Abstract
Each family of signal transduction systems requires specificity determinants that link individual signals to the correct regulatory output. In Bacillus subtilis, a family of four anti-terminator proteins controls the expression of genes for the utilisation of alternative sugars. These regulatory systems contain the anti-terminator proteins and a RNA structure, the RNA anti-terminator (RAT) that is bound by the anti-terminator proteins. We have studied three of these proteins (SacT, SacY, and LicT) to understand how they can transmit a specific signal in spite of their strong structural homology. A screen for random mutations that render SacT capable to bind a RNA structure recognized by LicT only revealed a substitution (P26S) at one of the few non-conserved residues that are in contact with the RNA. We have randomly modified this position in SacT together with another non-conserved RNA-contacting residue (Q31). Surprisingly, the mutant proteins could bind all RAT structures that are present in B. subtilis. In a complementary approach, reciprocal amino acid exchanges have been introduced in LicT and SacY at non-conserved positions of the RNA-binding site. This analysis revealed the key role of an arginine side-chain for both the high affinity and specificity of LicT for its cognate RAT. Introduction of this Arg at the equivalent position of SacY (A26) increased the RNA binding in vitro but also resulted in a relaxed specificity. Altogether our results suggest that this family of anti-termination proteins has evolved to reach a compromise between RNA binding efficacy and specific interaction with individual target sequences.
INTRODUCTION
The development of new genetic properties occurs usually by duplication of existing genes that adapt to new functions rather than by de novo ‘invention’ of genes and proteins. This mode of evolution resulted in large families of enzymes that act on similar but distinct substrates and that carry out similar functions. Similarly, the regulatory repertoire of all organisms is made up of members of a rather small number of regulator families. Usually, the activity of members of one family is controlled in a similar way and the proteins interact with similar regulatory targets. Well-studied examples for such families of regulators are the different families of two-component regulatory systems or the LacI-GalR family of transcription regulators (1,2). This evolution is still going on, and it can be observed by studying the degradation of artificial pollutants and other xenobiotics (3).
We are interested in a family of bacterial RNA-binding regulatory proteins, the BglG/SacY family. These proteins control the expression of genes and operons required for the utilization of specific carbohydrates such as glucose, sucrose, lactose and β-glucosides. They are composed of a N-terminal RNA-binding domain (also called co-anti-terminator, CAT) and two reiterated regulatory domains that receive signals from the phosphoenolpyruvate:sugar phosphotransferase system (PTS) (4,5,6). The Gram-positive soil bacterium Bacillus subtilis possesses four regulatory systems that involve RNA-binding proteins of this family. The best studied of these proteins, LicT, controls the expression of the licS gene and the bglPH operon (7,8). Transcription of these genes is constitutively initiated but stops at a terminator structure upstream of the coding region unless β-glucosides are present and preferred carbon sources such as glucose are absent. Transcription beyond the terminator structure requires binding of the anti-terminator protein LicT to an mRNA sequence that partially overlaps the terminator (9). This RNA sequence, also called RNA anti-terminator (RAT) can adopt a secondary structure that is mutually exclusive with the formation of the transcription terminator. However, the terminator structure is much more stable, and thus the RAT structure can only form upon binding of the anti-terminator protein LicT. The activity of LicT is controlled by phosphorylation events in the PTS regulatory domains (PRDs). In the absence of β-glucosides, the β-glucoside permease of the PTS, encoded by bglP, was proposed to phosphorylate and thereby inactivate LicT on conserved histidine residues in the first PRD (10). If β-glucosides are available, the phosphate groups are drained to the substrate and the PRD-I of LicT is non-phosphorylated. Under these conditions, the availability of glucose decides whether LicT is active or not: if glucose or other preferred sugars are absent, the HPr protein of the PTS is phosphorylated on its His-15 (11), and this form of HPr can phosphorylate the PRD-II of LicT and thereby activate the protein (12). In the presence of glucose, there is not sufficient HPr (His-P) present, and LicT cannot be activated by HPr-dependent phosphorylation. The phosphorylation state of the PRDs is relayed to the CAT domain of LicT by a structural transition of the linker region between the CAT and PRD-I: this transition results in the stabilization of the CAT dimer and allows RNA binding (13).
In addition to LicT, the GlcT anti-terminator protein controls the expression of the ptsG gene encoding the glucose permease of the PTS, and SacT and SacY regulate the sacPA and sacB genes, respectively, that are involved in sucrose utilization [for a review see (6)]. As described for LicT, the cognate sugar-specific PTS permeases and HPr phosphorylate, these proteins thus control their activity (14). If properly phosphorylated, they bind to their respective RAT structures in the ptsG or sacPA and sacB mRNAs and cause transcriptional anti-termination. These four regulatory systems share multiple levels of similarity: (i) the anti-termination proteins are conserved, (ii) the PTS components that phosphorylate the PRD-I are similar to each other and (iii) the RAT structures recognized by the CATs of the four anti-terminator proteins also share extensive similarity (Figure 1). Thus, it is not surprising that cross-talk between the anti-termination proteins and non-cognate RAT structures was observed (15,16). The complex formed between the CAT of LicT and the bglPH RAT has been studied by NMR, and it turned out that LicT contacts bases in the two internal loops (or bulges) of the RAT. The basic stretch at the N-terminus of the CAT (residues 5–10) and the residues Gly-26, Arg-27, Phe-31 and Gln-32 are involved in these contacts (17).
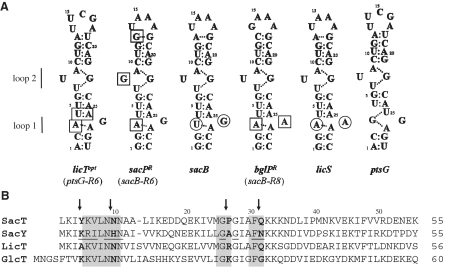
Figure 1.
Conservation of the regulatory components of the anti-termination systems of the BglG/ SacY family. (A) Summary of the relevant RAT structures. Boxes indicate nucleotides that differ from the cognate wild-type RAT. For licTopt RAT positions 3, 4 and 26 are mutated leading to a complete LicT dependent RAT structure. sacB was mutated that the RAT structure resembles sacP (sacB-R6) and bglP (sacB-R8) (18). These recombinant RAT structures are designated sacPR and bglPR, respectively. Circles indicate the 2 nt that are not conserved in the sacB RAT and licS RAT used in this study. The position of the asymmetric internal loops (1 and 2) characterizing the RAT hairpin is indicated. (B) Alignment of the CAT domains of the four anti-termination proteins of the BglG/SacY family in B. subtilis. Boxes indicate the amino acids involved in RNA recognition according to the structure of the LicT CAT/RAT complex (PDB ID code 1L1C) (17). Underlined residues in SacY CAT indicate the RNA-contacting region as mapped by NMR titration (5). Non-conserved amino acids that have been targeted for mutagenesis are labelled by arrows.
As in all other families of conserved regulatory systems, the straightness of signal transduction, i.e. the avoidance of cross-talk is also a major issue in the BglG/SacY family. Previous studies have shown that a structural difference in the lower loop of the RAT distinguishes the GlcT/ptsG system from all other systems of the family in B. subtilis (16). In addition, subtle differences between the RAT structures are specificity determinants responsible for preferred interaction of a given anti-termination protein with its cognate RAT (15). Moreover, the sugar specificity of the PTS permeases and additional levels of control of their expression (such as carbon catabolite repression) contribute to avoiding unfavourable cross-talk (18). Finally, the structures of the RNA-binding domains were proposed to contribute to RNA recognition specificity: the CAT dimer of LicT is more open than that of SacY, and the variable residue Arg-27 in LicT was found to be important for proper recognition of the cognate RAT (19).
