Abstract
Heterocyclic diamidines are compounds with antiparasitic properties that target the minor groove of kinetoplast DNA. The mechanism of action of these compounds is unknown, but topological changes to DNA structures are likely to be involved. In this study, we have developed a polyacrylamide gel electrophoresis-based screening method to determine topological effects of heterocyclic diamidines on four minor groove target sequences: AAAAA, TTTAA, AAATT and ATATA. The AAAAA and AAATT sequences have the largest intrinsic bend, whereas the TTTAA and ATATA sequences are relatively straight. The changes caused by binding of the compounds are sequence dependent, but generally the topological effects on AAAAA and AAATT are similar as are the effects on TTTAA and ATATA. A total of 13 compounds with a variety of structural differences were evaluated for topological changes to DNA. All compounds decrease the mobility of the ATATA sequence that is consistent with decreased minor groove width and bending of the relatively straight DNA into the minor groove. Similar, but generally smaller, effects are seen with TTTAA. The intrinsically bent AAAAA and AAATT sequences, which have more narrow minor grooves, have smaller mobility changes on binding that are consistent with increased or decreased bending depending on compound structure.
INTRODUCTION
Heterocyclic diamidines, and related compounds, with an appropriate structure can bind strongly to the DNA minor groove and exhibit important biological effects including clinical antiparasitic activity (1–4). Recent results show that the compounds can compete with transcription factor binding in both the minor and major grooves (5,6). The compounds can also target AT-rich sequences in mitochondrial kinetoplast minicircle DNA (kDNA) in eukaryotic parasites that cause human disease (1–4), and they have entered Phase III clinical trials against Human African Sleeping Sickness (4). The kinetoplast is composed of maxicircles that contain the mitochondrial genomic DNA and thousands of 1000–2000 bp catenated DNA minicircles that code for RNAs that guide editing of the maxicircle RNA transcripts (7). The kDNA minicircles have a number of unusual properties and structural features, such as short, curved A-tract sequences that are in phase with the helix repeat so that they always appear on the same side of the double helix and cause global bending of the helix (7–9). Given these unusual structural features, it seems likely that topological changes in the DNA minicircles induced by minor groove binding compounds can lead to kinetoplast destruction during opening of minicircles for transcription and replication and, thus, play a part in the antiparasitic activity of the compounds (1). Binding-induced changes in DNA topology are also important in the regulation of transcription of many genes, and modulation of bending can enhance or reduce transcription (10–12). Small molecules that can affect DNA bending in different ways at selected sites could, thus, provide tunable controls on the expression of the target genes.
In order to test the hypothesis that compound-induced topological changes in kDNA are important for their antiparasitic activity, a polyacrylamide gel electrophoresis (PAGE) ligation ladder assay (13–17), with phased AAAAA bent sequence and ATATA relatively unbent sequences, was established to determine whether heterocyclic compounds can cause topological changes in DNA (18). Out of phase ligation ladders of the same AT sequences were used as controls. Ligation ladders with repeated identical binding sites in phase with the helix twist amplify and simplify the analysis of compound topological effects at a specific sequence and different binding sites can be evaluated in the same context. The results in the initial assay showed quite clearly that the A-tract sequences were slightly straightened by DB75, while the compound significantly bent the alternating AT DNA sequence (18). Compounds with single atom changes in DB75 were also evaluated by replacing the furan with thiophene (DB351) and selenophene (DB1213) groups and compound-specific effects were observed. In these experiments the gel electrophoresis was done with the compound included in the gel at a concentration to maintain saturation of binding sites during the experiment (Method 1). The method of putting compound in the gel to help prevent dissociation has been successfully used with other compounds (19,20). The heterocyclic diamidines investigated here are positively charged at the typical buffer pH of 8.3, and thus migrate in a direction opposite that of the DNA during electrophoresis. Including the compound in the gel (Method 1), however, limits the experiment to analysis of one compound at one concentration per gel.
The interesting DNA topological changes observed on binding of DB75 and analogs suggested that it would be worthwhile to evaluate additional compounds as well as DNA sequences to determine the generality of the observations. Such studies will also help to establish relationships for compound structural effects on the topology of kDNA and other biologically important sequences. In order to increase the number of compounds and DNA sequences that can be evaluated per gel, a method that does not use compound in the gel is needed. Here a different PAGE method (Method 2), without compound in the gel, has been tested. Method 2 was tested with compounds of varied structure (Figure 1), and binding constants were determined to establish the limits for application of the method. The PAGE method without compound in the gel is able to qualitatively reproduce the more quantitative results with Method 1, and with Method 2 it is possible to study several compounds at different ratios to DNA sites with different DNAs on one gel.
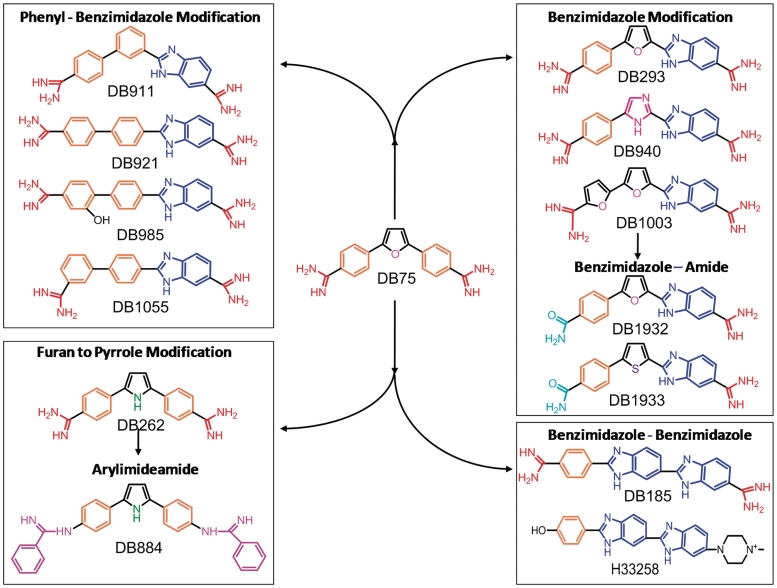
Figure 1.
Compounds grouped by structural similarity.
