Abstract
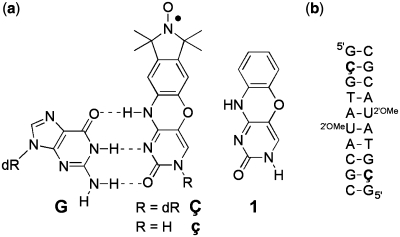
Previously, we developed the deoxycytosine analog Ç (C-spin) as a bi-functional spectroscopic probe for the study of nucleic acid structure and dynamics using electron paramagnetic resonance (EPR) and fluorescence spectroscopy. To understand the effect of Ç on nucleic acid structure, we undertook a detailed crystallographic analysis. A 1.7 Å resolution crystal structure of Ç within a decamer duplex A-form DNA confirmed that Ç forms a non-perturbing base pair with deoxyguanosine, as designed. In the context of double-stranded DNA Ç adopted a planar conformation. In contrast, a crystal structure of the free spin-labeled base ç displayed a ∼20° bend at the oxazine linkage. Density function theory calculations revealed that the bent and planar conformations are close in energy and exhibit the same frequency for bending. These results indicate a small degree of flexibility around the oxazine linkage, which may be a consequence of the antiaromaticity of a 16-π electron ring system. Within DNA, the amplitude of the bending motion is restricted, presumably due to base-stacking interactions. This structural analysis shows that the Ç forms a planar, structurally non-perturbing base pair with G indicating it can be used with high confidence in EPR- or fluorescence-based structural and dynamics studies.
INTRODUCTION
Interconnected with the central importance of the macromolecular structural scaffold, the dynamics or movements of structural elements play a key role in all biological processes. Electron paramagnetic resonance (EPR) spectroscopy can provide information on both the dynamics as well as the global structure of biological molecules (1–4). For such studies on nucleic acids, several nitroxide spin labels have been developed (5–11). Most of these reporters contain flexible linkers with one or more rotatable bonds between the nitroxide spin label and the nucleic acid. Flexible linkers introduce uncertainty in the measurement of dynamics, or determination of distances between two nitroxide spin labels due to movement of the probe independent of the nucleic acid. Therefore, we designed and synthesized the rigid nitroxide spin-labeled cytidine analog Ç (C-spin) for use in studies of the structure and dynamics of nucleic acids by EPR spectroscopy (12). The nitroxide moiety of this reporter can be reduced with a mild reducing reagent such as sodium sulfide (Na2S) to produce a fluorescent nucleoside (Çf), which was used for detection of mismatches in DNA (13,14). Therefore, both EPR and fluorescence spectroscopies can be used with the same spectroscopic label as illustrated in folding studies of the cocaine DNA aptamer (15) and dynamics of DNA hairpin loops (16). Pulsed electron–electron double resonance (PELDOR) has also been used to determine precise distances and angular orientations within Ç-labeled DNAs (17,18). Thus, this bi-functional reporter allows for the study of nucleic acid structure and dynamics via the complementary spectroscopic techniques of fluorescence and EPR.
As with any reporter group used in biophysical experiments, it is important that the probe does not perturb the structure of the biological system of interest. If the reporter group is structurally perturbing, the results obtained from biophysical experiments would falsely report the nature of the macromolecule. Therefore, the effect of the reporter on the structure of the biopolymer must be carefully analyzed. Despite the widespread use of spectroscopic probes in biophysical studies, there are very few crystal structures containing covalently bound spectroscopic probes aside from 2-aminopurine for nucleic acids (19–21) and nitroxide spin labels for proteins (22–25). Obtaining a high-resolution crystal structure of a nucleic acid containing a spectroscopic probe remains challenging, and thus most researchers have relied on a variety of biophysical techniques to provide indirect evidence as to whether or not a probe alters the structure of the macromolecule of interest. For example, melting temperature analysis of DNAs containing Ç showed only subtle changes in the melting temperatures relative to unlabeled DNAs (16). In addition, EPR studies of dynamics (12,16) and distance measurements (18) aligned well with predicted models. Despite these encouraging biophysical data, no direct structural evidence exists to demonstrate whether or not the reporter Ç is structurally perturbing, forms a proper base pair with deoxyguanosine and what may be the preferred conformation. Furthermore, one concern with regards to the structure of the spin-labeled nucleoside was the fact that the phenoxazine-derived nucleobase contains 16 π-electrons. This number of electrons in a cyclic π-system indicates antiaromaticity, which could result in non-planarity of the ring system. Because a bend in the nucleobase might affect the use of this probe for biophysical studies, we undertook a detailed analysis of its high-resolution structure.
Here we report a detailed crystallographic characterization of Ç. This analysis includes small molecule crystal structures of the ç nucleobase, which has been used for non-covalent spin labeling of nucleic acids containing an abasic site (26), and its phenoxazine analog (1) (Figure 1) as well as a 1.7 Å resolution crystal structure of a decamer DNA duplex containing Ç. The high-resolution nucleic acid structure demonstrates that within the context of the nucleic acid, the nitroxide spin label Ç adopts a planar conformation while forming a standard three hydrogen bond base pair with deoxyguanosine. These results validate the interpretation of distance and orientation measurements between two Ç reporters described previously and provide the basis for further structural and dynamics studies on oligonucleotides with unknown folds.