In this study, we addressed the role of non-conserved amino acids in the CAT domains of LicT, SacT and SacY. For this purpose, we made use of a RAT variant that is exclusively recognized by LicT to isolate mutant SacT CATs that have gained the ability to bind this non-cognate RAT. Interestingly, a randomly isolated mutation affected the non-conserved residue Pro-26 in SacT, corresponding to Arg-27 in LicT. This position, together with position 31, are the most versatile amino acid positions of the RNA-binding site. This led us to investigate the role of residues 26 and 31 for RNA recognition in more detail. A series of SacT CAT mutants with different amino acids at these two positions was found to have lost their RNA recognition specificity. Instead, these CAT variants bind, to different extent, to all RAT structures, even to the structurally very different ptsG RAT. Moreover, the exchange of the corresponding residues between LicT and SacY indicated that residue 26 is an important determinant for both specificity and affinity. Our observations suggest that the amino acids naturally present at the two positions 26 and 31 result from the selective pressure to obtain RAT recognition specificity.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The B. subtilis strains used in this study are shown in Table 1. All B. subtilis strains are derivatives of the wild-type strain 168. Escherichia coli DH5α, BL21(DE3) (20) and XL1-red (Stratagene) were used for cloning experiments, for the expression of recombinant proteins, and for in vivo mutagenesis, respectively.
Table 1.
Bacillus subtilis strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| GP61a | trpC2 ΔlicT::cat amyE::(licTopt-lacZ aphA3) | see ‘Materials and Methods’ section |
| GP109 | trpC2 ΔglcT8 amyE::(ΔLA ptsG′-′lacZ aphA3) | 27 |
| GP408a | trpC2 amyE::(licTopt-lacZ aphA3) | 18 |
| GP440 | trpC2 ΔsacT::spc amyE::(sacB-lacZ aphA3) | 18 |
| GP487b | trpC2 ΔsacT::spc amyE::(bglPR -lacZ aphA3) | 18 |
| GP538c | trpC2 ΔsacT::spc amyE::(sacPR -lacZ aphA3) | 18 |
| SA501 | sacBΔ23 sacXYΔ3 sacTΔ4 ΔlicT ::aphA3 SPβ ::(sacB′-lacZ cat) | 5 |
| SA504 | sacBΔ23 sacXYΔ3 sacTΔ4 ΔlicT ::aphA3 SPβ ::(sacB′3A/26A/t-lacZ cat) | 5 |
aIn the original publication, the transcriptional fusion present in this strain is referred to as ΔLA ptsG-R6′-′lacZ.
bIn the original publication, the transcriptional fusion present in this strain is referred to as sacB-R8-lacZ.
cIn the original publication, the transcriptional fusion present in this strain is referred to as sacB-R6-lacZ.
Bacillus subtilis was grown in SP medium or in CSE minimal medium (21). The media were supplemented with auxotrophic requirements (at 50 mg/l), carbon sources and inducers as indicated. Escherichia coli was grown in LB medium and transformants were selected on plates containing ampicillin (100 µg/ml). LB and SP plates were prepared by the addition of 17 g Bacto agar/l (Difco) to LB or SP medium, respectively.
Transformation and characterization of the phenotype
Bacillus subtilis was transformed with plasmid DNA according to the two-step protocol described previously (22). Transformants were selected on SP plates containing kanamycin (Km 5 µg/ml), chloramphenicol (Cm 5 µg/ml), spectinomycin (Spc 100 µg/ml) or erythromycin plus lincomycin (Em 2 µg/ml and Lin 25 µg/ml).
Quantitative studies of lacZ expression in B. subtilis in liquid medium were performed as follows: cells were grown in CSE medium supplemented with ribose as the carbon source. Cells were harvested at OD600 0.6–0.8. Cell extracts were obtained by treatment with lysozyme and DNase. β-Galactosidase activities were determined as previously described using o-nitrophenyl-galactoside as a substrate (22). One unit is defined as the amount of enzyme that produces 1 nmol of o-nitrophenol per minute at 28°C.
DNA manipulation
Transformation of E. coli and plasmid DNA extraction were performed using standard procedures (20). Restriction enzymes, T4 DNA ligase and DNA polymerases were used as recommended by the manufacturers. DNA fragments were purified from agarose gels using the QIAquick gel extraction kit (Qiagen®, Hilden, Germany). Pfu DNA polymerase was used for the polymerase chain reaction (PCR) as recommended by the manufacturer. The combined chain reaction for site-specific mutagenesis (23) was performed with Pfu DNA polymerase and thermostable DNA ligase (Ampligase®, Epicentre, Wisconsin, USA). DNA sequences were determined using the dideoxy chain-termination method (20). Chromosomal DNA of B. subtilis was isolated as described (22).
Construction of a licT mutant strain by allelic replacement
To construct a licT mutant strain, the long flanking homology PCR (LFH-PCR) technique was used (24). Briefly, a cassette carrying the cat resistance gene was amplified from the plasmids pGEM-cat (25) using the primer pair cat-fwd/cat-rev (18). DNA fragments of about 1000 bp flanking the licT region at its 5′- and 3′-end were amplified. The 3′-end of the upstream fragment as well as the 5′-end of the downstream fragment extended into the licT gene in a way that all expression signals of genes up- and downstream of licT remained intact. The primers were designed in a way that the reverse primer of the upstream fragment and the forward primer of the downstream fragment are complementary to the end of the cat resistance cassette obtained with cat-fwd/cat-rev. The joining of the two fragments to the resistance cassette was performed in a second PCR using the forward primer of the upstream fragment and the reverse primer of the downstream fragment as described previously (18). The PCR product was directly used to transform B. subtilis GP408. The integrity of the regions flanking the integrated resistance cassette was verified by sequencing PCR products of about 1000 bp amplified from chromosomal DNA of the resulting mutant strain GP61 (ΔlicT::cat).
Mutagenesis of the RNA-binding domain of SacT
To study the effect of point mutations in the RNA-binding domain of SacT, a plasmid encoding the CAT of SacT was subjected to random mutagenesis using the E. coli mutator strain XL1-red. For this, the fragment of the sacT gene coding for the CAT domain was amplified by PCR using the oligonucleotides SHU59 (5′ aaaGGATCCcaaattggcgggagagataacctc) and SHU65 (5′ aaaAAGCTTtcacttttcattctcgtcgcgcac), digested with BamHI and HindIII and cloned into the shuttle vector pBQ200 (26). The resulting plasmid was pGP446. Plasmid pGP446 was used to transform E. coli XL1-red, and five independent cultures were incubated for 2 days to allow the occurrence of mutations. Plasmid DNA from the individual pools was isolated and used to transform the indicator strain B. subtilis GP61. The transformants were incubated on CSE plates containing X-Gal to allow the detection of the expression of the lacZ fusion. Blue colonies were isolated and subjected to detailed analyses.
As a control, we used the plasmids pGP118 (27) and pGP447 expressing the RNA-binding domains of GlcT and LicT, respectively. Plasmid pGP447 was constructed by amplification of the region of the licT gene corresponding to the CAT domain using the primers SHU57 (5′ aaaaGGATCCgtagatttggagggacatgcc) and SHU64 (5′ aaaAAGCTTtcatgatacatccttgttatcgagc). The PCR product was digested and cloned into pBQ200 as described above for SacT.
In a second approach, we focused on the role of the amino acids Pro-26 and Gln-31 of SacT for RNA recognition specificity. For this purpose, a semi-random mutagenesis of these two sites was performed by applying the combined chain reaction (23) with the external primers SHU59 and SHU65, and the phosphorylated mutagenesis primer SHU78 (5′ P-cgtgatgggaNNNggaatcgcttttNNNaaaaagaaaaatgatctcatccc) (N: any base). This oligonucleotide allows the incorporation of any base at the positions of the two amino acids, Pro-26 and Gln-31. The PCR product was cloned into pBQ200 and the resulting plasmids were screened for anti-termination activity in B. subtilis GP61 as described above.
Expression and purification of the mutant RNA-binding domains
To fuse the mutant CAT domains of SacT to a Strep tag at their C termini, DNA fragments corresponding to amino acids 1–57 of SacT were amplified by PCR using the plasmids carrying the mutations as the template and the primer pair OS97/OS98 (18). The PCR products were digested with NdeI and BamHI, and the resulting fragments were cloned into the expression vector pGP574 (18). For the expression of the wild-type CATs of GlcT and SacT, we used the plasmids pGP575 and pGP577, respectively (18).