Compound selection
Compounds were selected for testing in the screening assay based on systematic modifications to each of the molecular units of DB75 (Figure 1). The compounds are grouped based on common structural motifs (Figure 1). The central furan of DB75 was replaced with a pyrrole (DB262) and from that structure the terminal amidines were changed to reversed amidines (DB884). One phenyl was changed to a benzimidazole (DB293) and from that compound the furan was changed to an imidazole (DB940), or the other flanking phenyl was replaced with a furan (DB1003). Another group had one amidine replaced with an amide (DB1932 and DB1933) such that they were monocations, not dications. DB185 is a different structure that is related to the well-known bisbenzimidazole, Hoechst 33258 (Figure 1). A final group of compounds has one phenyl of DB75 changed to benzimidazole and the furan converted to a phenyl. In this group DB911 has the substituents in a meta position at the central phenyl to give a classically curved minor groove binding shape, while an isomer has the central phenyl para substituted (DB921) to give a relatively linear compound. Two analogs of DB921, one with the addition of a hydroxyl group (DB985) and one with the position of the terminal phenyl amidine group in a meta position (DB1055), are included in this group.
MATERIALS AND METHODS
DNA binding compounds
Compounds in Figure 1 were synthesized by previously described methods (21–26), while the synthesis of DB940, DB1003, DB1932 and DB1933 will be published elsewhere. All the compounds were prepared in a 1-mM stock solution and stored at 4°C away from light.
DNA oligonucleotides
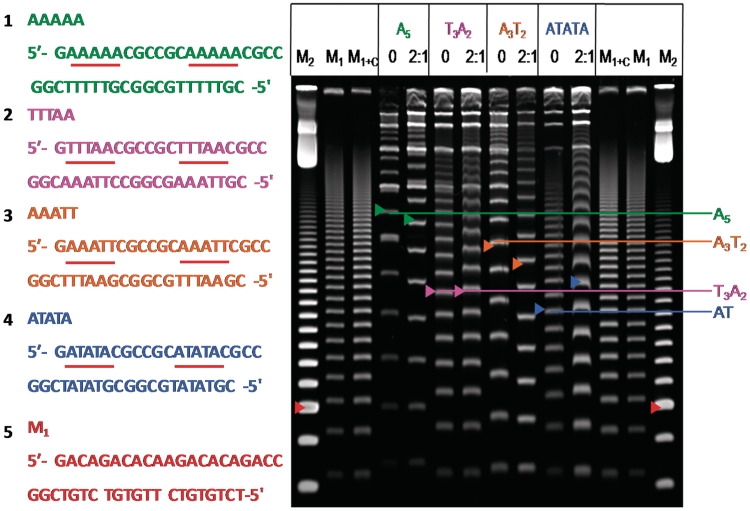
Two 21-mer duplexes (AAAAA and ATATA in Figure 2) from Tevis et al. (18) and two new duplexes (TTTAA and AAATT in Figure 2) were used in this study. The marker, M1, is also a 21 mer with the same base composition as the test duplexes, however; there is no AT binding site (18). Marker, M2, is a commercially available 20 bp ladder (Bayou Biolabs, Harahan, LA, USA) with a double intensity band at 100 bp. High-performance liquid chromatography (HPLC) purified and lyophilized DNA oligomers were purchased from Integrated DNA Technologies, Inc. (IDT, Coraville, IA, USA). The concentration of single-strand DNA was brought to ∼1 mM using water. The exact concentration was then determined using the extinction coefficient calculated by the nearest neighbor approximation. Then the complementary strands (100 µM) were combined in a 1:1 ratio based on the determined concentrations and annealed in 1× ligation buffer (New England BioLabs, Ipswich, MA, USA) containing 50 mM Tris–HCl, 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP and 25 mg/ml bovine serum albumin.
Figure 2.
Sequence of DNA duplexes used (left) and 8% PAGE gel of DB75 with the duplexes (right). The colored arrows indicate the 189-bp duplex for each ladder. The red arrow of M2 indicates 100 bp, while the red arrow of M1 indicates 189 bp of the random sequence marker. Electrophoresis was done for 2 h 20 min at 200 V and compound was not included in the gel. For each duplex, 0 indicates DNA that is not incubated with compound prior to electrophoresis, and the ratio 2:1 (compound to binding site) indicates DNA incubated with compound prior to electrophoresis. The brightest bands near the top of the lanes are circular products.
Ligation ladders
The ligation ladder protocol is the same as reported by Maher et al. (15,27,28) and Tevis et al. (18). Annealed duplexes (100 µM) were 5′-phosphorylated using T4 polynucleotide kinase (New England Biolabs) and then ligated using T4 DNA ligase (New England Biolabs) in the buffer provided. The kinase reactions were 200 µl of 6 µM DNA with 80 U of T4 polynucleotide kinase. The reaction was incubated for 30 min at 37°C followed by enzyme deactivation for 30 min at 65°C. Ligation reactions were 200 µl using 1200 U of ligase and ligation was done at room temperature for 20 min followed by enzyme deactivation for 25 min at 65°C.
Gel electrophoresis
Ligation ladders were separated using 8% native polyacrylamide (1.5 mm thick, 20 cm long) prepared from a 40% acrylamide stock solution (29:1 bis-acrylamide:acyrlamide; EMD, Gibbstwon, NJ, USA) in 1× TBE buffer (0.089 M Tris, 0.089 M boric acid, 2.0 mM EDTA, pH 8.3). Each sample had a final volume of 20 µl, which contained the following: 10 µl of 2 µM DNA, 4 µl of 6× load dye (Promega, Madison, WI, USA), the volume of compound needed for the desired ratio of compound to binding site and double distilled water to bring to the final volume, if needed. For the 1:1 and 2:1 ratios 3 and 6 µl of compound (13.3 µM) were added, respectively. Electrophoresis was done at 200 V (10 V/cm) for 140 min at 25°C ± 0.1°C in 1× TBE buffer. The quite different observed mobilities with compounds in adjacent lanes indicate that when using the 140-min time period, there is no significant interference from inter-lane diffusion when different compounds are used in the gel lanes. The lack of interference is also supported by the observation that compounds have the same mobilities when studied alone (Figures 2 and 3) or on a gel with other compounds (Figure 4).
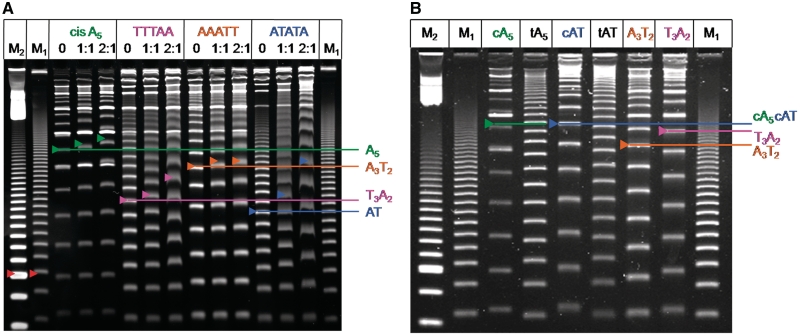
Figure 3.
