagonize Set2-mediated H3K36 methylation and promote transcription elongation in constitutively highly expressed genes, and such equilibrated events take place in the coding region of genes. Here, we studied a DNA damage-inducible gene, PHR1, which is repressed under normal growth conditions. Different from previous findings, our results reveal that H3K36 demethylase Rph1 functions as a repressor and associates at the promoter of PHR1 in the absence of DNA damage signals. Defective H3K36 methylation, lack of Set2 and overexpression of Rph1 (39,40) increased the sensitivity to UV irradiation. However, deletion of SET2 did not affect the expression of PHR1 under our experimental conditions, whereas overexpression of Rph1 and H3K36A mutant strains decreased the expression level of PHR1.
Three possibilities may serve to explain our observations. The first is that Set2-mediated H3K36 methylation is not required to initiate basal transcription of PHR1 under the normal condition (−UV). The genome-wide distribution profile of H3K36 methylation at promoters is relatively lower than that in coding regions. Therefore, an increase in level of H3K36 methylation at specific promoters is likely due to a decreased activity of histone demethylase, such as Rph1 (Figure 2A), rather than the recruitment of HMT activity (Set2) to add methyl groups to H3K36. The second explanation is based on the recruitment of RNA Pol II at the promoter region. Pol II was found to occupy inactive promoter regions of signal-inducible genes, named PRGs (primary response genes). These genes are regulated at the transition between de-repression and transcriptional initiation in the basal state by an interplay with a co-repressor complex. Most PRG promoters have a high basal level of H3K4me and H3K9Ac (58). Indeed, we demonstrated that Pol II binding at the PHR1 promoter is decreased in the H3K36A mutant but not the set2Δ strain (Figure 3E), which implies a distinct role of Set2 in Pol II recruitment at the promoter and coding regions. H3K36A mutant may change the level of histone H3K9/14 acetylation by an unknown mechanism (Figure 3D) and consequently affect Pol II recruitment at the PHR1 promoter. The third possibility is a temporal dependency of other modifications on H3K36 such as lysine acetylation. Recent studies demonstrated that H3K36 can be modified by Set2-mediated methylation and Gcn5-dependent acetylation (59). Acetylation at H3K36 is localized predominantly at the promoters of RNA polymerase II-transcribed genes and functions as a prelude to transcriptional initiation (59,60). The set2Δ eliminated only H3K36 methylation, and H3K36A wiped out both methylation and acetylation at the PHR1 promoter. These observations suggest that the transition between H3K36ac and H3K36me represents a novel ‘dual-modification chromatin switch’ that controls the regulation of gene transcription at the PHR1 promoter. The functional complexity of both acetylation and methylation, but not simultaneously, on H3K9 is an example of the ‘dual-modification chromatin switch’. The recruitment of chromodomain protein HP1 to initiate the formation of heterochromatin depends on H3K9 methylation in mammals and fission yeast (61,62). We hypothesize that the interplay among acetylation, deacetylation, methylation and demethylation at the same site can also occur on H3K36 at the PHR1 promoter in budding yeast. None of the three suggested mechanisms above are mutually exclusive and all could act cooperatively.
Set2-dependent H3K36 methylation is required for Rpd3C(S) recruitment co-transcriptionally to the coding region for transcriptional activation (63). Surprisingly, our results indicate that Set2 plays a minor, if any, role in transcriptional activation of PHR1. However, our finding is not an isolated case. The expression of the starvation-induced genes ARG1 and HIS4 does not depend on Set2-mediated methylation at H3K36 (64,65). Our results may add PHR1 to this list.
Multiple lines of evidence suggest that chromatin modifications play a complex role in the regulation of transcription. The crosstalk between histone modifications can facilitate or repress chromatin-mediated processes (3). Here, we show that the histone H3K36 demethylation is linked to reduced histone acetylation involving the histone deacetylase Rpd3 at the PHR1 promoter (Figure 3B and C). Furthermore, Rph1 and Rpd3 can associate with URSPHR1, which may form the Ume6-Rpd3-Rph1 co-repressor complex and consequently block the UASPHR1 for basal trans-activation. Moreover, Rph1-mediated H3K36 demethylation at URSPHR1 may specify a histone mark to recruit a co-repressor complex or prevent the recruitment of a co-activator that consequently silences PHR1 expression in the absence of UV damage.
Alternatively, the variation in histone acetylation may be due to the recruitment of HATs. Many studies of gene expression involving genome-wide approaches or focusing on individual genes suggest that the histone acetylases Gcn5 (SAGA complex) and Esa1 (NuA4 complex) are generally recruited to the promoters of protein-coding active genes (66). Here, we observed that Rph1 was dissociated from the URSPHR1 region and cooperatively increased acetylation of histone H3/H4 after UV irradiation (Figure 4A). The dissociation of a putative Rph1/co-repressor complex (Figure 4C), as well as deletion of RPH1 (Figure 1C), may provide a more accessible region for the HAT complex to target at the URSPHR to enrich acetylations on chromatin for subsequent transcriptional activation. These observations imply that dissociation of demethylase at the promoter influences the chromatin dynamics.