Escherichia coli BL21(DE3)/pLysS was used as host for the overexpression of recombinant proteins. Expression was induced by the addition of IPTG (final concentration 1 mM) to exponentially growing cultures (OD600 of 0.8). Cells were lysed using a French press. After lysis the crude extracts were centrifuged at 15 000 g for 30 min and then passed over a Streptactin column (IBA, Göttingen, Germany). The recombinant protein was eluted with desthiobiotin (Sigma, final concentration 2.5 mM). After elution the fractions were tested for the desired protein using 15% SDS–PAGE gels. The relevant fractions were combined and dialysed overnight. The protein concentration was determined according to the method of Bradford using the Bio-rad dye-binding assay and bovine serum albumin (BSA) as the standard. GST-fusion proteins encoded by pGEX-2T derivatives were produced in E. coli BL21(DE3) and purified by affinity chromatography on glutathione sepharose as previously described (19).
Assay of interaction between the CAT domains and RAT RNAs
To obtain templates for the in vitro synthesis of the different RAT RNAs, the primer pairs OS25/OS26 (16) and OS86/OS87 (18) were used to amplify RAT variants based on the ptsG and the sacB RATs, respectively. As templates served pGP66 (ptsG, 28), pGP556 (licTopt), pGP564 (sacB), pGP595 (sacPR) and pGP587 (bglPR) (18). The presence of a T7 RNA polymerase promoter on primers OS25 and OS86 allowed the use of the PCR product as a template for in vitro transcription with T7 RNA polymerase (Roche Diagnostics). The integrity of the RNA transcripts was analyzed by denaturating agarose gel electrophoresis (29).
Binding of the CAT domains to RAT–RNA was analysed by gel retardation experiments as described previously (18). Briefly, the RAT–RNA (in water) was denatured by incubation at 90°C for 2 min and renatured by dilution 1:1 with ice cold water and subsequent incubation on ice. Purified protein was added to the RAT–RNA and the samples were incubated for 10 min at room temperature in TAE buffer in the presence of 300 mM NaCl. After this incubation, glycerol was added to a final concentration of 10% (w/v). The samples were then analysed on 10% tris–acetate PAA gels.
In vivo and in vitro assays with the SacY- and LicT-derived RNA-binding domains
Construction of the B. subtilis and E. coli strains encoding the mutant SacY and LicT CAT domains were performed using the procedures described previously (30). In order to increase the expression levels of the recombinant sacY and licT genes in B. subtilis, plasmid pRL23 was constructed by replacing the Pspac promoter of pND23 (30) by the strong constitutive degQ36 promoter (31). Anti-termination activities were tested in vivo by introducing the pRL23 constructs into strains SA501 and SA504 expressing a lacZ reporter gene under the control of the sacB or licS RAT. Cells were cultured in minimum medium supplemented with glucose (1% w/v) and phleomycin (0.2 mg/ml) and β-galactosidase activities were measured as described above.
Surface plasmon resonance (SPR) studies were carried out on a BIAcore X optical biosensor (GE Healthcare, USA) having two microflow cells that can be run simultaneously. Experiments were performed using GST-fusion proteins and sacB RAT or licS RAT RNA following a procedure that was previously described in details (19). GST protein solutions at 0.1 mM were injected onto a CM5 sensor chip with immobilized GST-antibododies (GE Healthcare, USA) with a flow-rate of 20 µl/min. GST alone was bound on the reference flowcell (FC1) and the GST–CAT fusions on the other flowcell (FC2). The injection was stopped manually and eventually repeated until 1900 responsive units (RU) remain bound on each flowcell. RNAs were injected onto both cells simultaneously for 1 min at a flow rate of 10 µl/min in running buffer [10 mM Tris pH 8, 300 mM NaCl, 0.0008% (w/v) sodium azide, 0.005% (w/v) Surfactant P20 (Biacore)] and the difference in RU (ΔRU) between the two cells was measured, allowing direct vizualisation of the amount of RNA specifically bound to the immobilized fusion protein in FC2. For titration experiments, RNA was injected at increasing concentrations for 1 or 2 min and the ΔRU was recorded when the equilibrium of the binding reaction was reached. Under the conditions used, the ΔRUmax measured or estimated at saturating RNA concentration was about 200 ΔRU. Equilibrium binding constants (KD) were determined grafically by plotting the ΔRU steady-state values versus the injected RNA concentration.
RESULTS
Establishment of a screening system for the isolation of specificity variants of the RNA-binding domains
In order to get an unbiased view of the determinants of recognition specificity of the RNA-binding domains, we designed a screen for the isolation of CAT variants that had lost their specificity. The isolation of specificity mutations in a CAT domain required a highly specific reporter system. However, with the exception of GlcT and the ptsG RAT structure, the conserved RNA-binding domains and the corresponding RAT structures in B. subtilis are not completely specific and cross-talk was observed (15,18). The absence of any cross-talk including GlcT or the ptsG RAT indicated that it might be very risky to aim at the isolation of CAT variants of the other anti-terminator proteins that recognize this RAT structure. Therefore, we made use of a RAT variant, licTopt, which is exclusively recognized by LicT (18).
To facilitate mutagenesis, we intended to use multi-copy plasmids. On the one hand, this allows an expression level sufficient for the dimer formation and in vivo activity of the isolated CATs (27), and on the other hand, this approach ensures that mutations may only occur in the relevant region of the gene. For this purpose, plasmids pGP447 and pGP446 expressing the CAT domains of LicT and SacT, respectively, were constructed. The activity of these artificially expressed RNA-binding domains was tested in reporter strains in which the expression of the lacZ gene depends on the licTopt or on the sacPR RAT structures. With the chromosomally encoded anti-terminator proteins, licTopt and sacPR are exclusively recognized by LicT and SacT, respectively (18). In the reporter strains, the genes encoding the cognate anti-terminator proteins were deleted to ensure that all β-galactosidase synthesis depends on the interaction of the plasmid-borne CAT domains with the RAT sequences. The bacteria were grown in CSE minimal medium and the activity of β-galactosidase was determined. The results of this analysis are shown in Table 2. As expected, the licTopt RAT was efficiently recognized by the RNA-binding domain of LicT but not by that of SacT. Similarly, the sacPR RAT was a specific target of the CAT of SacT. These observations confirm that the CAT domains are highly specific for these RAT structures even when they are overexpressed. We concluded that this system was well suited for the isolation of mutations in the RNA-binding domain of SacT that bind the licTopt RAT structure.
Table 2.
Interaction between the RNA-binding domains of SacT and LicT with different RAT structures
| Strain | Plasmid | Relevant genotype | β-Galactosidase activity (U/mg protein)a |
|---|---|---|---|
| GP61 | pGP446 | ΔlicT, licTopt-lacZ, sacT-CAT | 20 |
| GP61 | pGP447 | ΔlicT, licTopt-lacZ, licT-CAT | 828 |
| GP538 | pGP446 | ΔsacT, sacPR -lacZ, sacT-CAT | 265 |
| GP538 | pGP447 | ΔsacT, sacPR -lacZ, licT-CAT | 2 |
aRepresentative values of lacZ expression.
All measurements were performed at least twice.
Isolation of a variant of the SacT RNA-binding domain that binds the licTopt RAT structure
The RNA-binding domain of SacT was mutagenized by propagating the expression plasmid pGP446 in the E. coli mutator strain XL1-red. Briefly, plasmid pools isolated from this strain were used to transform the reporter strain B. subtilis GP61 and screened for the expression of the licTopt-lacZ fusion on plates containing X-Gal. Due to the absence of the cognate anti-terminator protein LicT, this strain forms normally white colonies. However, we isolated one clone from our mutant plasmid pool that formed blue colonies, indicating that the corresponding SacT variant was able to bind the licTopt RAT structure. The sacT allele in this plasmid, pGP448, was sequenced, and a single base pair exchange (C → T) at position 76 of the sacT coding sequence was observed. This mutation results in a replacement of Pro-26 in SacT by a serine residue. Interestingly, we had already proposed in an earlier study that this site might be important for the specificity of CAT–RAT interaction (19).