(A) PAGE (8%) of cis-ligated sequences with DB921 not in the gel. Electrophoresis was done as in Figure 2. For each duplex, 0 indicates DNA that is not incubated with compound prior to electrophoresis, and the ratios (compound to binding site) indicate DNA is incubated with compound prior to electrophoresis. The colored arrows indicate 189 bp for each duplex. The red arrow of M2 indicates 100 bp, while the red arrow of M1 indicates 105 bp of the random sequence marker. (B) PAGE (8%) was done for 2 h 20 min at 200 V and compound was included in the gel. Experiments were done with the same cis sequences as in (A) and with trans-ligated DNA sequences of AAAAA (tA5) and ATATA (tAT). The colored arrows indicate 189 bp and the double intensity band of M2 is 100 bp.
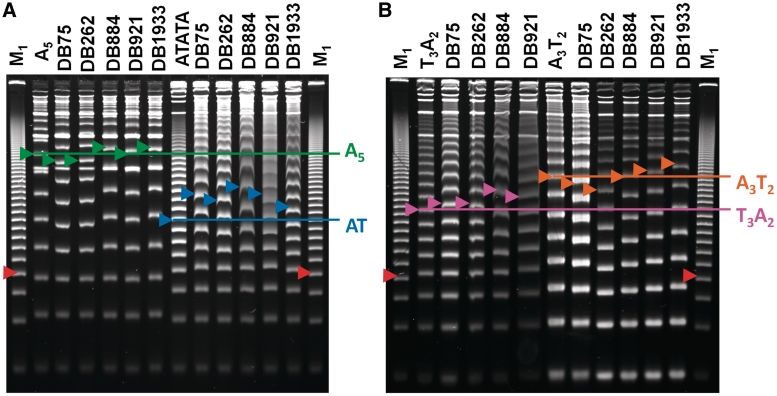
Figure 4.
PAGE (8%) of the DNA sequences (Figure 2) with multiple DB compounds. (A) The duplexes AAAAA and ATATA with five selected compounds, and (B) TTTAA and AAATT with the same five compounds. For each duplex, DNAs are incubated with compound prior to electrophoresis at a ratio of 2:1 (compound to binding site). The colored arrows indicate 189 bp for each duplex. The red arrow M1 indicates 105 bp of the random sequence marker. Electrophoresis was done as in Figure 2 and compound was not included in the gel.
After electrophoresis, gels were stained with SYBR Gold Nucleic Acid stain (Invitrogen, Carlsbad, CA, USA) at the concentration and time recommended by the manufacturer. The gels were imaged using an UltraLum Omega 10 gD Molecular Imaging System (UltraLum, Claremont, CA, USA).
Calculating RL
The analysis of topology changes was done with ImageQuantTL (Amersham Biosciences, Piscataway, NJ, USA). From the base pair (bp) assignments the parameter RL, where RL = Lapparent/Lactual, was calculated (18,27,28). The mobility of each band of the standard, M1, was determined and correlated with its length, in bp (Lact) and molecular weight. The mobilities of each band of the test DNAs and their complexes with diamidines were next determined and correlated to apparent bp lengths (Lapparent) and molecular weights based on the standard values. The RL value for each unbound DNA duplex was normalized to 1.00 so that the changes induced by compound binding could be more easily compared within each sequence.
Biosensor-surface plasmon resonance studies
AAAAA, ATATAT, AAATT and TTTAA hairpin duplexes with 5′-biotin groups were purchased from IDT (Integrated DNA Technology) with HPLC purification and mass spectrometry characterization. The oligomer sequences are all as follows: GCGWWWWWCGTCTCCGWWWWWCGC, where the W sequences are AAAAA·TTTTT, ATATAT·ATATAT, AAATT·AATTT, TTTAA·TTAAA and the hairpin loop is underlined. A 1-mM stock solution of each compound was prepared in water and diluted to working concentrations with buffer immediately before use. Surface plasmon resonance (SPR) measurements were performed with a four-channel Biacore 2000 optical biosensor system by using published procedures (29,30). For the duplex binding determinations, the 5′-biotin-labeled DNA samples were immobilized in separate flow cells on streptavidin-coated sensor chips (Biacore SA) with one flow cell left blank as a control. The SPR experiments were performed at 25°C in filtered, degassed 10 mM cacodylic acid buffer (pH 6.25) containing 100 mM NaCl, 1 mM EDTA and 0.005% surfactant P20. Compound solutions were prepared by serial dilution from the stock solution and injected from 7-mm plastic vials with pierceable plastic crimp caps at the flow rate of 25 µl/min. The compound was then replaced by buffer flow to monitor dissociation of the complex. The reference response from the blank cell was subtracted from the response in each cell containing DNA to give a signal (response units, RU) that is directly proportional to the amount of bound compound. Sensorgrams, RU versus time, at different concentrations for binding of each compound to DNA were obtained, and the RU in the steady-state region was determined by linear averaging over a selected time span. The number of binding sites and the equilibrium constant were obtained from fitting plots of RU versus Cfree. Binding results from the SPR experiments were fit with either a single site model (K2 = 0) or with a two-site model:
| (1) |
where r represents the moles of bound compound per mole of DNA hairpin duplex, K1 and K2 are macroscopic binding constants and Cfree is the free compound concentration in equilibrium with the complex.
RESULTS
Topological screening methods: comparison of two assays
In Figure 2 the A5 sequences show increased mobility (green arrows), while the ATATA sequences have a reduced mobility (blue arrows) relative to the same DNAs without DB75, and the results are very similar to those previously reported for the same sequences with the compound in the gel (18). Two new duplexes, TTTAA and AAATT, are also included in Figure 2 and the pink arrows show that DB75 does not have a detectable topological effect on TTTAA under these conditions, while the orange arrows show that DB75 increases the mobility of AAATT to an even greater extent than with A5. There are two alternative explanations for the mobility changes observed for DB75 and the other compounds in this study; the compounds could be changing the bend angles of the DNAs or they may be slightly changing the helical repeat, resulting in improved or decreased phasing of the AT repeats. Because the trans A5 sequence is already significantly dephased, an increase in the mobility of the A5 ladder is probably due to a decrease in the curvature of the A5 A-tracts. In the same manner the dramatic reduction in mobility of the ATATA sequence is highly unlikely to result from a phasing change because the sequence is straight no matter whether it is phased or not (18 and cis-trans in Figure 3). It seems likely then that bending rather than dephasing of helical repeats is responsible for the observed changes in mobility for these DNAs and compounds. Additional experiments with 20- and 22-bp duplexes would be required in order to definitively differentiate between phasing and bending or straightening of the AT sequences to account for the mobility changes.