Rad53 kinase activity and S652 phosphorylation of Rph1 are required for the dissociation of Rph1 in response to DNA damage
We demonstrate that the activated Rad53 complex mediates the Rph1 phosphorylation in response to DNA damage. Rad53 regulates the chromatin binding of Rph1 as well as enrichment of H3K36 methylation at the URSPHR1 region (Figures 5 and 6). In addition, we provide in vivo evidence of the functional role of phosphorylation in a histone demethylase, Rph1. Abolishment of the phosphorylation at S652 (rph1-S652A) had important biological impacts, as indicated by the significant differences in UV sensitivity, PHR1 repression and Rph1 binding to PHR1 promoter (Figure 6B and C). Phosphorylation is linked to protein function, such as conformational change, stability and activity. Phosphorylation on transcription factors can regulate the chromatin association and biological functions. A recent study of the Methyl-CpG Binding Protein 2 (MECP2) in mouse cortical neurons demonstrated that MeCP2-S80A mutation attenuated chromatin association affinity at candidate gene promoters and caused subtle gene expression changes (67). The mechanistic regulation of histone demethylase function is not clear yet. Two possible regulatory mechanisms are mediated by PTM and association with auxiliary factors (68). A recent study demonstrated that the H3K4 demethylase Jhd2 is modified by polyubiquitination to control the protein level of Jhd2 through proteasome-mediated degradation (69). Rosenfeld’s group currently reported that phosphorylation on PHF8, a histone H4K20 demethylase, was required for its chromatin dismissal in prophase and was involved in regulating cell cycle progression (70). The observations in PHF8 and Rph1 strongly support that phosphorylation may be evolutionally linked to the function of histone demethylases responding to diverse cellular signals. Because Rph1 contains multiple putative phosphorylation sites, further studies are required to precisely define the roles of PTMs and putative regulatory factors that are critical for the regulation of histone demethylases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Academia Sinica and a grant from the National Science Council (NSC 97-2311-B-001-018-MY3 to W.-S.L.), Taiwan. Funding for open access charge: Academia Sinica.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Lorraine Pillus for critically reading the manuscript and Drs Z.F. Chang, L.J. Juan and H.M. Shih for helpful discussion.
REFERENCES
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Millar CB, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- 3.Suganuma T, Workman JL. Crosstalk among Histone Modifications. Cell. 2008;135:604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 4.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 5.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 6.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z, Zang J, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao Q, Pan CH, Dai S, et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 8.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 9.Agger K, Christensen J, Cloos PA, Helin K. The emerging functions of histone demethylases. Curr. Opin. Genet. Dev. 2008;18:159–168. doi: 10.1016/j.gde.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 11.Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 12.Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 13.Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- 14.Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tu S, Bulloch EM, Yang L, Ren C, Huang WC, Hsu PH, Chen CH, Liao CL, Yu HM, Lo WS, et al. Identification of histone demethylases in Saccharomyces cerevisiae. J. Biol. Chem. 2007;282:14262–14271. doi: 10.1074/jbc.M609900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 17.Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 20.Kim T, Buratowski S. Two Saccharomyces cerevisiae JmjC domain proteins demethylate histone H3 Lys36 in transcribed regions to promote elongation. J. Biol. Chem. 2007;282:20827–20835. doi: 10.1074/jbc.M703034200. [DOI] [PubMed] [Google Scholar]
- 21.Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 23.Jang YK, Wang L, Sancar GB. RPH1 and GIS1 are damage-responsive repressors of PHR1. Mol. Cell. Biol. 1999;19:7630–7638. doi: 10.1128/mcb.19.11.7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sebastian J, Sancar GB. A damage-responsive DNA binding protein regulates transcription of the yeast DNA repair gene PHR1. Proc. Natl Acad. Sci. USA. 1991;88:11251–11255. doi: 10.1073/pnas.88.24.11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sancar GB, Ferris R, Smith FW, Vandeberg B. Promoter elements of the PHR1 gene of Saccharomyces cerevisiae and their roles in the response to DNA damage. Nucleic Acids Res. 1995;23:4320–4328. doi: 10.1093/nar/23.21.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sweet DH, Jang YK, Sancar GB. Role of UME6 in transcriptional regulation of a DNA repair gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:6223–6235. doi: 10.1128/mcb.17.11.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sebastian J, Kraus B, Sancar GB. Expression of the yeast PHR1 gene is induced by DNA-damaging agents. Mol. Cell. Biol. 1990;10:4630–4637. doi: 10.1128/mcb.10.9.4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 31.Yuen T, Zhang W, Ebersole BJ, Sealfon SC. Monitoring G-protein-coupled receptor signaling with DNA microarrays and real-time polymerase chain reaction. Methods Enzymol. 2002;345:556–569. doi: 10.1016/s0076-6879(02)45047-1. [DOI] [PubMed] [Google Scholar]