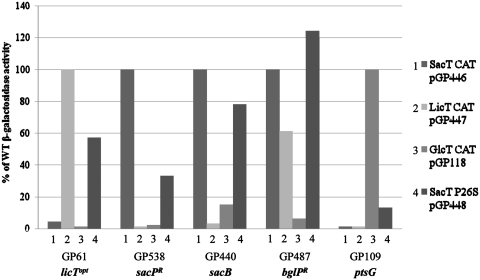
Next, we wished to study whether the mutation in the CAT of SacT results in a complete switch of specificity, or in a relaxed RNA recognition. For this purpose, we used a set of reporter strains with lacZ fusions under the control of the different RATs. The results are shown in Figure 2. As expected, the licTopt RAT structure present in strain GP61 was recognized by LicT but not by SacT or GlcT. However, the isolated P26S variant of SacT was able to cause anti-termination at this structure. Similarly and in agreement with the data shown in Table 2, the sacPR RAT was specifically recognized by SacT. Interestingly, the SacT P26S variant CAT was able to anti-terminate at this structure; however, the activity was reduced to about one-third as compared to the wild-type CAT of SacT. The RAT structure of the sacB gene present in GP440 was a target of SacT. This RNA was also efficiently recognized by the SacT P26S CAT domain. The bglPR RAT is identical to the natural RNA structure of the bglPH operon. As shown previously, this RAT was recognized by both the SacT and the LicT CATs (20) whereas the CAT of GlcT was unable to cause anti-termination at this RNA structure. Interestingly, the SacT P26S CAT allows even higher anti-termination at this RAT than the wild-type RNA-binding domains. The last RAT structure in this study was that of ptsG present in B. subtilis GP109. This RAT was recognized by GlcT but neither by LicT nor by SacT. This is in good agreement with previous reports (16,18). The analysis of the activity of the SacT P26S CAT at this structure revealed that the mutant form of SacT is able to cause some anti-termination at this RAT (Table 2). This is the first time that a CAT domain different from that of GlcT was found to bind a ptsG RAT structure.
Figure 2.
Analysis of the SacT P26S mutant. The isolated SacT P26S variant was tested against all RAT structures in B. subtilis. The wild-type RNA-binding domains of SacT, LicT and GlcT were used as controls. The β-galactosidase activity is shown in percentage of the wild type activity. In the case of bglPR RAT SacT was set 100% because in this artificial system SacT has a higher activity towards this RAT structure. 1: SacT 2: LicT 3: GlcT 4: SacT P26S.
Taken together, our data indicate that the SacT P26S variant is able to interact with all RAT structures that are present in B. subtilis and demonstrate that the proline residue at position 26 of the SacT RNA-binding domain is an important specificity determinant.
Identification of residues important for RNA recognition specificity
A comparison of the RNA-binding domains of the four anti-terminator proteins of the BglG/SacY family in B. subtilis revealed a high conservation (between 28% and 46% identical amino acids, see Figure 1B). Specifically, the amino acids known to be involved in direct interactions to the RAT RNA [based on the structure of the LicT CAT–RAT complex, (17)] are highly conserved. However, two of these positions, corresponding to Pro-26 and Gln-31 in SacT, are variable and are therefore candidates that may be implicated in RNA recognition specificity. The finding that replacements of residue 26 (or 27 in LicT) result in relaxed interaction specificity strongly supports this idea.
To address the role of Pro-26 and Gln-31 of SacT in RNA recognition in more detail, we performed a semi-random mutagenesis in which these two amino acids could be replaced by any couple of amino acids. As described above, mutations that allowed interaction of the variant SacT CATs with the the licTopt RAT structure of strain GP61 were detected as blue colonies on plates containing X-Gal. This approach resulted in the isolation of 22 independent mutants. The sacT alleles of these clones were sequenced, and nine different combinations of amino acids at the two positions were detected (Table 3).
Table 3.
In vivo recognition of the different RAT structures by the CAT variants
| Plasmid | Mutation pos. 26/31 | β-Galactosidase activity (U/mg protein) |
||||
|---|---|---|---|---|---|---|
| GP61 licTopt | GP538 sacPR | GP440 sacB | GP487 bglPR | GP109 ptsG | ||
| GlcT RBD pGP118 | K/G | 26 (10) | 35 (3) | 7 (5) | 7 (4) | 1195 (276) |
| SacT RBD pGP446 | P/Q | 38 (11) | 467 (117) | 53 (26) | 185 (28) | 12 (1) |
| LicT RBD pGP447 | R/Q | 806 (208) | 3 (1) | 3 (1) | 122 (26) | 11 (3) |
| pGP448 | S/Q | 503 (105) | 221 (29) | 35 (26) | 158 (37) | 141 (51) |
| pGP449 | S/R | 185 (38) | 503 (33) | 33 (8) | 90 (21) | 89 (14) |
| pGP450 | C/L | 1382 (461) | 215 (77) | 237 (80) | 146 (18) | 693 (181) |
| pGP451 | R/R | 652 (230) | 208 (13) | 144 (6) | 198 (45) | 545 (117) |
| pGP452 | C/R | 546 (85) | 146 (24) | 219 (14) | 128 (3) | 615 (47) |
| pGP453 | S/G | 970 (170) | 517 (179) | 297 (12) | 185 (20) | 839 (209) |
| pGP454 | V/G | 146 (15) | 188 (42) | 268 (18) | 259 (9) | 702 (48) |
| pGP455 | C/S | 387 (49) | 33 (9) | 92 (26) | 36 (3) | 195 (25) |
| pGP456 | A/K | 81 (22) | 337 (88) | 97 (22) | 185 (24) | 52 (2) |
| pGP457 | R/A | 796 (72) | 205 (70) | 414 (6) | 119 (13) | 649 (151) |
All measurements were performed at least twice. Standard deviation values are indicated within parenthesis.
As described for the SacT P26S CAT, we assayed the activity of the mutant forms using the set of reporter strains in which the lacZ expression depends on the different RAT structures. For this purpose, the strains carrying the plasmids with the different wild-type and mutant CATs were grown in CSE minimal medium and the β-galactosidase activities were determined. The results are summarized in Table 3.
Since all CAT variants were initially screened for their activity to cause anti-termination at the licTopt RAT, it was not surprising that all variants allowed β-galactosidase synthesis when the lacZ gene was expressed under the control of this RAT. However, the actual activity levels differed significantly. The CAT variant in pGP450 (Cys-26, Leu-31) allowed a higher level of expression than the cognate CAT of LicT (1382 versus 806 units of β-galactosidase). In contrast, the CAT domain present on plasmid pGP456 (Ala-26, Lys-31) exhibited only 10% of LicT activity. It is interesting to note, that a second CAT variant with a rather low activity towards the licTopt RAT (present in pGP454, Val-26, Gly-31) does also have an uncharged amino acid at position 26. These two plasmids harboured the only CATs with uncharged residues at position 26.
When the activity of the mutant CATs at the sacPR RAT was tested, only three CAT variants caused anti-termination similar to the cognate wild-type SacT CAT. These variants were present on pGP449 (Ser-26, Arg-31), pGP453 (Ser-26, Gly-31) and pGP456 (Ala-26, Lys-31). In contrast, the CAT encoded on pGP455 (Cys-26, Ser-31) was nearly inactive on the sacPR RAT structure, suggesting that these mutations prevent the productive interaction with the corresponding sacPA RAT structure.
The sacB RAT structure is recognized by the SacT and SacY anti-terminator proteins, however, anti-termination at this RAT is rather inefficient. In contrast, sacB RAT mutant derivatives with otherwise identical expression signals (e.g. the sacPR RAT) confer much better anti-termination (18). Interestingly, nearly all mutant CATs isolated in this study allow better anti-termination at the sacB RAT than the wild-type CAT of SacT. This is very intriguing for the CAT encoded on pGP457 (Arg-26, Ala-31). This protein gives rise to a 5-fold increase of β-galactosidase as compared to the SacT CAT (414 versus 53 U of β-galactosidase). The other extreme is defined by the CAT variant encoded on pGP449 (Ser-26, Arg-31) that exhibits only a very weak activity at the sacB RAT structure.