The brightest bands near the top of each lane are circular products that appear for all DNAs as the molecular weight increases. The circular bands appear at lower molecular weights for the bent DNAs, AAAAA and AAATT than for the more linear duplexes. As can be seen in the figure, the compounds have very little effect on the mobilities of the circular DNAs. As compounds are added, the linear bands can change markedly in mobility while there are no detectable changes for the circular DNAs. The potential significance of this for biological activity is presented in the ‘Discussion’ section.
Figure 3 shows an additional comparison of gels done with DB921 by Method 1, which has DB921 in the gel (Figure 3B), and the new Method 2, where DB921 is added only to the separate DNAs and is not in the gel (Figure 3A). DB921 has a very different structure from classical minor groove binders with a more linear shape that incorporates an interfacial water molecule to bind in the minor groove (31). As can be seen, DB921 reduced the mobility of all four of the cis-ligated duplexes of interest by similar amounts in both assays and the results are quite different from those with DB75 (Figure 1; 18). This example shows that both methods give equivalent results for compounds of quite different structure. Interestingly, for the TTTAA and ATATA sequences there is a smearing of the bands in Figure 3A that is not present in Figure 3B and this smearing appears to be the result of partial dissociation of the DNA-compound complex during the gel experiment. In order to help clarify this difference, binding constants (see below) were determined for DB921 and show that it binds more weakly to TTTAA and ATATA than to AAAAA and AAATT. These results explain the smearing of DB921 bands in Figure 3A because it will more easily dissociate from the TTTAA and ATATA DNAs during the gel experiment. Note, however, that the general effects of DB921 on all the DNAs are comparable in both methods and can be determined from the screening method.
For comparison with our previous results (18), Figure 3 also includes the trans AAAAA and ATATA sequences. With DB75 we observed much smaller effects on both the bent AAAAA sequence and the straight ATATA sequence. DB921 reduces the mobility of the trans duplexes, but the effects are much less than with the cis sequences, as expected.
Effects of compound structure on DNA topology
In Figure 4, PAGE results for two DNAs and multiple compounds at a fixed ratio are compared. Topological effects of the compounds used in these gels agree with the results in Figures 2 and 3 and Supplementary Data. The screening method with multiple compounds, thus, allows for rapid testing and direct comparisons of topological changes for compounds in a library. In Figure 4 the 2:1 ratio is used, because smaller effects are seen at a 1:1 ratio and going to a 3:1 ratio did not generally cause large changes in mobility with these DNA sequences and compounds. The PAGE results for all the other compounds in Figure 1 can be seen in Supplementary Data. In the gels for DB1003 and DB185 (Supplementary Figures SM4 and SM10, respectively) additional ratios were included and none of the extra ratios changed the conclusions. To accommodate the extra ratios, the ATATA sequence was omitted for DB1003 and DB185. These two compounds were selected for this test because DB1003 is a relatively weak binder (32), while DB185 is a very strong minor groove binding agent (33).
Using RL to compare topological changes
RL values for each DNA and complex were calculated relative to the mobility of the standard as described in the ‘Materials and Methods’ section. Table 1 compares RL values of several compounds that were determined in both assays. The topological changes shown in Table 1 are similar by both assays, however; for some compounds and sequences the effects on RL are larger by Method 1. This difference is not unexpected from the results in Figure 3 and as noted above, is due to partial dissociation of the complex in the screening assay (see binding results below). The trend and general magnitude of the changes, however, can be determined without compound in the gel. For quantitative determination of bending angle changes on compound binding, however, Method 1 should be used.
Table 1.
RL values for both assays for each DNA sequence
| AAAAA |
TTTAA |
AAATT |
ATATA |
|||||
|---|---|---|---|---|---|---|---|---|
| Drug | C | No C | C | No C | C | No C | C | No C |
| DB75 | 0.77 | 0.82 | 1.20 | 1.01 | 0.75 | 0.86 | 1.31 | 1.19 |
| DB262 | 0.92 | 0.95 | 1.25 | 1.06 | 0.76 | 0.81 | 1.48 | 1.27 |
| DB884 | 1.22 | 1.11 | 1.16 | 1.08 | 1.05 | 1.03 | 1.19 | 1.13 |
| DB921 | 1.1 | 1.12 | 2.1 | 1.33 | 1.09 | 1.08 | 2.5 | 1.9 |
C indicates compound in the gel matrix (Method 1), and No C indicates no compound in the gel matrix (Method 2). The Method 2 comparison uses the RL values at the 2:1 ratio of compound to binding site.
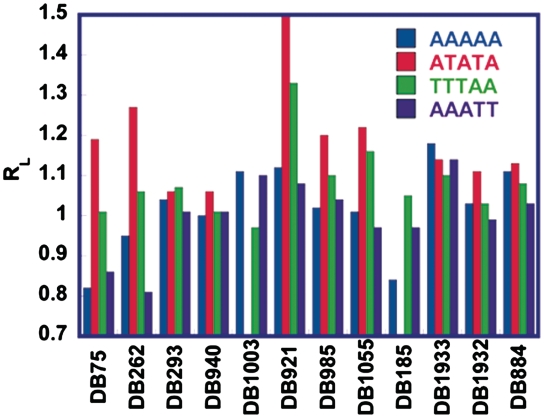
To assist in comparing results for different DNAs, all RL values were normalized by dividing them by the RL value of the free DNA, and the results are plotted in Figure 5. A compound with an RL value greater than 1 indicates the sequence had lower mobility than the free DNAs and is consistent with bending, while a value below one is consistent with straightening of the sequence. Interestingly, the ATATA sequence was bent by all compounds tested although the magnitude of bending varied with compound structures. DB921 and DB911 show the largest induced bending, followed by DB262 and DB75. All three of the monocations caused a small reduction of mobility and a small bend with the ATATA sequence.
Figure 5.