- 32.Kuras L, Kosa P, Mencia M, Struhl K. TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000;288:1244–1248. doi: 10.1126/science.288.5469.1244. [DOI] [PubMed] [Google Scholar]
- 33.Lo WS, Gamache ER, Henry KW, Yang D, Pillus L, Berger SL. Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. EMBO J. 2005;24:997–1008. doi: 10.1038/sj.emboj.7600577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurdistani SK, Robyr D, Tavazoie S, Grunstein M. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 2002;31:248–254. doi: 10.1038/ng907. [DOI] [PubMed] [Google Scholar]
- 35.Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- 36.Usui T, Ogawa H, Petrini JH. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell. 2001;7:1255–1266. doi: 10.1016/s1097-2765(01)00270-2. [DOI] [PubMed] [Google Scholar]
- 37.Yadav AK, Renfrow JJ, Scholtens DM, Xie H, Duran GE, Bredel C, Vogel H, Chandler JP, Chakravarti A, Robe PA, et al. Monosomy of chromosome 10 associated with dysregulation of epidermal growth factor signaling in glioblastomas. JAMA. 2009;302:276–289. doi: 10.1001/jama.2009.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klose RJ, Gardner KE, Liang G, Erdjument-Bromage H, Tempst P, Zhang Y. Demethylation of histone H3K36 and H3K9 by Rph1: a vestige of an H3K9 methylation system in Saccharomyces cerevisiae? Mol. Cell. Biol. 2007;27:3951–3961. doi: 10.1128/MCB.02180-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Game JC, Williamson MS, Spicakova T, Brown JM. The RAD6/BRE1 histone modification pathway in Saccharomyces confers radiation resistance through a RAD51-dependent process that is independent of RAD18. Genetics. 2006;173:1951–1968. doi: 10.1534/genetics.106.057794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin LJ, Minard LV, Johnston GC, Singer RA, Schultz MC. Asf1 can promote trimethylation of H3 K36 by Set2. Mol. Cell. Biol. 2010;30:1116–1129. doi: 10.1128/MCB.01229-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Psathas JN, Zheng S, Tan S, Reese JC. Set2-dependent K36 methylation is regulated by novel intratail interactions within H3. Mol. Cell. Biol. 2009;29:6413–6426. doi: 10.1128/MCB.00876-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL. Snf1–a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 2001;293:1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- 44.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Moazed D, Gygi SP. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 2002;277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- 47.Hampsey M, Reinberg D. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- 48.Kim EM, Jang YK, Park SD. Phosphorylation of Rph1, a damage-responsive repressor of PHR1 in Saccharomyces cerevisiae, is dependent upon Rad53 kinase. Nucleic Acids Res. 2002;30:643–648. doi: 10.1093/nar/30.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Branzei D, Foiani M. The Rad53 signal transduction pathway: Replication fork stabilization, DNA repair, and adaptation. Exp. Cell Res. 2006;312:2654–2659. doi: 10.1016/j.yexcr.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 50.Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 51.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 52.Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl Acad. Sci. USA. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl Acad. Sci. USA. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X, Gerber SA, Rudner AD, Beausoleil SA, Haas W, Villen J, Elias JE, Gygi SP. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 56.Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 57.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 58.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morris SA, Rao B, Garcia BA, Hake SB, Diaz RL, Shabanowitz J, Hunt DF, Allis CD, Lieb JD, Strahl BD. Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J. Biol. Chem. 2007;282:7632–7640. doi: 10.1074/jbc.M607909200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stewart MD, Li J, Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol. Cell. Biol. 2005;25:2525–2538. doi: 10.1128/MCB.25.7.2525-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SI. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol. Cell. 2005;20:173–185. doi: 10.1016/j.molcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 63.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 64.Merker JD, Dominska M, Greenwell PW, Rinella E, Bouck DC, Shibata Y, Strahl BD, Mieczkowski P, Petes TD. The histone methylase Set2p and the histone deacetylase Rpd3p repress meiotic recombination at the HIS4 meiotic recombination hotspot in Saccharomyces cerevisiae. DNA Repair. 2008;7:1298–1308. doi: 10.1016/j.dnarep.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, Rolfe A, Workman JL, Gifford DK, Young RA. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tao J, Hu K, Chang Q, Wu H, Sherman NE, Martinowich K, Klose RJ, Schanen C, Jaenisch R, Wang W, et al. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proc. Natl Acad. Sci. USA. 2009;106:4882–4887. doi: 10.1073/pnas.0811648106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 69.Mersman DP, Du HN, Fingerman IM, South PF, Briggs SD. Polyubiquitination of the demethylase Jhd2 controls histone methylation and gene expression. Genes Dev. 2009;23:951–962. doi: 10.1101/gad.1769209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.