Figure 1.
(a) Structures of phenoxazine-derived nitroxide spin labels Ç and ç, and the unmodified phenoxazine derivative 1. The spin labels are shown base-paired with guanine (G), with hydrogen bonds indicated by dashed lines (b) Sequence and secondary structure of the duplex DNA used to obtain a high-resolution crystal structure of a Ç-containing DNA helix. dR = 2′-deoxyribose. 2′OMeU = 2′-O-methyluridine.
MATERIALS AND METHODS
Small molecule crystallization and structure determination
The nitroxide spin-labeled nucleobase ç was prepared as previously described (26), and the synthesis of phenoxazine 1 is described in the Supplementary Data. Yellow crystals of the ç spin label were obtained by slow evaporation from ethanol. Yellowish-brown crystals of 1 were obtained by slow evaporation from 3:1 dichloromethane:methanol solution. Crystals of ç and 1 were mounted on a Rigaku MM007/Mercury X-ray diffractometer (confocal optics Mo Kα radiation, 0.71073 Å). X-ray diffraction experiments were performed at 93 K. Intensity data were collected using accumulated area detector frames spanning at least a hemisphere of reciprocal space for all structures. Data were integrated using Crystal Clear. All data were corrected for Lorentz, polarization and long-term intensity fluctuations. Absorption effects were corrected on the basis of multiple equivalent reflections. The structures were solved by direct methods. Hydrogen atoms bound to carbon were idealized. Structural refinements were obtained with full-matrix least-squares based on F2 by using the program SHELXTL (27). The theta(max) resolution of the small molecule structure of 1 was 27.52 and the theta(max) resolution of the small molecule structure of ç was 25.3.
Density function theory calculations
Density function theory (DFT) calculations on the spin-labeled nucleobase ç were performed with the B3LYP functional, the 6-31G* basis set and unrestricted spin-wave functions using Gaussian03 (28). To obtain a geometry optimized structure and single point energy for the bent spin label, the atoms of the spin label’s phenoxazine moiety were frozen to the position obtained from the small molecule crystal structure and hydrogen atoms were added to the vacant positions. The constraints on the phenoxazine moiety were then relaxed to obtain the energy optimized structure and single point energy of the unbent spin label. The vibrational frequencies for both geometry-optimized structures were all positive, indicating the structures represent an energy minimum. The single point energy for the bent and planar spin label was −2 795 044.283 kJ/mol and −2 795 045.594 kJ/mol respectively. The unbent structure is 1.31 kJ/mol less in energy. The frequency for the bending motion around the oxazine linkage is 18.8 and 28.3 cm-1 for the planar and the bent conformation, respectively.
DNA crystallization and structure determination
A 10 nt-long DNA containing Ç at position 2 and 2′-O-methyl U at position 6 (Figure 1b) was prepared via solid-phase chemical synthesis as described previously (12,16). The decamer duplex DNA sample containing the nitroxide spin-labeled nucleotide Ç was crystallized as described for a similar phenoxazine-containing cytosine analog (29). Crystals were grown at 16°C using the sitting drop vapor diffusion method from 0.4 µl of DNA (2.4 mM) mixed with 0.4 µl of 10% (±)-2-methane-2,4-pentanediol (MPD), 40 mM sodium cacodylate pH 6.0, 12 mM spermine·HCl, 80 mM NaCl, 12 mM KCl, 12 mM MgCl2 and equilibrated against a reservoir of 35% MPD. Crystals of the 10 nt-long DNA containing 2′-O-methyl U and Ç grew over several months. Most crystals grew as clusters, although one drop contained two single crystals (Supplementary Data). A single crystal was vitrified by plunging into liquid nitrogen and a data set was collected using a Rigaku FR-E+ SuperBright rotating anode X-ray generator with VariMax HF optics and a Saturn 944+ CCD detector. The crystal diffracted beyond the resolution limits of the detector at the minimum detector distance (Supplementary Data), and thus a 2θ swing of −15° was used to enhance the resolution limits given the in house X-ray system. Nevertheless, the crystal diffracted beyond the resolution limits of the in house X-ray detection system, as determined by the strong signal in the highest resolution bin (I/σ = 20; Table 1). The data were reduced with XDS and XSCALE (30). The structure was solved by molecular replacement using PHASER (31) from the CCP4 suite (32) and contained two DNA molecules (i.e. a self-complementary duplex) in the asymmetric unit. The structure was refined with numerous reiterative rounds of refinement in REFMAC (33) and manual building in Coot (34). The final crystallographic model was produced after two additional rounds of refinement in Phenix (35) which utilized a CIF file containing the modified nucleotide linkage definitions generated using the program Jligand (G.N. Murshudov et al., unpublished data).
Table 1.