Histogram showing RL values for all compounds with AAAAA, ATATA, TTTAA and AAATT sequences. The blue represents AAAAA, red ATATA, the green TTTAA and the purple AAATT. Each group represents a structural group from Figure 1. All the data shown are for a 2:1 ratio of compound to binding site.
The changes in RL for the AAAAA sequence were both positive and negative and were generally smaller than the changes with ATATA. The following compounds cause a decrease in mobility, consistent with increased bending of the AAAAA sequence: DB1933, DB921, DB1003, DB911 and DB884. DB75 and DB185, on the other hand, show an increase in mobility and DB262, the pyrrole analog of DB75 increase mobility but to a lesser extent. Interestingly, the monocation, DB1933, causes the largest decrease in mobility in AAAAA. DB884, with a reverse amidine cation, also decreased the mobility of the intrinsically bent AAAAA sequence. The compounds DB293, DB940, DB985, DB1055 and DB1932 did not have a significant topological effect on the AAAAA sequence. In general, the results with AAATT are similar to those with AAAAA.
For the TTTAA sequence the group containing the phenyl-benzimidazole modification (Figure 1) shows the largest effect on bending. DB921 and DB911 increase bending more than DB1055 that bends more than DB985. DB262 increased the degree of bending more than DB75. For the benzimidazole modification group, DB293 caused the largest bending effect with TTTAA and the bis-benzimidazole DB185 shows a similar effect. The effects with DB75, DB940, DB1003 and DB1932 are not significant for the sequence.
Biosensor-SPR determination of equilibrium binding constants (K)
Binding assays were conducted for the compounds from the topological studies to answer two important questions: (i) what is the minimum K value that is necessary before Method 2 can be used without compound in the gel and (ii) is there any correlation of the K and RL values. Because three biotin-labeled DNAs can be immobilized on four-channel Biacore streptavidin-coated sensor chips with one flow cell left as a blank (29,30), two sensor chips were used to evaluate K values for the four DNA AT-binding sequences used in the topology-ligation assays. This provided repeat values for two DNAs to give a better evaluation of reproducibility of K values determined with completely different experimental sensor chips, samples, etc. The variation in K is <10% in the repeat assays.
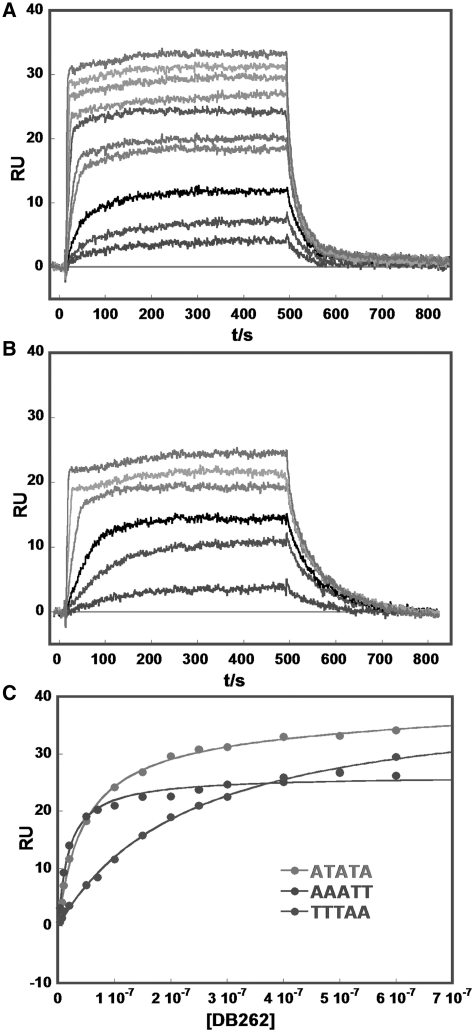
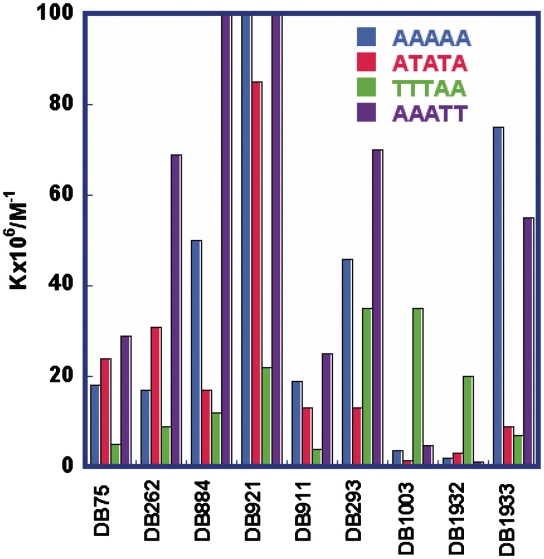
Example sensorgrams for DB262, which has similar structure and DNA binding to DB75, show that the compound has relatively fast association and dissociation kinetics for binding to ATATA and AAATT (Figure 6A and B). In this case K values can be determined by fitting the kinetic constants for association (ka) and dissociation (kd) or from the steady-state response where the rates of association and dissociation are equal (Figure 6A and B). The steady-state method is not subject to any mass transport issues at the sensor surface, and steady-state RU values for DB262 and three DNA sequences are plotted in Figure 6C. K values were determined by fitting the plots in Figure 6C to one or two site functions as described in the ‘Materials and Methods’ section. In all cases the K1 values are significantly larger than the K2 values when a two-site fit was required, for example, with the ATATA and TTTAA DNAs (Figure 6C). A simple two-site binding function can be used in these cases because the primary binding is for complex formation at the AT target sequences while the much weaker, secondary binding is generally a non-specific electrostatic interaction (34). Results for DB921, which has slower association and dissociation rates, are shown in Supplementary Figure SM15, Supplementary Data. K values for DB921 and related compounds were determined by global fitting of the sensorgrams to determine ka and kd values as a function of the compound concentration in the flow solution. DB921, DB985 and DB1055 are generally quite strong minor groove binding compounds. For visualization, primary binding constants (K1) are compared for representative compounds as a histogram in Figure 7, and the actual K values are tabulated in Supplementary Figure SM15.
Figure 6.