The RAT structure of the bglPH operon (here exemplified by the bglPR RAT) is well recognized by the CATs of LicT and SacT. As stated above, this RAT was efficiently used by the P26S CAT. Similarly, it is a very good target for the CAT domains encoded on pGP451 (Arg-26, Arg-31), pGP453 (Ser-26, Gly-31), pGP454 (Val-26, Gly-31) and pGP456 (Ala-26, Lys-31). In contrast, a replacement of Pro-26 to cysteine and Gln-31 to serine (pGP455) resulted in a severe loss of interaction with this RAT structure.
Finally, we investigated whether the mutant CAT variants were able to cause anti-termination at the ptsG RAT structure. While there is excessive cross-talk between the anti-terminator proteins and RAT structures of the bgl- and sac-type, a cross-talk involving the ptsG RAT structure is not possible in nature. As reported above, the P26S variant of the SacT CAT does allow a weak expression of the reporter fusion that is controlled by the ptsG RAT suggesting that this border can be crossed. Indeed, the CAT encoded on pGP453 (Ser-26, Gly-31) was nearly as efficient on this RAT as was the cognate CAT of GlcT (839 versus 1195 U of β-galactosidase). As observed for the other RAT structures, the efficiency of the different CATs varied over a broad range. The CAT domain carrying an alanine at position 26 and a lysine at position 31 (pGP456, see Table 3) showed the weakest activity with the ptsG RAT structure.
Binding of the SacT CAT variants to the different RAT structures
The experiments with reporter constructs described above allow concluding on the interaction between a CAT domain and a specific RAT structure based on the expression of the lacZ gene that is controlled by this regulatory system. In order to get more direct evidence for the effect of the mutations on the protein–RNA interaction, we performed electrophoretic mobility shift assays using the purified CAT domains and the different RAT RNAs. The RNA-binding domains of SacT and of GlcT served as controls.
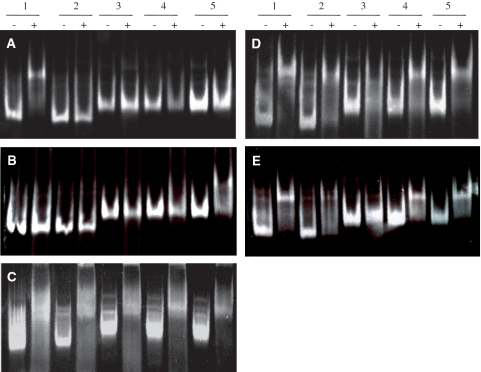
The RNA-binding domain of GlcT did only recognize its cognate RAT, ptsG (Figure 3A). This is in excellent agreement with our previous observations. As shown in Figure 3B, the CAT of SacT was able to retard the migration of its cognate sacPA RAT. In addition, weak retardation of the sacB RAT was observed. This is in good agreement with the observation that the sacB RAT structure is a poor target for the naturally occurring anti-terminator proteins (see above). The three selected CAT variants (originally encoded on pGP451, pGP452 and pGP453; see Table 3) retarded all the tested RAT RNAs (Figure 3C–E). This confirms the loss of RNA recognition specificity already observed in the in vivo anti-termination assay.
Figure 3.
Electrophoretic mobility shift analysis of the interaction between the variants of the SacT CAT with the different RAT structures. The different CAT domains [(A) GlcT (B) SacT (C) SacT P26R, Q31R (D) SacT P26C, Q31R (E) SacT P26S, Q31] were tested against the RAT RNAs (1: ptsG; 2: licTopt; 3: sacB; 4: bglPR 5: sacPR). A 100 pmol of the RAT–RNAs were used. In lanes labelled with ‘+’ 250 pmol of the RNA-binding domain was added to the RNA as indicated prior to electrophoresis.
Structure-based mutagenesis of the RNA-binding domains of SacY and LicT
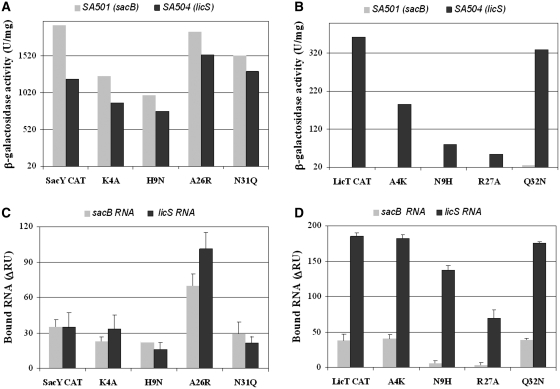
In a complementary approach, we have undertaken site-directed mutagenesis of the CAT domains of SacY and LicT based on the structural information available for these proteins. The structure of the LicT CAT-RAT anti-termination complex was solved by NMR, and the residues making direct interaction with the RAT hairpin were identified (17). For SacY CAT the protein–RNA contact region has been mapped by NMR foot printing (5) and it overlaps very well with that of LicT CAT at the dimer interface (Figure 1B). At the RNA level, the sacB RAT and licS RAT sequences differ by only 2 nt (Figure 1A), therefore the 3D structures of the anti-terminator stem–loop are expected to be very similar. In spite of these very strong structural similarities, SacY CAT and LicT CAT display very different affinity and specificity towards their cognate RAT targets (19). The origin of these differences was investigated by introducing point mutations into the RNA-binding domains of SacY and LicT. Four non-conserved residues within the RNA-contacting region of the SacY CAT were targeted (Lys-4, His-9, Ala-26 and Asn-31) and replaced with the amino acid side-chain found at the corresponding positions in the LicT CAT (Ala-4, Asn-9, Arg-27 and Gln-32, respectively). The genes encoding the resulting variants of SacY CAT (K4A, H9N, A26R and N31Q, respectively), or the reciprocal variants of LicT CAT (A4K, N9H, R27A and Q32N, respectively) were introduced into B. subtilis or E. coli expression vectors, and the effect of the mutations on the recognition of sacB RAT and licS RAT was tested both in vivo and in vitro (Figure 4).
Figure 4.
Effect of reciprocal point mutations in SacY CAT and LicT CAT on RNA recognition in vivo and in vitro. (A and B) Relative anti-termination activity of the wild-type RNA binding domains (SacY CAT and LicT CAT) and their variants carrying the indicated amino acid substitution, in B. subtilis reporter strains SA501 and SA504 expressing the lacZ gene under the control of sacB RAT (grey bars) or licS RAT (black bars). β-Galactosidase activities are expressed in units/mg of proteins, above the background level (about 20 U/mg) measured for transformants of strain SA501 or SA504 harbouring the empty pRL23 cloning vector (with no CAT gene). Note that in this in vivo assay, the anti-termination activity of SacY CAT at the sacB RAT locus appears about 5-fold higher than that of LicT CAT at the licS RAT locus. All measurements were performed on two different transformants and two different extracts from the same bacterial culture. (C and D) Relative RNA binding activity of the wild-type and mutant CATs measured by SPR. The amount of sacB RAT (grey bars) or licS RAT (black bars) RNA bound at equilibrium is expressed as the ΔRU measured using GST alone in the reference flow-cell. The mean value and standard deviation are shown for each GST-CAT fusion, obtained from two independent experiments on different sensor chips (only one experiment for the H9N variant). All measurements were performed on two different transformants and two different extracts from the same bacterial culture. Standard deviations were <10%, except for weak activities (below 50 U/mg) where they were up to 30%.
The anti-termination activity of the wild-type and mutant SacY and LicT CATs was compared in B. subtilis strains SA501 and SA504 expressing a lacZ reporter gene under the control of the sacB or licS RAT, respectively (Figure 4A and B). β-Galactosidase synthesis was high in all strains encoding SacY CAT or its variants, indicating that these RNA-binding domains were all efficient in anti-termination. As previously observed (17), SacY CAT was active on both the sacB- and licS RAT structure, confirming the poor RNA recognition specificity of this RNA-binding domain. In contrast, the LicT CAT as well as all its variants displayed a very strong preference for the licS RAT target present in B. subtilis SA504. The β-galactosidase activities were always lower than with the SacY CATs, but this resulted from a lower expression level of the LicT constructs in the reporter strains used in this study rather than from reduced RNA binding activity. In this in vivo assay, all the mutations introduced in the RNA-binding site of either SacY or LicT appeared to reduce the anti-termination activity at their cognate RAT, yet to different extent. In SacY CAT, it is at position His-9 that the loss of activity is more pronounced, although it does not exceed 50%. The mutation at this position in LicT CAT was also deleterious but to a higher extent (about 80% activity loss compared to wild-type in B. subtilis SA504). The most severe mutation was observed at position Arg-27 of LicT CAT (about 15% residual activity for R27A) whereas the reciprocal variant of SacY CAT (A26R) retained anti-termination activity similar to that of the wild-type parent in B. subtilis SA501. Interestingly, this A26R variant appeared to anti-terminate more efficiently than wild-type SacY CAT in B. subtilis SA504 carrying the non-cognate licS RAT reporter fusion.