Crystallographic statistics for a DNA containing Ç
| Space group | P212121 |
| Unit cell | a = 24.71 Å, b = 44.55 Å, c = 45.94 Å; α = β = γ = 90° |
| Vm | 1.95 Å3/Daa |
| Solvent content | 57.4% |
| Resolution | 50–1.7 Å (1.74–1.70 Å)b |
| I/σ | 54.0 (20.0) |
| Completeness | 94.3% (82.9%) |
| Rmerge | 0.022 (0.070) |
| Multiplicity | 8.4 (6.4) |
| Reflections | 5613 (354) |
| Refinement | |
| Rcryst | 15.9% (0.138) |
| Rfree | 19.5% (0.218) |
| Mean B-factor | 14.5 Å2 |
| Number of atoms | |
| DNA | 436 |
| Solvent | 83 |
| Total | 519 |
aMatthews co-efficient and solvent content calculated based on a predicted duplex DNA MW of 6494 Da.
bValues in parenthesis indicated highest of 20 resolution bins for data reduction and highest of four resolution bins for refinement.
RESULTS AND DISCUSSION
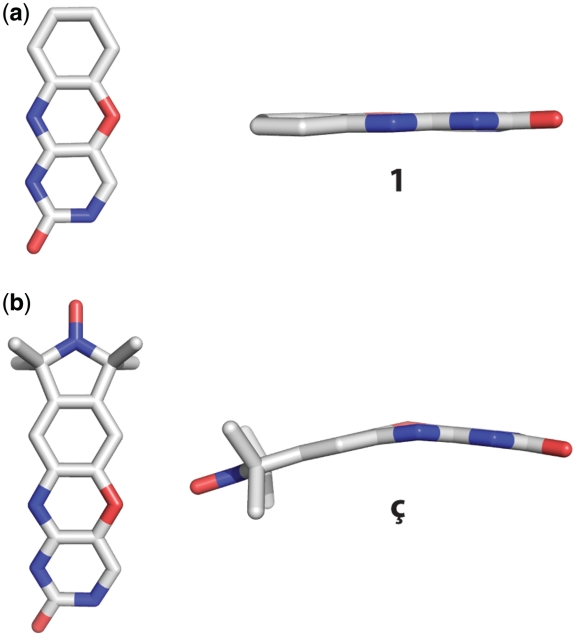
Small molecule crystal structures were determined for the ç nucleobase as well as the phenoxazine analog 1 (Figure 2a and Supplementary Data). The phenoxazine nucleobase 1 adopts a planar conformation with almost no bend at the oxazine linkage between the cytosine and benzene rings (Figure 2a). In contrast, ç adopts a non-planar geometry with a bend of ∼20° at the oxazine linkage between the cytosine and benzene rings (Figure 2b and Supplementary Data). In order to rationalize the conformational differences of the phenoxazine moiety, DFT calculations have been performed on ç in the bent conformation as found in the small molecule crystal structure and on ç in the planar conformation as found in the small molecule crystal structure of 1. These calculations revealed that the bent form is only 1.31 kJ/mol higher in energy than the planar form and that all vibrational frequencies for both conformations are positive. Thus, both conformations are similar energy minima or might actually belong to the same energy minimum. This is supported by the finding that the frequencies for the bending motion differ by only 10 cm−1 (18 and 28 cm−1 for the planar and bent conformation, respectively). Taken together, we interpret these results as both conformations belonging to the same energy minimum with a low energy bending motion around the oxazine linkage. Because bending the phenoxazine moiety costs little energy, the surrounding environment such as crystal packing can drive it into either conformation. This raises the question as to whether ç is bent or planar when incorporated into an oligonucleotide structure. Thus, we proceeded to obtain a high-resolution structure of a DNA containing Ç to establish the preferred conformation of Ç within the context of a nucleic acid.
Figure 2.
Small molecule crystal structures of (a) phenoxazine (1) and (b) the nitroxide spin-labeled nucleobase ç.
Initially, we selected two previously reported duplex DNA crystal forms, one A-form (36) and one B-form (37), for incorporation of Ç and subsequent crystallographic structure determination. Each of these crystal forms was chosen because it contained a cytosine residue that in principle had sufficient space within the crystal lattice to accommodate replacement with Ç. We incorporated Ç at positions 4 or 6 of the self-complementary octamer A-form duplex DNA as well as position 9 of the self-complementary dodecamer B-form duplex DNA. Despite producing large, well diffracting crystals using unmodified DNA samples, we observed only thin plate crystals that were unsuitable for structure determination using DNA samples containing Ç (data not shown). Therefore, we sought a different crystal form for incorporation of Ç and structural characterization.