Example biosensor-SPR sensorgrams (instrument response in RU values versus time) for binding of DB262 to (A) ATATA and (B) AAATT at 25°C. The unbound compound concentrations in the flow solutions range from 0.005 µM in the lowest curve to 1 µM in the top curve. Experiments were conducted in cacodylic acid buffer with 0.1 M NaCl. (C) SPR binding plots for DB262 in the presence of ATATA (top), AAATT (middle) and TTTAA (lower). The RU values from the steady-state region of SPR sensorgrams are plotted versus the unbound compound concentration. The RUs and the concentrations of DB262 are from repeat experiments. The lines in the Figure are for one or two-site fitting (‘Methods’ section). K values for AAATT were obtained by using a one-site model and for ATATA and TTTAA with a two-site model to get K1 and K2. K2 was much lower than K1 and only K1 values were used in the comparison plot in Figure 7.
Figure 7.
Histograms of equilibrium constants, K (M−1) for representative compounds binding to ATATA, AAAAA, AAATT and TTTAA.
As can be seen, there is a large variation in binding constants for the compounds with different DNAs. In general, the highest binding constants were observed with the AAATT site and the lowest with the reversed sequence TTTAA. Significant outliers in this pattern are DB1003 and DB1932, which bind best to TTTAA as expected for compounds with that structure (32). The compounds with the largest K values in this group are DB921 and analogues, but they also have relatively low K values for TTTAA and the highest values for AAAAA and AAATT. The two pyrrole compounds, DB262 and DB884, have quite large values for AAATT relative to their Ks for the other three DNAs. Most compounds have larger K values for the non-alternating, AAAAA, than for the alternating ATATA sequence. Interestingly, the diamidines, DB75 and DB262, with a diphenyl-5-member ring structure, have larger values for ATATA. As will be discussed below and as shown in Supplementary Figure SM16, there is no direct correlation of binding constants with the mobility changes for the compounds and DNAs.
DISCUSSION
Experiments with ligation ladders from DNA sequences with AAAAA and ATATA binding sites have shown that DB75 and some close analogs induce topological changes in DNA that are sensitive to DNA sequence/structure and to compound structure and properties (18). At present there is considerable knowledge of the effects of protein binding on DNA topology but much less information about small molecules that target DNA. Such compounds are being designed for therapeutic applications that range from antiparasitic to anticancer activities (1–4), and in order to develop these agents in a rational way we need to understand the topological effects at the target sequences. Topological changes on binding of the heterocyclic diamidines of Figure 1 to the mitochondrial kinetoplast DNA of disease-causing eukaryotic parasites may be a key step in destruction of the kinetoplast and the biological mechanism of action of the compounds (3). To clarify this question, more topological information on the effects of the compounds on kinetoplast target sequences is required.
Method 1, with compound in the gel to prevent the cationic compounds from dissociating from DNA during electrophoresis, has been developed for quantitative evaluation of compound-induced topological changes on complex formation (18). This method, however, allows only one compound at one concentration (ratio per DNA binding sites) on each gel. In this work, Method 2, in which the compounds are added to DNA at different ratios but are not in the gel, was tested. This method allows the possibility of different compounds on each gel and on a single gel with 20 lanes, for example two ratios of different compounds can be tested with up to four different DNAs, free DNA and appropriate standards. Figure 3, with one compound, and Table 1 with four compounds and four DNAs compare results by Methods 1 and 2. In Table 1 the changes observed by Method 1 are generally greater than those with Method 2, as expected if there is some compound dissociation and loss during electrophoresis. Partial dissociation is supported by results in Figure 3, where some smearing is seen in gel A (Method 2) with the TTTAA and ATATA sequences, but no smearing is observed in gel B (Method 1). The smearing effect is observed with compounds and DNAs where there is a large difference in RL with a relatively low K value. The smearing, for example, occurs with the TTTAA and ATATA sequences, and DB884, which has lower binding constants with these DNAs than with AAAAA and AAATT (Figure 7 and Supplementary Figure SM1). The encouraging aspect of these comparisons, however, is that in all cases the direction of the changes and general relative magnitude is reported identically by the two methods. These results indicate that Method 2 can be used for qualitative analysis of induced topological changes by small cations that target DNA. For compounds that are found to be particularly interesting from these results, more quantitative, but slower, studies can be done by Method 1 as necessary.
With these encouraging results for Method 2, several groups of minor groove binding compounds of quite different structures and properties (Figure 1) were selected for a more extensive, comparative analysis of induced topological effects on complex formation. The set of compounds and DNAs is satisfactory to determine if the induced changes are dominated by either compound or DNA structure and properties, and also to look for any patterns in the results. A qualitative conclusion from the results that are summarized in Figure 5 for RL and Figure 7 for K is that there is no direct correlation between the magnitude and direction of the induced topological changes and the binding affinity, as shown in Supplementary Figure SM16. As will be described below, the observed topological changes appear to be best correlated with DNA intrinsic minor groove width as well as with compound shape and properties.
The AAAAA minor groove is intrinsically narrow with ordered water and cations stabilizing the close approach of phosphate anions across the groove (35,36) and as a result, this sequence is the most intrinsically bent in our set of four DNAs (27,28). The groove is of the correct width to bind most compounds of Figure 1 with little change in width. The compounds can cause a slight increase in bending by stabilizing a slightly more narrow minor groove with increased bending into that groove. Decreased bending, which can be caused by the opposite effect of a slight widening of the groove, might be generally expected, and for this compound set it is observed for most complexes. Effects on the AAATT sequence, which also has A-tract properties and significant bending for the free DNA, are generally similar to those for AAAAA and can be either in the direction of increased or decreased bending.
The ATATA sequence is the least intrinsically bent DNA in our set of four (18,37–42) and this DNA has a wider groove than AAAAA or AAATT. The groove is too wide to bind tightly to the compounds of Figure 1 without some structural changes. Minor groove binding that decreases the groove to better fit the general width of the compounds should bend the DNA into the minor groove. All compounds of Figure 1 whether mono or dications, with or without a benzimidazole group, and with amidines or a reversed structure (DB884) bend ATATA indicating that topological effects with this sequence, are dominated by the DNA structure. The benzimidazole-biphenyl isomers, DB921 and DB911, cause the most bending, and the related compounds, DB985 and DB1055, cause a smaller but significant amount of bending. The biphenyl motif of these compounds is intrinsically twisted by 30–40° and this may explain the relatively large bending effects of the compound family. Replacing the central phenyl by a furan, DB293, or with an imidazole, DB940, results in a planar system with smaller induced bends in ATATA. The diphenyl furan, DB75 and pyrrole, DB262, compounds are also planar, however; and cause significant bending. Clearly compound shape and perhaps other features are important in addition to compound twist. The clear pattern with ATATA is that all minor groove binders studied to date cause bending of that sequence. The magnitude of bending, however, depends on the interaction of the compound structure and chemical groups with the ATATA groove. The TTTAA sequence also has a wider groove than the A-tract sequences and it has a much lower degree of bending (41,42). Compounds also generally show bending on complex formation with this DNA, but the bending magnitude is lower than with ATATA. This could be due to the fact that compounds, such as DB1003 and DB293, have been shown to bind to the TTAA sequence as a dimer (32), which will maintain a wider groove.