The wild-type and mutant CAT domains were then purified as GST fusion proteins and their interaction with oligoribonucleotides containing either the sacB or licS RAT sequence was monitored by SPR. The binding capacity of the different GST–CAT fusions immobilized on sensor chips was compared by injecting the sacB RAT or licS RAT RNAs at 1 µM and measuring the amplitude of the SPR signal (Figure 4C and D). Interaction with both RNAs was observed for SacY CAT and all its variants, in good agreement with the results of the in vivo assay. In the case of LicT CAT, the SPR assay confirmed that, despite the relatively low anti-termination activity measured with the reporter system (Figure 4B), this CAT exhibits in fact much better RNA binding properties as compared to SacY CAT, in good agreement with previous comparative in vitro studies (19). The comparison of Figure 4A/B and C/D reveals some discrepancies in the RNA recognition patterns obtained in vivo and in vitro. The most conspicuous is the A26R variant of SacY for which the SPR signal was about 2- to 3-fold higher than that of wild-type SacY CAT. In contrast, the R27A LicT variant carrying the reciprocal mutation at position Arg-27 exhibited severely altered RNA binding capacities, highlighting the importance of this amino acid position for the specific recognition of the RAT structures.
RNA affinity and specificity changes in SacY and LicT CATs
Titration experiments were then performed by SPR in order to better quantify the relative affinity and specificity of the SacY- and LicT-derived CATs for their cognate or non-cognate RAT structures. Dissociation constants (KD) were determined by injecting the sacB or licS RNAs at different concentrations onto the immobilized GST fusions and measuring the SPR signal reached at equilibrium (Table 4). Similar affinity constants in the micromolar range were estimated for the interaction of SacY CAT with both RNAs, again evidencing the relatively weak affinity and the absence of specificity of this CAT for both RAT targets. As previously observed (19), LicT CAT interacted about a 100-fold more strongly and more specifically with its cognate RAT, the KD values being estimated here at around 0.05 µM for licS RAT as compared to 5 µM for sacB RAT.
Table 4.
Relative affinity and specificity of wild-type and mutant SacY-CAT and LicT-CAT for sacB RAT and licS RAT determined by SPR
|
sacB RAT |
licS RAT |
Specificity factorc | |||
|---|---|---|---|---|---|
| KD (µM)a | Rel. aff.b | KD (µM) | Rel. aff.a | ||
| SacY CAT | 6.2 ± 2.5 | 1 | 6.0 ± 2.4 | 1 | 1.0 |
| K4A | 8.2 ± 3.3 | 0.8 | 9.0 ± 3.6 | 0.7 | 1.1 |
| H9N | 12 ± 4.8 | 0.5 | 22 ± 8.8 | 0.3 | 1.8 |
| A26R | 2.0 ± 0.8 | 3.1 | 0.6 ± 0.2 | 10 | 0.3 |
| N31Q | 9.0 ± 3.6 | 0.7 | 12 ± 4.8 | 0.5 | 1.3 |
| LicT CAT | 5.5 ± 2.2 | 1 | 0.05 ± 0.02 | 1 | 110 |
| A4K | 6.1 ± 2.4 | 0.9 | 0.05 ± 0.02 | 1 | 120 |
| N9H | 32 ± 12 | 0.2 | 0.19 ± 0.08 | 0.3 | 170 |
| R27A | 120 ± 48 | 0.05 | 1.80 ± 0.72 | 0.03 | 70 |
| Q32N | 7.1 ± 2.8 | 0.8 | 0.05 ± 0.02 | 1 | 140 |
aDetermined graphically considering a systematic error of ±40%. Values shown in italics were estimated by extrapolating values from single point measurements.
bRelative affinity for sacB RAT or licS RAT of mutant SacY and LicT CATs compared to their cognate wild-type parent = KD (wild-type)/KD (mutant).
cFor SacY CAT and variants = KD(licS RAT)/KD(sacB RAT); for LicT CAT and variants = KD(sacB RAT)/KD(licS RAT).
In both the SacY and LicT CATs, the amino acid substitutions at positions Lys-4/Ala-4 and Asn-31/Gln-32 induced no or little alteration in the RNA recognition mode of the variants as compared to their wild-type parents. The cross mutations introduced at position 9 had a general deleterious effect on the relative affinities (KD of the wild-type/KD of the variant) but not on the specificity factor (KD of non-cognate RAT/KD of cognate RAT). The most significant and interesting result of this analysis concerns the mutational effects observed at position Ala26/Arg27. When comparing relative affinities, that of the SacY CAT A26R variant was increased by a factor 3 for sacB RAT and by a factor of 10 for licS RAT. Hence, this variant is not only a better RNA binder than wild-type SacY CAT, but it can also better discriminate between the two RAT structures and preferentially interacts with the non-cognate target. Inversely, replacement of arginine with alanine at the corresponding position in LicT resulted in over 90% loss of affinity for both RNAs and a drop in the specificity factor of about 40%. Hence, the arginine side-chain at position 27 of LicT is a key contributor to both the high affinity and marked specificity of this anti-terminator protein for its RNA targets.
DISCUSSION
In a previous work, we have shown that subtle changes in the RAT structures may cause a shift in recognition specificity from one RNA anti-terminator to the other (18). This suggested that the RNA-binding domains of the anti-terminator proteins are similar enough to recognize other RAT RNAs upon introduction of only a few mutations. Indeed, this work demonstrates that this hypothesis is true: several mutations at the two non-conserved positions of SacT that are thought to be in contact with the RAT RNA resulted in binding of the CAT to non-cognate RAT structures; and in LicT and SacY, a single amino acid replacement within the RNA-binding site is sufficient to drastically alter the specificity and affinity of these proteins for their natural RNA targets.
Our findings on SacT provide clear evidence that those two residues (Pro-26 and Gln-31) that contact the RNA but that are not conserved among the anti-terminator proteins are major specificity determinants of these proteins. We have isolated a large set of CAT variants of SacT that contain different pairs of amino acids at positions 26 and 31. All of the mutations result not only in binding to the licTopt RAT but with few exceptions, also to the other RAT structures that were tested.
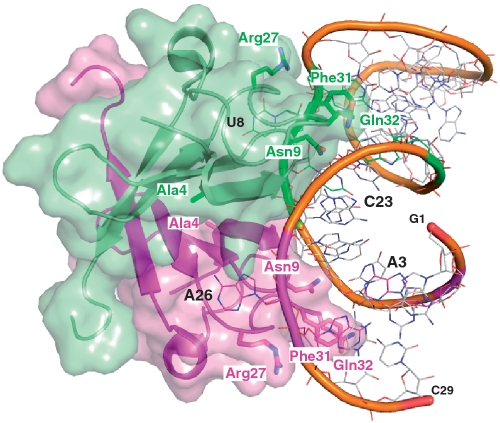
Examination of the homologous LicT CAT–RAT complex structure (Figure 5) shows that the corresponding residues in LicT (Arg-27 and Gln-32) are located on the outer edges of the RNA-binding surface of the protein dimer. The side-chains of these polar residues form like two grips, each contacting one strand of the RNA stem flanking the two internal loops (1 and 2) characterizing the RAT hairpin (see also Figure 1A). Although different in sequence, these loops present analogous 3D structures and can therefore be recognized in a similar way by symmetry-related elements of the LicT CAT dimer (15). In particular, A26 in loop 1 and U8 in loop 2, which are major specificity determinants of the RAT RNAs, are both expelled from the core of the RNA helix and their base is similarly accommodated within a cavity formed on each side of the dimer interface. The side-chain of Arg-27 contributes to the formation of these cavities and adopts a slightly different conformation in the two CAT monomers in order to optimize contacts with the bulged-out pyrimidine or purine, as well as with the sugar phosphate backbone. On the other side of the RNA minor groove Gln-32 is interacting with the phosphate group of U4 and C23, but also with the aromatic side-chain of the strictly conserved phenylalanine at position 31, which is crucial for the formation and stabilization of the sheared base pairs in both loops 1 and 2.