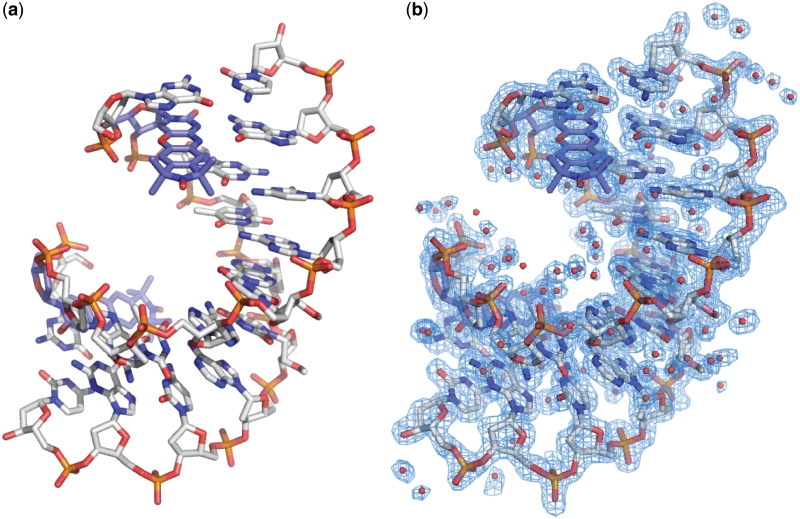
Egli et al. (38) reported crystal structures of an A-form decamer DNA duplex with either cytosine or the phenoxazine-derived cytosine analog ‘G-clamp’ (29). These crystal structures contain a 2′-O-methoxymethyl T at position 6 that promotes formation of A-form DNA in solution rather than B-form, which is standard for DNA. To our knowledge, the 2′-O-methoxymethyl T phosphoramidite is not commercially available, and thus we first produced crystals with a C at position 2 and a 2′-O-methyl U at position 6. These crystals diffracted X-rays to better than 2.3 Å resolution in house (data not shown). Next, a DNA sample was prepared with the nitroxide spin label Ç incorporated into position 2 and a 2′-O-methyl U at position 6 (Figure 1b) and resulted in a crystal from which we determined a 1.7 Å resolution crystal structure (Figure 3, Table 1 and Supplementary Data). Crystals suitable for structure determination failed to grow from a sample containing Ç at position 2 with a dT at position 6 (i.e. no 2′-O-methyl). EPR spectra of these two Ç-containing samples, one with a 2′-O-methyl U at position 6 that should be A-form in solution and one with a dT at position 6 that should be B-form in solution, were found to be nearly identical (Supplementary Data). Thus, there is likely to be little difference in the mobility of the probe in comparison of A- and B-form DNA in solution. Inspection of the electron density maps from our 1.7 Å resolution structure obtained from the DNA sample containing Ç at position 2 and a 2′-O-methyl U at position 6 clearly showed the 2′-O-methyl group of U6 (Figure 3 and Supplementary Data).
Figure 3.
Overall crystal structure of a DNA containing Ç. (a) Stick figure representation of an A-form duplex DNA crystal structure containing Ç solved at 1.7 Å resolution. For clarity, Ç is shown in light blue carbon backbone and the remainder of the DNA is shown in gray carbon backbone. (b) Final crystallographic model containing waters overlaid with the 2|Fo|−|Fc| electron density map shown in blue mesh and contoured at 1.0 σ.
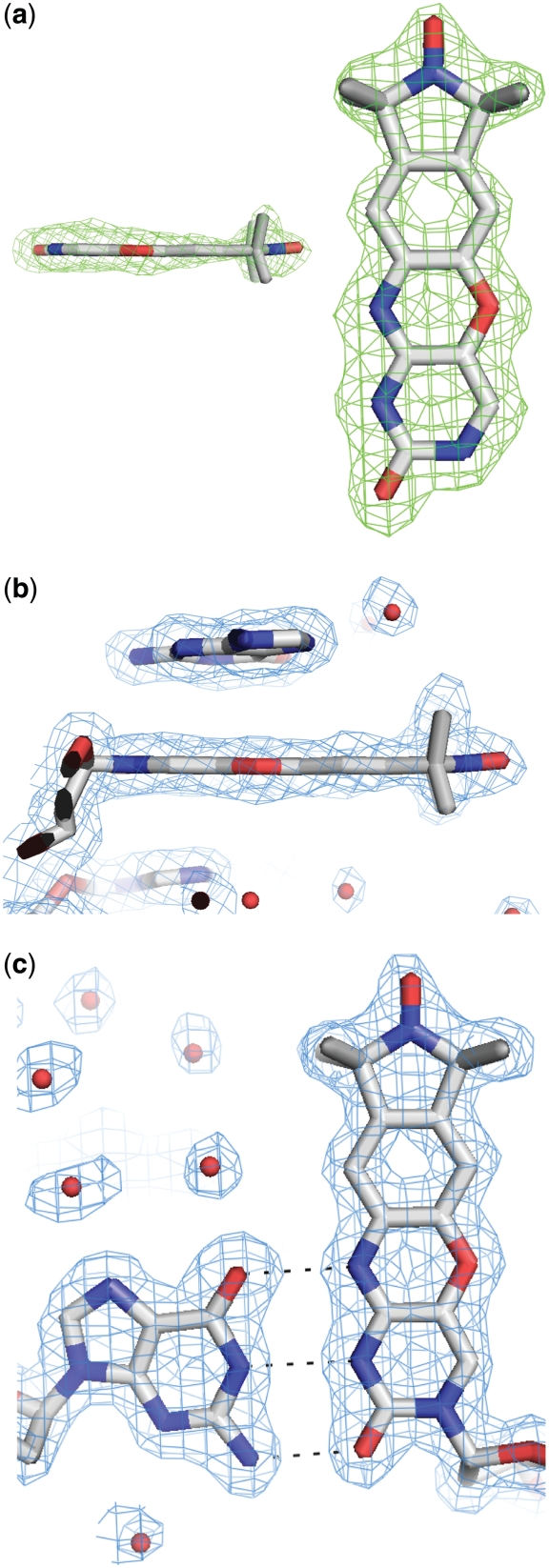
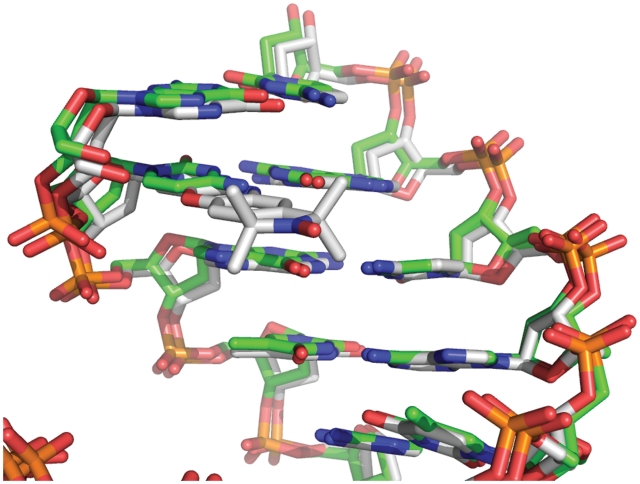
The 1.7 Å resolution crystal structure of the decamer duplex DNA containing Ç at position 2 was initially refined with an abasic site at this position. The resulting omit |Fo|−|Fc| electron density map showed unambiguous electron density for the Ç nucleobase (Figure 4a). The simulated-annealing omit 2|Fo|−|Fc| electron density map calculated with phases from the refined model was of excellent quality and revealed unambiguously that Ç forms a standard base pair with dG9 of the opposite strand with three ∼2.8 Å long hydrogen bonds (Figure 4b and c). The structure of the nucleic acid helix containing Ç superimposes closely on a previously reported structure containing a deoxycytidine residue at this position (38) (Figure 5), indicating that Ç does not perturb the DNA structure relative to a standard deoxycytidine residue. EPR spectroscopic analysis showed that Ç had decreased mobility in duplex DNA relative to single-stranded (12,16) or bulged (39) sites and could be used to measure single strand to duplex transitions during folding (15), implying Ç forms a base pair with dG. Furthermore, thermal denaturation experiments showed that the Ç•dG pairing had a similar melting temperature to a dC•dG pair, but showed a decrease in the melting temperature of 10–15°C when Ç was paired with dA, dT or dC (12). Combined with the EPR spectroscopy and thermal denaturation results, the high-resolution crystal structure described here shows that Ç is a non-perturbing cytosine analog that forms a Watson–Crick base pair with dG as designed.