It is a very interesting result that the circular DNA bands in Figures 2–4 (the bright bands in the upper part of the gels, see Figure 2) do not change mobility by any significant amount when compound is added. This is true even though they have the same sequence as the linear DNAs and even when the mobilities of the linear bands change by a large amount. Clearly constraining the ends of these sequences is the dominant feature of their shape and mobility in PAGE. Because the kinetoplast DNAs are circular, this observation suggests that the compounds would have relatively little effect on structure until the DNAs were opened for transcription or replication by mitochondrial enzymes. At this point their topology could change significantly and closing the DNAs to reform the kinetoplasts would be inhibited. Given the thousands of minicircles in each kinetoplast (7), even a small amount of inhibition could lead to very significant destruction of the kinetoplast structure and network.
In summary, the major features of both induced topological changes and of compound binding constants for minor groove complex formation are dominated by the groove characteristics of DNA AT sequences. There are, however, major variations within a DNA sequence for changes in compound structure and properties. The screening method described in this report can now be used with a quite large set of DNA binding agents to determine correlations with molecular structure and induced topological changes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIAID AI64200 to W.D.W. and D.W.B.). Funding for open access charge: National Institutes of Health (NIAID AI64200).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Wilson WD, Tanious FA, Mathis A, Tevis D, Hall JE, Boykin DW. Antiparasitic compounds that target DNA. Biochimie. 2008;90:999–1014. doi: 10.1016/j.biochi.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathis AM, Bridges AS, Ismail MA, Kumar A, Francesconi I, Anbazhagan M, Hu Q, Tanious FA, Wenzler T, Saulter J, et al. Diphenyl furans and aza analogs: effects of structural modification on in vitro activity, DNA binding, and accumulation and distribution in trypanosomes. Antimicrob. Agents Chemother. 2007;51:2801–2810. doi: 10.1128/AAC.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soeiro MN, de Castro SL, de Souza EM, Batista DG, Silva CF, Boykin DW. Diamidine activity against trypanosomes: the state of the art. Curr. Mol. Pharmacol. 2008;2:151–161. doi: 10.2174/1874467210801020151. [DOI] [PubMed] [Google Scholar]
- 4.Paine MF, Wang MZ, Generaux CN, Boykin DW, Wilson WD, De Koning HP, Olson CA, Pohlig G, Burri C, Brun R, et al. Diamidines for human African trypanosomiasis. Curr. Opin. Investig. Drugs. 2010;11:876–883. [PubMed] [Google Scholar]
- 5.Peixoto P, Liu Y, Depauw S, Hildebrand MP, Boykin DW, Bailly C, Wilson WD, David-Cordonnier MH. Direct inhibition of the DNA-binding activity of POU transcription factors Pit-1 and Brn-3 by selective binding of a phenyl-furan-benzimidazole dication. Nucleic Acids Res. 2008;36:3341–3353. doi: 10.1093/nar/gkn208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miao Y, Cui T, Leng F, Wilson WD. Inhibition of high-mobility-group A2 protein binding to DNA by netropsin: a biosensor-surface plasmon resonance assay. Anal. Biochem. 2008;374:7–15. doi: 10.1016/j.ab.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu B, Liu Y, Motyka SA, Agbo EC, Englund PT. Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol. 2005;21:363–369. doi: 10.1016/j.pt.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Marini JC, Levene SD, Crothers DM, Englund PT. Bent helical structure in kinetoplast DNA. Proc. Natl Acad. Sci. USA. 1982;79:7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marini JC, Effron PN, Goodman TC, Singleton CK, Wells RD, Wartell RM, Englund PT. Physical characterization of a kinetoplast DNA fragment with unusual properties. J. Biol. Chem. 1984;259:8974–8979. [PubMed] [Google Scholar]
- 10.Garcia HG, Grayson P, Han L, Inamdar M, Kondev J, Nelson PC, Phillips R, Widom J, Wiggins PA. Biological consequences of tightly bent DNA: the other life of a macromolecular celebrity. Biopolymers. 2007;85:115–130. doi: 10.1002/bip.20627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chenoweth DM, Dervan PB. Allosteric modulation of DNA by small molecules. Proc. Natl Acad. Sci. USA. 2009;106:13175–13179. doi: 10.1073/pnas.0906532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crothers DM. DNA curvature and deformation in protein-DNA complexes: a step in the right direction. Proc. Natl Acad. Sci. USA. 1998;95:15163–15165. doi: 10.1073/pnas.95.26.15163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diekmann S. Analyzing DNA curvature in polyacrylamide gels. Methods Enzymol. 1992;212:30–46. doi: 10.1016/0076-6879(92)12004-a. [DOI] [PubMed] [Google Scholar]
- 14.Crothers DM, Drak J. Global features of DNA structure by comparative gel electrophoresis. Methods Enzymol. 1992;212:46–71. doi: 10.1016/0076-6879(92)12005-b. [DOI] [PubMed] [Google Scholar]
- 15.Ross ED, Den RB, Hardwidge PR, Maher LJ., III Improved quantitation of DNA curvature using ligation ladders. Nucleic Acids Res. 1999;27:4135–4142. doi: 10.1093/nar/27.21.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maki AS, Kim T, Kool ET. Direct comparison of A- and T-strand minor groove interactions in DNA curvature at A tracts. Biochemistry. 2004;43:1102–1110. doi: 10.1021/bi035340m. [DOI] [PubMed] [Google Scholar]
- 17.Zewail-Foote M, Hurley LH. Ecteinascidin 743: a minor groove alkylator that bends DNA toward the major groove. J. Med. Chem. 1999;42:2493–2497. doi: 10.1021/jm990241l. [DOI] [PubMed] [Google Scholar]