Figure 5.
Structure of the LicT-CAT/licS-RAT complex showing amino acid residues targeted for mutagenesis. The dimeric structure of the LicT N-terminal domain (residues 1–56) determined by NMR (17, PDB entry code 1L1C) is shown in cartoon and surface representation, with one monomer coloured in pink and the other in green. The amino acid side-chains of the residues targeted for site-directed mutagenesis in this study as well as a key residue of CAT–RAT interaction (Phe31) are labelled and shown in sticks. The licS RAT RNA is shown in wire frame with the phosphate backbone cartooned in pink for internal loop 1, green for internal loop 2 and orange elsewhere. In the LicT CAT-RAT structure, A26 in loop 1 and U8 in loop 2 are bulged out from the RNA helix core and are recognized by symmetry-related elements of the LicT-CAT dimer interface.
Together with structural information, there are a few principles in CAT–RAT recognition that can be deduced from the present mutational results. Polar amino acids at position 26 are preferred for the recognition of the licTopt RAT by SacT, whereas non-charged amino acids do not seem to be optimal for this RAT. Two of the SacT variants capable of very strong interaction with the licTopt RAT have an arginine at position 26. This amino acid is also found at this position in the CAT of LicT. These results are supported by the analysis of the reciprocal variants of SacY and LicT: A replacement of the arginine-27 residue in LicT by alanine led to a severe reduction of binding to the cognate RAT. In contrast, replacing Ala-26 by Arg in SacY resulted in an increased affinity to both its cognate sacB RAT and the non-cognate licS RAT. Thus, an arginine at position 26 might generally facilitate RAT binding, probably through electrostatic interaction between the positively charged amino acid side-chain and the phosphodiester backbone. Our observations indicate that an arginine at this position is also a major contributor for the specific recognition of LicT-dependent RAT structures: In the LicT context, it is the only position where a single mutation was found to decrease the specificity factor; more remarkably, we observed better binding of the SacY A26R CAT to the licS RAT structure as compared to the sacB RAT. Since these two RNAs differ only for 2 nt in the lower internal loop of the RAT hairpin, it can be concluded that the interactions established between this structural feature and the Arg-27 side-chain of LicT are key for the discrimination process. It should be noted that SacT with a proline at position 26 does not interact efficiently with licTopt, but is capable of binding a structure that corresponds to the wild-type bglP RAT normally recognized by LicT. The major difference between these two RAT structures is also the nucleotide in the lower loop: In the licTopt RAT there is a G at position 27, whereas an A is present at the corresponding position of the bglPR RAT (Figure 1). This difference in the RAT sequences may explain the differential recognition by SacT.
A common feature of SacT and LicT is the glutamine at position 31, one of the residues that are in contact with the RNA. This amino acid may be involved in the cross-recognition of the sacP and bglP RATs by SacT. In SacY, an anti-terminator protein that is known to recognize non-cognate RAT structures (15,18), an asparagine is present at the corresponding position whereas GlcT that does not bind any of the non-cognate RAT sequences has a glycine at this position (Figure 1B). The exchange of the glutamine and asparagines residues of LicT and SacY did affect neither specificity nor affinity for the RAT structures suggesting that these similar amino acids are not involved in differential RNA binding. In contrast, two of the mutants SacT CATs have a glycine at position 31, yet they exhibit a loss of specificity with a slight preference for the RAT of ptsG, which is normally recognized by GlcT. More generally, our analysis of the double mutants of SacT CAT shows that the RNA recognition pattern is influenced by the nature of the amino acid present at both positions 26 and 31. However, no clear-cut conclusions for the individual contribution of each position in RNA recognition can be drawn from this analysis. Instead, the two amino acids that hold like a clamp the region surrounding the two internal loops might together contribute to CAT specificity. The combination that is found at these positions would determine which RAT structures are recognized, depending upon the nucleotides present in loops 1 and 2. Since CAT is a symmetrical dimer, the same combination is used for the recognition of both loops. Improved interactions with one loop may be detrimental for the recognition of the other loop or increase non-specific binding to all similar RATs. The subtle balance of these interactions determines the strength and specificity of a given CAT-RAT complex.
In conclusion, our results suggest that the amino acid combinations at positions 26 and 31 (SacT numbering) in the B. subtilis anti-terminator proteins influence both their affinity and specificity of binding to the different RAT structures. A particular protein context determines the extent to which each position contributes to the recognition of a specific RNA. Because most of the mutations at these positions lead to relaxed specificity, the residues found in the wild-type proteins can be considered as ‘anti-determinants’ of the cross-talk between the conserved anti-termination systems. Evolution seems to have selected for residues that prevent ‘wrong’ interactions with non-cognate targets rather than for residues that strengthen ‘correct’ interactions with the right target. Indeed, in both SacY and SacT, we could engineer CAT domains that are much better RNA binders than their parent CATs but all displayed a degenerated specificity towards their cognate RAT. Hence, in the wild-type proteins, a compromise seems to have been reached between RNA binding efficacy and specific interaction with individual RAT sequences.
This conclusion is likely to apply in other bacteria containing more than one anti-termination systems of the BglG/SacY family. Interestingly, natural selection seems to be in favour of the amino acid combinations found at positions 26 and 31 in the wild-type anti-terminator proteins of B. subtilis. Indeed, the combinations present in the four B. subtilis anti-terminator proteins occur in 96 anti-terminator proteins in the databases. Another combination, K26/N31, is present in 30 CATs, among them the β-glucoside-specific anti-terminator protein BvrA from Listeria monocytogenes (32). A few other combinations are found in no more than five proteins. Most of the combinations identified in our mutagenesis screen do not occur in natural proteins. Exceptions are the A26/K31 combination with one hit, and S26/Q31 and R26/R31 with each two proteins. This strong bias of the amino acids in the critical positions demonstrates that the evolution of these proteins is directed to providing the systems with specificity. Combinations that result in extensive cross-recognition of non-cognate RAT structures may be tolerated only if an organism contains only a single anti-termination system. Alternatively, cross-talk as observed with SacY may not cause a problem since the genes with the other RATs targeted by SacY are subject to carbon catabolite repression and therefore not expressed under conditions when SacY is active. The contribution of catabolite repression to the straightness of signalling in these conserved anti-termination systems is well documented (18).
This work and previous studies demonstrate that there are several mechanisms that together allow keeping the signalling chains in the PTS-dependent anti-termination systems straight. The major contribution is made by the recognition specificity that is determined by the RAT structures (16,18) and their interaction partners, the RNA-binding domains, especially by the amino acid pair at positions 26 and 31. Moreover, sugar transport specificity of the PTS permeases prevents activation of an anti-terminator protein in response to the ‘wrong’ sugar. Finally, the general mechanism of carbon catabolite repression contributes to the specificity in the system (18).
Specificity in signal transduction cascades is an important issue not only for the anti-terminator proteins studied here but also for two-component systems, sigma factors and classical repressors sharing strong structural similarities. The selective pressure towards a compromise between binding efficacy and interaction specificity that we have discovered in this study might be a general principle in all families of conserved regulatory systems.
FUNDING
Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie (to J.S.). Funding for open access charge: DFG funding.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Christina Herzberg for the help with some experiments and to Michel Kochoyan for providing synthetic RNAs.