Figure 4.
Examination of the nitroxide spin-labeled nucleotide Ç within the high-resolution DNA crystal structure. (a) Side- and top-down views of Ç superimposed with the |Fo|−|Fc| omit electron density map shown in green mesh contoured at 3.0 σ. (b) Side view of Ç superimposed with the 2|Fo|−|Fc| electron density map, shown in blue mesh contoured at 1.0 σ. (c) Top-down view of the base pair formed by deoxyguanosine and Ç superimposed with the 2|Fo|−|Fc| electron density map shown in blue mesh contoured at 1.0 σ. Hydrogen bonds are depicted as dashed lines.
Figure 5.
Overlay of the 1.7 Å resolution crystal structure of a DNA containing the nitroxide spin-labeled deoxycytosine analog Ç (gray carbon backbone) with a crystal structure containing deoxycytosine at the same position (PDB ID 1DPL, green carbon backbone) (38).
The crystal structure shows that within the context of a nucleic acid, Ç adopts a planar geometry (Figure 4b) rather than the bent geometry observed in the crystal structure of ç (Figure 2b). At the oxazine linkage, N4 of Ç is involved in a 2.9 Å hydrogen bond with dG9 of the opposite strand and O5 does not form hydrogen bonds with any water molecules, but rather packs 3.7 Å away from O2P of the Ç phosphate and also C3′ of residue G1. This is similar to what was observed for the small molecule crystal structure of phenoxazine 1 alone or the phenoxazine-derived ‘G-clamp’ modified nucleoside (29). At the current resolution limits, no water-mediated interactions were observed off the nitroxide of Ç in the major groove. However, if one extends the model 5′ of dG1, it is possible that the nitroxide could make water mediate mediated interactions with the phosphate or base two residues away on the 5′ side of Ç. If that residue were a purine, we speculate that additional packing interactions may be observed with the nitroxide ring. Interestingly, examination of the high-resolution DNA crystal structure shows that the average B-factor of the pyrimidine ring (12.9 Å2) is lower than either the benzene ring (19.3 Å2) or the nitroxide ring (25.2 Å2). This demonstrates greater static or dynamic disorder of the nitroxide ring relative to the pyrimidine ring, which could be reflective of the bending motion observed in DFT calculations for the free label ç. Alternatively, the increased B-factors could be reflective of rigid body motion of the entire nucleobase relative to the sugar-phosphate backbone. Nevertheless, the preferred conformation of the nucleobase of Ç in DNA is planar; the amplitude of the bending motion is clearly more restricted within the DNA duplex than observed in the crystal structure of the free label ç, presumably due to the benefit of van der Waals packing interactions with the 5′ deoxyguanosine base (Supplementary Data and below). Although the phenoxazine ring system is formally antiaromatic, it has been shown with 20 and 24 π electron N,N-dihydrodiazatetracenes by X-ray crystallography, cyclic voltammetry and nucleus independent chemical shift calculations that they have reduced aromaticity, rather than antiaromaticity (40).