- 18.Tevis DS, Kumar A, Stephens CE, Boykin DW, Wilson WD. Large, sequence-dependent effects on DNA conformation by minor groove binding compounds. Nucleic Acids Res. 2009;37:5550–5558. doi: 10.1093/nar/gkp558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cons BM, Fox KR. Effects of sequence selective drugs on the gel mobility of a bent DNA fragment. Biochem. Biophys. Res. Commun. 1990;171:1064–1070. doi: 10.1016/0006-291x(90)90792-l. [DOI] [PubMed] [Google Scholar]
- 20.Persil O, Santani CT, Jain SS, Hud NV. Assembly of an antiparallel homo-adenine DNA duplex by small-molecule binding. J. Am. Chem. Soc. 2004;126:8644–8645. doi: 10.1021/ja0492891. [DOI] [PubMed] [Google Scholar]
- 21.Das BP, Boykin DW. Synthesis and antiprotozoal activity of 2,5-bis(4-guanylphenyl)furans. J. Med. Chem. 1977;20:531–536. doi: 10.1021/jm00214a014. [DOI] [PubMed] [Google Scholar]
- 22.Dann O, Fick H, Pietaner B, Walkenhorst E, Fernbach R, Zeh D. Trypanocidal diamidines with three isolated ring systems. Justus Liebigs Annalen der Chemie. 1975;1:160–194. [Google Scholar]
- 23.Munde M, Lee M, Neidle S, Arafa R, Boykin DW, Liu Y, Bailly C, Wilson WD. Induced fit conformational changes of a “reversed amidine” heterocycle: optimized interactions in a DNA minor groove complex. J. Am. Chem. Soc. 2006;129:5688–5698. doi: 10.1021/ja069003n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hopkins KT, Wilson WD, Bender BC, McCurdy DR, Hall JE, Tidwell RR, Kumar A, Bajic M, Boykin DW. Extended aromatic furan amidino derivatives as anti-pneunocystis carinii agent. J. Med. Chem. 1998;41:3872–3878. doi: 10.1021/jm980230c. [DOI] [PubMed] [Google Scholar]
- 25.Ismail MA, Batista-Parra A, Miao Y, Wilson WD, Wenzler T, Brun R, Boykin DW. Dicationic near-linear biphenyl benzimidazole derivatives as DNA-targeted antiprotozoal agents. Bioorg. Med. Chem. 2005;13:6718–6726. doi: 10.1016/j.bmc.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 26.Czarny A, Wilson WD, Boykin DW. Synthesis of mono-cationic and dicationic analogs of Hoechst 33258. J. Hetero. Chem. 1996;33:1393–1397. [Google Scholar]
- 27.Hardwidge PR, Maher LJ., III Experimental evaluation of the Liu-Beveridge model of DNA structure. Nucleic Acids Res. 2001;29:2619–2625. doi: 10.1093/nar/29.12.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardwidge PR, Den RB, Ross ED, Maher LJ., III Relating independent measures of DNA curvature: electrophoretic anomaly and cyclization efficiency. J. Biomol. Struct. Dyn. 2000;18:219–230. doi: 10.1080/07391102.2000.10506660. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen B, Tanious FA, Wilson WD. Biosensor-surface plasmon resonance: quantitative analysis of small molecule-nucleic acid interactions. Methods. 2007;42:150–161. doi: 10.1016/j.ymeth.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Wilson WD. Quantitative analysis of small molecule-nucleic acid interactions with a biosensor surface and surface plasmon resonance detection. Methods Mol. Biol. 2010;613:1–23. doi: 10.1007/978-1-60327-418-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao Y, Lee MP, Parkinson GN, Batista-Parra A, Ismail MA, Neidle S, Boykin DW, Wilson WD. Out-of-shape DNA minor groove binders: induced fit interactions of heterocyclic dications with the DNA minor groove. Biochemistry. 2005;44:14701–14708. doi: 10.1021/bi051791q. [DOI] [PubMed] [Google Scholar]
- 32.Munde M, Kumar A, Nhili R, Depauw S, David-Cordonnier MH, Ismail MA, Stephens CE, Farahat AA, Batista-Parra A, Boykin DW, et al. DNA minor groove induced dimerization of heterocyclic cations: compound structure, binding affinity, and specificity for a TTAA site. J. Mol. Biol. 2010;402:847–864. doi: 10.1016/j.jmb.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanious FA, Hamelberg D, Bailly C, Czarny A, Boykin DW, Wilson WD. DNA sequence dependent monomer-dimer binding modulation of asymmetric benzimidazole derivatives. J. Am. Chem. Soc. 2004;126:143–153. doi: 10.1021/ja030403+. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen B, Wilson WD. The effects of hairpin loops on ligand-DNA interactions. J. Phys. Chem. B. 2009;113:14329–14335. doi: 10.1021/jp904830m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hud NV, Polak M. DNA-cation interactions: the major and minor grooves are flexible ionophores. Curr. Opin. Struct. Biol. 2001;11:293–301. doi: 10.1016/s0959-440x(00)00205-0. [DOI] [PubMed] [Google Scholar]
- 36.Hamelberg D, Williams LD, Wilson WD. Influence of the dynamic positions of cations on the structure of the DNA minor groove: sequence-dependent effects. J. Am. Chem. Soc. 2001;123:7745–7755. doi: 10.1021/ja010341s. [DOI] [PubMed] [Google Scholar]
- 37.Fox KR. Probing the conformations of eight cloned DNA dodecamers; CGCGAATTCGCG, CGCGTTAACGCG, CGCGTATACGCG, CGCGATATCGCG, CGCAAATTTGCG, CGCTTTAAAGCG, CGCGGATCCGCG and CGCGGTACCGCG. Nucleic Acids Res. 1992;20:6487–6493. doi: 10.1093/nar/20.24.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abu-Daya A, Brown PM, Fox KR. DNA sequence preferences of several AT-selective minor groove binding ligands. Nucleic Acids Res. 1995;23:3385–3392. doi: 10.1093/nar/23.17.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burkhoff AM, Tullius TD. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987;48:935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- 40.Hud NV, Feigon J. Characterization of divalent cation localization in the minor groove of the A(n)T(n) and T(n)A(n) DNA sequence elements by (1)H NMR spectroscopy and manganese(II) Biochemistry. 2002;41:9900–9910. doi: 10.1021/bi020159j. [DOI] [PubMed] [Google Scholar]
- 41.Stefl R, Wu H, Ravindranathan S, Sklenar V, Feigon J. DNA A-tract bending in three dimensions: solving the dA4T4 vs. dT4A4 conundrum. Proc. Natl Acad. Sci. USA. 2004;101:1177–1182. doi: 10.1073/pnas.0308143100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.