REFERENCES
- 1.Szurmant H, Hoch JA. Interaction fidelity in two-component signalling. Curr. Opin. Microbiol. 2010;13:190–197. doi: 10.1016/j.mib.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen CC, Saier MH., Jr Phylogenetic, structural and functional analyses of the LacI–GalR family of bacterial transcription factors. FEBS Lett. 1995;377:98–102. doi: 10.1016/0014-5793(95)01344-x. [DOI] [PubMed] [Google Scholar]
- 3.Galvao TC, Mencia M, de Lorenzo V. Emergence of novel functions in transcriptional regulators by regression to stem protein types. Mol. Microbiol. 2007;65:907–919. doi: 10.1111/j.1365-2958.2007.05832.x. [DOI] [PubMed] [Google Scholar]
- 4.Langbein I, Bachem S, Stülke J. Specific interaction of the RNA-binding domain of the Bacillus subtilis transcriptional antiterminator GlcT with its target RNA, RAT. J. Mol. Biol. 1999;293:795–805. doi: 10.1006/jmbi.1999.3176. [DOI] [PubMed] [Google Scholar]
- 5.Manival X, Yang Y, Strub MP, Kochoyan M, Steinmetz M, Aymerich S. From genetic to structural characterization of a new class of RNA-binding domain within the SacY/BglG family of antiterminator proteins. EMBO J. 1997;16:5019–5029. doi: 10.1093/emboj/16.16.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stülke J. Control of transcription termination in bacteria by RNA-binding proteins that modulate RNA structures. Arch. Microbiol. 2002;177:433–440. doi: 10.1007/s00203-002-0407-5. [DOI] [PubMed] [Google Scholar]
- 7.Le Coq D, Lindner C, Krüger S, Steinmetz M, Stülke J. New β-glucoside (bgl) genes in Bacillus subtilis: the bglP gene product has both transport and regulatory functions similar to those of BglF, its Escherichia coli homolog. J. Bacteriol. 1995;177:1527–1535. doi: 10.1128/jb.177.6.1527-1535.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schnetz K, Stülke J, Gertz S, Krüger S, Krieg M, Hecker M, Rak B. LicT, a Bacillus subtilis transcriptional antiterminator of the BglG family. J. Bacteriol. 1996;178:1971–1979. doi: 10.1128/jb.178.7.1971-1979.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krüger S, Gertz S, Hecker M. Transcriptional analysis of bglPH expression in Bacillus subtilis: evidence for two distinct pathways mediating carbon catabolite repression. J. Bacteriol. 1996;178:2637–2644. doi: 10.1128/jb.178.9.2637-2644.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tortosa P, Declerck N, Dutartre H, Lindner C, Deutscher J, Le Coq D. Sites of positive and negative regulation in the Bacillus subtilis antiterminators LicT and SacY. Mol. Microbiol. 2001;41:1381–1393. doi: 10.1046/j.1365-2958.2001.02608.x. [DOI] [PubMed] [Google Scholar]
- 11.Singh KD, Schmalisch MH, Stülke J, Görke B. Carbon catabolite repression in Bacillus subtilis: A quantitative analysis of repression exerted by different carbon sources. J. Bacteriol. 2008;190:7275–7284. doi: 10.1128/JB.00848-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindner C, Galinier A, Hecker M, Deutscher J. Regulation of the activity of the Bacillus subtilis antiterminator LicT by multiple, enzyme I- and HPr-catalysed phosphorylation. Mol. Microbiol. 1999;31:995–1006. doi: 10.1046/j.1365-2958.1999.01262.x. [DOI] [PubMed] [Google Scholar]
- 13.Déméné H, Ducat T, de Guillen K, Birck C, Aymerich S, Kochoyan M, Declerck N. Structural mechanism of signal transduction between the RNA-binding domain and the phosphotransferase system regulation domain of the LicT antiterminator. J. Biol. Chem. 2008;283:30838–30849. doi: 10.1074/jbc.M805955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmalisch M, Bachem S, Stülke J. Control of the Bacillus subtilis antiterminator protein GlcT by phosphorylation: Elucidation of the phosphorylation chain leading to inactivation of GlcT. J. Biol. Chem. 2003;278:51108–51115. doi: 10.1074/jbc.M309972200. [DOI] [PubMed] [Google Scholar]
- 15.Aymerich S, Steinmetz M. Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family. Proc. Natl Acad. Sci. USA. 1992;89:10410–10414. doi: 10.1073/pnas.89.21.10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schilling O, Langbein I, Müller M, Schmalisch M, Stülke J. A protein-dependent riboswitch controlling ptsGHI operon expression in Bacillus subtilis: RNA structure rather than sequence provides interaction specificity. Nucleic Acids Res. 2004;32:2853–2864. doi: 10.1093/nar/gkh611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, Declerck N, Manival X, Aymerich S, Kochoyan M. Solution structure of the LicT-RNA antitermination complex: CAT clamping RAT. EMBO J. 2002;21:1987–1997. doi: 10.1093/emboj/21.8.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schilling O, Herzberg C, Hertrich T, Vörsmann H, Jessen D, Hübner S, Titgemeyer F, Stülke J. Keeping signals straight in transcription regulation: specificity determinants for the interaction of a family of conserved bacterial RNA-protein couples. Nucleic Acids Res. 2006;34:6102–6115. doi: 10.1093/nar/gkl733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Declerck N, Vincent F, Hoh F, Aymerich S, van Tilbeurgh H. RNA recognition by transcriptional antiterminators of the BglG/SacY family: functional and structural comparison of the CAT domain from SacY and LicT. J. Mol. Biol. 1999;294:389–402. doi: 10.1006/jmbi.1999.3256. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 21.Wacker I, Ludwig H, Reif I, Blencke HM, Detsch C, Stülke J. The regulatory link between carbon and nitrogen metabolism in Bacillus subtilis: regulation of the gltAB operon by the catabolite control protein CcpA. Microbiol. 2003;149:3001–3009. doi: 10.1099/mic.0.26479-0. [DOI] [PubMed] [Google Scholar]
- 22.Kunst F, Rapoport G. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 1995;177:2403–2407. doi: 10.1128/jb.177.9.2403-2407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bi W, Stambrook PJ. Site-directed mutagenesis by combined chain reaction. Anal. Biochem. 1998;256:137–140. doi: 10.1006/abio.1997.2516. [DOI] [PubMed] [Google Scholar]
- 24.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 25.Youngman P. Use of transposons and integrational vectors for mutagenesis and construction of gene fusions in Bacillus subtilis. In: Harwood CR, Cutting SM, editors. Molecular Biological Methods for Bacillus. Chichester: John Wiley & sons; 1990. pp. 221–266. [Google Scholar]
- 26.Martin-Verstraete I, Débarbouillé M, Klier A, Rapoport G. Interactions of wild type and truncated LevR of Bacillus subtilis with the upstream activating sequence of the levanase operon. J. Mol. Biol. 1994;241:178–192. doi: 10.1006/jmbi.1994.1487. [DOI] [PubMed] [Google Scholar]
- 27.Bachem S, Stülke J. Regulation of the Bacillus subtilis GlcT antiterminator protein by components of the phosphotransferase system. J. Bacteriol. 1998;180:5319–5326. doi: 10.1128/jb.180.20.5319-5326.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stülke J, Martin-Verstraete I, Zagorec M, Rose M, Klier A, Rapoport G. Induction of the Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol. Microbiol. 1997;25:65–78. doi: 10.1046/j.1365-2958.1997.4351797.x. [DOI] [PubMed] [Google Scholar]
- 29.Ludwig H, Homuth G, Schmalisch M, Dyka FM, Hecker M, Stülke J. Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol. Microbiol. 2001;41:409–422. doi: 10.1046/j.1365-2958.2001.02523.x. [DOI] [PubMed] [Google Scholar]
- 30.Declerck N, Minh NL, Yang Y, Bloch V, Kochoyan M, Aymerich S. RNA recognition by transcriptional antiterminators of the BglG/ SacY family: mapping of SacY RNA binding site. J. Mol. Biol. 2002;319:1035–1048. doi: 10.1016/S0022-2836(02)00373-X. [DOI] [PubMed] [Google Scholar]
- 31.Msadek T, Kunst F, Klier A, Rapoport G. DegS-DegU and ComP-ComA modulator-effector pairs control expression of the Bacillus subtilis pleiotropic regulatory gene degQ. J. Bacteriol. 1991;173:2366–2377. doi: 10.1128/jb.173.7.2366-2377.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brehm K, Ripio MT, Kreft J, Vázquez-Boland JA. The bvr locus of Listeria monocytogenes mediates virulence gene repression by beta-glucosides. J. Bacteriol. 1999;181:5024–5032. doi: 10.1128/jb.181.16.5024-5032.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]