Crystal structures with other phenoxazine-containing compounds include those of the antibiotic actinomycin, which reveal a slight bend at the oxazine linkage (41), and of actinomycin bound to DNA which reveal a planar conformation of the phenoxazine moiety (42–44). We note that there is significant π-stacking interaction of the phenoxazine component of Ç with dG1 (Supplementary Data), as was observed in the G-clamp crystal structure (29) and is consistent with π-stacking in the actinomycin-DNA crystal structures (42–44). Previously, we examined the melting temperature of a series of DNAs to determine the effect of Ç, and in some cases, a +5°C shift in the melting temperature of duplex DNAs containing Ç has been observed (16), which could be consistent with the π-stacking observed in the crystal structure presented here and with other phenothiazine- and phenoxazine-labeled DNA structures (45,46). Increased melting temperatures do not necessarily correlate with π-stacking, but could be due to simple van der Waals packing as observed in crystal structures of a modified deoxycytosine base with a non-aromatic two-ring system (47). Given that the antibiotic actinomycin and ç/Ç are both observed in planar and bent forms and our DFT calculations show that the bent and planar conformations of ç are similar in energy and frequency, we predict that other phenoxazine-derived molecules would exhibit a similar general pattern. In summary, the high-resolution crystal structure of a DNA containing Ç showed that the spin label adopts a planar geometry with indicators of modest mobility at the oxazine ring, consistent with small molecule crystal structures, DFT geometry calculations and previously published crystal structures of molecules containing phenoxazine-based compounds, such as actinomycin.
Only a handful of crystal structures of biological macromolecules containing spectroscopic probes have hitherto been reported. We have performed a detailed crystallographic analysis of the nitroxide spin-labeled deoxycytosine analog Ç. Consistent with previous EPR and fluorescence spectroscopic and thermal stability results, the results presented here demonstrate that Ç is a non-perturbing cytosine analog that forms a Watson–Crick base pair with dG. These results increase the accuracy and interpretation of distance and orientation measurements made with this spectroscopic probe and provide a benchmark for structural characterization of spectroscopic probes.
DATA BANK ACCESSION CODES
Coordinates and structure factors files have been deposited into the Protein Data Bank with accession code 3OT0.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Icelandic Research Fund (60028021); Research Councils of the UK with a RCUK fellowship (to O.S.); BBSRC (SBSO YBB 016 to O.S. and G.W.) doctoral fellowships (to P.C.); University of Iceland (to S.A.S.); W.M. Keck Foundation, partial (to A.R.F.); Damon Runyon Cancer Research Foundation (DRG-1844-04 to T.E.E.); Emerald BioStructures (to T.E.E.). Funding for open access charge: Icelandic Research Fund (60028021).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Roy and M. Buehl for help with the DFT calculations, A. M. Z. Slawin for X-ray structure determination of nitroxide spin-labeled nucleobase ç and phenoxazine derivative and A. Gardberg for assistance with JLigand and refinement in Phenix. A.R.F. was a Distinguished Young Scholar in Medical Research of the W.M. Keck Foundation and is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Columbus L, Hubbell WL. A new spin on protein dynamics. Trends Biochem. Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 2.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 3.Qin PZ, Dieckmann T. Application of NMR and EPR methods to the study of RNA. Curr. Opin. Struct. Biol. 2004;14:350–359. doi: 10.1016/j.sbi.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Schiemann O, Prisner TF. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Q. Rev. Biophys. 2007;40:1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 5.Edwards TE, Sigurdsson ST. Site-specific incorporation of nitroxide spin-labels into 2′-positions of nucleic acids. Nat. Protoc. 2007;2:1954–1962. doi: 10.1038/nprot.2007.273. [DOI] [PubMed] [Google Scholar]
- 6.Jakobsen U, Shelke SA, Vogel S, Sigurdsson ST. Site-directed spin-labeling of nucleic acids by click chemistry: detection of abasic sites in duplex DNA by EPR spectroscopy. J. Am. Chem. Soc. 2010;132:10424–10428. doi: 10.1021/ja102797k. [DOI] [PubMed] [Google Scholar]
- 7.Okamoto A, Inasaki T, Saito I. Nitroxide-labeled guanine as an ESR spin probe for structural study of DNA. Bioorg. Med. Chem. Lett. 2004;14:3415–3418. doi: 10.1016/j.bmcl.2004.04.076. [DOI] [PubMed] [Google Scholar]
- 8.Schiemann O, Piton N, Plackmeyer J, Bode BE, Prisner TF, Engels JW. Spin labeling of oligonucleotides with the nitroxide TPA and use of PELDOR, a pulse EPR method, to measure intramolecular distances. Nat. Protoc. 2007;2:904–923. doi: 10.1038/nprot.2007.97. [DOI] [PubMed] [Google Scholar]
- 9.Sicoli G, Wachowius F, Bennati M, Hobartner C. Probing secondary structures of spin-labeled RNA by pulsed EPR spectroscopy. Angew Chem. Int. Ed. Engl. 2010;49:6443–6447. doi: 10.1002/anie.201000713. [DOI] [PubMed] [Google Scholar]
- 10.Sowa GZ, Qin PZ. Site-directed spin labeling studies on nucleic acid structure and dynamics. Prog. Nucleic Acid Res. Mol. Biol. 2008;82:147–197. doi: 10.1016/S0079-6603(08)00005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang XJ, Cekan P, Sigurdsson ST, Qin PZ. Methods in Enzymology. Vol. 469. Elsevier; 2009. pp. 303–328. http://www.sciencedirect.com/science/bookseries/00766879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barhate N, Cekan P, Massey AP, Sigurdsson ST. A nucleoside that contains a rigid nitroxide spin label: a fluorophore in disguise. Angew Chem. Int. Ed. Engl. 2007;46:2655–2658. doi: 10.1002/anie.200603993. [DOI] [PubMed] [Google Scholar]
- 13.Cekan P, Sigurdsson ST. Single base interrogation by a fluorescent nucleotide: each of the four DNA bases identified by fluorescence spectroscopy. Chem. Commun. 2008:3393–3395. doi: 10.1039/b801833b. [DOI] [PubMed] [Google Scholar]
- 14.Gardarsson H, Sigurdsson ST. Large flanking sequence effects in single nucleotide mismatch detection using fluorescent nucleoside C(f) Bioorg. Med. Chem. 2010;18:6121–6126. doi: 10.1016/j.bmc.2010.06.060. [DOI] [PubMed] [Google Scholar]
- 15.Cekan P, Jonsson EO, Sigurdsson ST. Folding of the cocaine aptamer studied by EPR and fluorescence spectroscopies using the bifunctional spectroscopic probe C. Nucleic Acids Res. 2009;37:3990–3995. doi: 10.1093/nar/gkp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cekan P, Smith AL, Barhate N, Robinson BH, Sigurdsson ST. Rigid spin-labeled nucleoside C: a nonperturbing EPR probe of nucleic acid conformation. Nucleic Acids Res. 2008;36:5946–5954. doi: 10.1093/nar/gkn562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marko A, Margraf D, Cekan P, Sigurdsson ST, Schiemann O, Prisner TF. Analytical method to determine the orientation of rigid spin labels in DNA. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2010;81:021911. doi: 10.1103/PhysRevE.81.021911. [DOI] [PubMed] [Google Scholar]
- 18.Schiemann O, Cekan P, Margraf D, Prisner TF, Sigurdsson ST. Relative orientation of rigid nitroxides by PELDOR: beyond distance measurements in nucleic acids. Angew Chem. Int. Ed. Engl. 2009;48:3292–3295. doi: 10.1002/anie.200805152. [DOI] [PubMed] [Google Scholar]
- 19.Myers JC, Shamoo Y. Human UP1 as a model for understanding purine recognition in the family of proteins containing the RNA recognition motif (RRM) J. Mol. Biol. 2004;342:743–756. doi: 10.1016/j.jmb.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 20.Shandrick S, Zhao Q, Han Q, Ayida BK, Takahashi M, Winters GC, Simonsen KB, Vourloumis D, Hermann T. Monitoring molecular recognition of the ribosomal decoding site. Angew Chem. Int. Ed. Engl. 2004;43:3177–3182. doi: 10.1002/anie.200454217. [DOI] [PubMed] [Google Scholar]
- 21.Lenz T, Bonnist EY, Pljevaljcic G, Neely RK, Dryden DT, Scheidig AJ, Jones AC, Weinhold E. 2-Aminopurine flipped into the active site of the adenine-specific DNA methyltransferase M.TaqI: crystal structures and time-resolved fluorescence. J. Am. Chem. Soc. 2007;129:6240–6248. doi: 10.1021/ja069366n. [DOI] [PubMed] [Google Scholar]
- 22.Fleissner MR, Cascio D, Hubbell WL. Structural origin of weakly ordered nitroxide motion in spin-labeled proteins. Protein Sci. 2009;18:893–908. doi: 10.1002/pro.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Z, Cascio D, Hideg K, Hubbell WL. Structural determinants of nitroxide motion in spin-labeled proteins: solvent-exposed sites in helix B of T4 lysozyme. Protein Sci. 2008;17:228–239. doi: 10.1110/ps.073174008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Z, Cascio D, Hideg K, Kalai T, Hubbell WL. Structural determinants of nitroxide motion in spin-labeled proteins: tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme. Protein Sci. 2007;16:1069–1086. doi: 10.1110/ps.062739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagelueken G, Ingledew WJ, Huang H, Petrovic-Stojanovska B, Whitfield C, ElMkami H, Schiemann O, Naismith JH. PELDOR spectroscopy distance fingerprinting of the octameric outer-membrane protein Wza from Escherichia coli. Angew Chem. Int. Ed. Engl. 2009;48:2904–2906. doi: 10.1002/anie.200805758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shelke SA, Sigurdsson ST. Noncovalent and site-directed spin labeling of nucleic acids. Angew Chem. Int. Ed. Engl. 2010;49:7984–7986. doi: 10.1002/anie.201002637. [DOI] [PubMed] [Google Scholar]
- 27.Sheldrick GM. A short history of SHELX. Acta Crystallogr. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 28.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Kudin KN, Burant JC, et al. Wallingford CT: Gaussian, Inc.; 2007. 2004. [Google Scholar]
- 29.Wilds CJ, Maier MA, Tereshko V, Manoharan M, Egli M. Direct observation of a cytosine analogue that forms five hydrogen bonds to guanosine: guanidino G-clamp. Angew Chem. Int. Ed. Engl. 2002;41:115–117. doi: 10.1002/1521-3773(20020104)41:1<115::aid-anie115>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 30.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. http://www.ncbi.nlm.nih.gov/pubmed/15299374. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bingman C, Li X, Zon G, Sundaralingam M. Crystal and molecular structure of d(GTGCGCAC): investigation of the effects of base sequence on the conformation of octamer duplexes. Biochemistry. 1992;31:12803–12812. doi: 10.1021/bi00166a014. [DOI] [PubMed] [Google Scholar]
- 37.Sines CC, McFail L, Howerton SB, Vanderveer D, Williams LD. Cations mediate B-DNA conformational heterogeneity. J. Am. Chem. Soc. 2000;122:11048–11056. [Google Scholar]
- 38.Egli M, Tereshko V, Teplova M, Minasov G, Joachimiak A, Sanishvili R, Weeks CM, Miller R, Maier MA, An H, et al. X-ray crystallographic analysis of the hydration of A- and B-form DNA at atomic resolution. Biopolymers. 1998;48:234–252. doi: 10.1002/(SICI)1097-0282(1998)48:4<234::AID-BIP4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 39.Smith AL, Cekan P, Brewood GP, Okonogi TM, Alemayehu S, Hustedt EJ, Benight AS, Sigurdsson ST, Robinson BH. Conformational equilibria of bulged sites in duplex DNA studied by EPR spectroscopy. J. Phys. Chem. B. 2009;113:2664–2675. doi: 10.1021/jp808260b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miao S, Brombosz SM, Schleyer PV, Wu JI, Barlow S, Marder SR, Hardcastle KI, Bunz UH. Are N,N-dihydrodiazatetracene derivatives antiaromatic? J. Am. Chem. Soc. 2008;130:7339–7344. doi: 10.1021/ja077614p. [DOI] [PubMed] [Google Scholar]
- 41.Schaefer M, Sheldrick GM, Bahner I, Lackner H. Crystal structures of actinomycin D and actinomycin Z3. Angew Chem. Int. Ed. Engl. 1998;37:2381–2384. doi: 10.1002/(SICI)1521-3773(19980918)37:17<2381::AID-ANIE2381>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 42.Hou MH, Robinson H, Gao YG, Wang AH. Crystal structure of actinomycin D bound to the CTG triplet repeat sequences linked to neurological diseases. Nucleic Acids Res. 2002;30:4910–4917. doi: 10.1093/nar/gkf619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson H, Gao YG, Yang X, Sanishvili R, Joachimiak A, Wang AH. Crystallographic analysis of a novel complex of actinomycin D bound to the DNA decamer CGATCGATCG. Biochemistry. 2001;40:5587–5592. doi: 10.1021/bi002859z. [DOI] [PubMed] [Google Scholar]
- 44.Takusagawa F, Takusagawa KT, Carlson RG, Weaver RF. Selectivity of F8-actinomycin D for RNA:DNA hybrids and its anti-leukemia activity. Bioorg. Med. Chem. 1997;5:1197–1207. doi: 10.1016/s0968-0896(97)00062-x. [DOI] [PubMed] [Google Scholar]
- 45.Engman KC, Sandin P, Osborne S, Brown T, Billeter M, Lincoln P, Norden B, Albinsson B, Wilhelmsson LM. DNA adopts normal B-form upon incorporation of highly fluorescent DNA base analogue tC: NMR structure and UV-Vis spectroscopy characterization. Nucleic Acids Res. 2004;32:5087–5095. doi: 10.1093/nar/gkh844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandin P, Borjesson K, Li H, Martensson J, Brown T, Wilhelmsson LM, Albinsson B. Characterization and use of an unprecedentedly bright and structurally non-perturbing fluorescent DNA base analogue. Nucleic Acids Res. 2008;36:157–167. doi: 10.1093/nar/gkm1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Magat Juan EC, Shimizu S, Ma X, Kurose T, Haraguchi T, Zhang F, Tsunoda M, Ohkubo A, Sekine M, Shibata T, et al. Insights into the DNA stabilizing contributions of a bicyclic cytosine analogue: crystal structures of DNA duplexes containing 7,8-dihydropyrido [2,3-d]pyrimidin-2-one. Nucleic Acids Res. 2010;38:6737–6745. doi: 10.1093/nar/gkq519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.