Abstract
U1 Adaptors are a recently reported novel approach for targeted reduction of mRNA transcripts. A U1 adaptor oligonucleotide comprising of a target-complimentary hybridization domain and a U1 recruitment domain, directs the U1 snRNP complex to the terminal exon of a targeted gene, subsequently inhibiting poly(A) tail addition and leading to degradation of that RNA species within the nucleus. Here, we present data demonstrating U1 adapter-mediated gene silencing can result in significant ‘off-target’ silencing effects as demonstrated by the reduction of multiple mRNA species that were not intended to be targeted. Our data suggest that a substantial portion of this U1 adaptor-mediated off-target mRNA reduction is the result of sequestration U1 snRNP at levels sufficient to affect splicing and processing of non-target transcripts.
INTRODUCTION
Recently, an approach for reducing gene expression based on ∼25-nt sequences called U1 adaptors was reported (1). U1 adaptors are comprised of a 5′ sequence or ‘target domain’, which hybridizes to the final exon of an mRNA target and a 3′ sequence or ‘U1 domain’ that binds to the U1 small nuclear RNA component of the U1 small nuclear ribonucleoprotein (U1 snRNP). Reportedly, tethering U1 snRNP to the target pre-mRNA inhibits poly(A)-tail addition, causing degradation of that RNA in the nucleus. U1 adaptors were reported to inhibit both endogenous and reporter genes in a sequence-specific manner and it was determined that the reductions in target RNAs were quite selective.
The U1snRNP is comprised of the 164 nt U1 small nuclear RNA (snRNA) and 10 associated polypeptides. A component of the cellular splicing apparatus, it is best-known for its role in recognizing the 5′-splice sites of introns through hybridization between these sequences and the 5′-end of U1 snRNA (2). Together with SF2/ASF and hnRNP A1, the U1 snRNP modulates alternative 5′-splice site selection (3,4). In addition, the U1 snRNP also inhibits polyadenylation of some pre-mRNAs by binding to a 5′-splice-site-like sequence in the 3′-untranslated region (3′-UTR), leading to degradation of the pre-mRNA (5,6).
It has been reported that sequestration of U1 snRNP by specific, transiently expressed RNA decoys, changes the splicing of reporter pre-mRNAs (7). Since U1 adaptors rely on the binding of endogenous U1 snRNP to the target RNA, sequestration of U1 snRNPs by a U1 adaptor might be expected to affect the splicing of multiple transcripts in the cell. In addition to inhibiting splicing, U1 snRNP knockdown has also recently been demonstrated to cause premature cleavage and polyadenylation in numerous pre-mRNAs at cryptic polyadenylation signals (8). U1 adaptors may, therefore, have unintended effects on splicing and polyadenylation, potentially limiting the utility of the approach.
In this article, we show that U1adaptors non-specifically and significantly reduce expression of non-targeted genes. Our data suggest that a significant proportion of the non-specific activity is the result of the sequestering of U1 snRNP by U1 adaptors. Using a minigene splicing system, we demonstrate significant effects of U1 adaptor treatment on splicing that are similar to those observed when U1 snRNA is intentionally reduced. We also demonstrate transcriptome-wide effects on gene expression and splicing, not related to specific reduction of the intended U1 adaptor target, but resulting instead from errors in processing of pre-mRNA. The magnitude of this activity appears to be a function of the sequence of the U1 adaptor and the mRNA targeted.
MATERIALS AND METHODS
Preparation of U1 adaptors and antisense oligonucleotides
The U1 adaptors targeting RAF1, PCSK9 and SMN2 were manufactured by Integrated DNA Technologies (IDT). The sequences of the RAF1 (UA25) and PCSK9 (UA31e) U1 adaptors have been previously described (1). Synthesis and purification of phosphorothioate/2′-MOE oligonucleotides was performed using an Applied Biosystems 380B automated DNA synthesizer as described previously (9). RNAse H-dependent antisense oligonucleotide (ASOs) used for target mRNA reduction were 18–20 bases in length, full phosphorothioate with 2′-O-methoxyethyl substitutions at the positions indicated by bold type. The sequences and chemistry of the U1 adaptors and ASOs used in the study are depicted in Table 1.
Table 1.
Oligonucleotides used in study
| Name/number | Chemistry | Target | Sequence (5′–3′) |
|---|---|---|---|
| U1A-RAF1 | U1 Adaptor | RAF1 | CCGCCTGTGACATGCATTcagguaaguau |
| U1A-SMN2 | U1 Adaptor | SMN2 | CGCTTCACATTCCAGATCTGcagguaaguau |
| U1A-PCSK9 | U1 Adaptor | PCSK9 | CTCGCAGGCCACGGTCACgccagguaaguau |
| 194166 | RNaseH ASO | RAF1 | TCCCGCCTGTGACATGCATT |
| 13649 | Full MOE ASO | RAF1 | TCCCGCCTGTGACATGCATT |
| 450880 | RNaseH ASO | SMN2 | CGCTTCACATTCCAGATCTG |
| 469892 | RNaseH ASO | PCSK9 | CTCGCAGGCCACGGTCAC |
| 469508 | RNaseH ASO | RNU1 | CTCCCCTGCCAGGTAAGTAT |
| 469509 | RNaseH ASO | RNU1 | ATCCGGAGTGCAATGGATAA |
| 469511 | RNaseH ASO | RNU1 | CTACCACAAATTATGCAGTC |
| 479333 | RNaseH ASO | RNU4 | GGTATTGGGAAAAGTTTTCA |
| 479338 | RNaseH ASO | RNU6 | CCATGCTAATCTTCTCTGTA |
For U1 adaptors upper case = DNA, bold = LNA, lower case = 2′OMe. For ASOs upper case = P=S DNA bold= 2′-O-methoxyethyl.
qRT/PCR primer/probes
The sequence for the RAF1 primer/probe set used in qRT/PCR reactions is AGCTTGGAAGACGATCAGCAA for the forward primer, AAACTGCTGAACTATTGTAGGAGAGATG for the reverse primer and AGATGCCGTGTTTGATGGCTCCAGC for the probe. The sequence for the SMN2 primer/probe set is CAGGAGGATTCCGTGCTGTT for the forward primer, TCAGTGCTGTATCATCCCAAATG for the reverse primer and CGGCACAGGCCAGAGCGATG for the probe. The sequence for the PTEN primer/probe set is AATGGCTAAGTGAAGATGACAATCAT for the forward primer, TGCACATATCATTACACCAGTTCGT for the reverse primer and TTGCAGCAATTCACTGTAAAGCTGGAAAGG for the probe. The sequence for the PCSK9 primer/probe set is CCTGCGCGTGCTCAACT for the forward primer, GCTGGCTTTTCCGAATAAACTC for the reverse primer and CCAAGGGAAGGGCACGGTTAGCG for the probe. The sequence for the YY1 primer/probe set GCGAATCCATACCGGAGACA for the forward primer, AGGTTAGTTGACTGAGCAAACTTCTTATT for the reverse primer and CCCTATGTGTGCCCCTTCGATGGTT for the probe. The sequence for the IL4R primer/probe set is AGACCCCCAAAATCGTGAACT for the forward primer, TGGATAAGCCCTAGTCCTCATCTG for the reverse primer and CATACATGAGGGTCTCTTAGGTGCAT for the probe. Probe Chemistry: 5′FAM, 3′TAMRA.
U1 adaptor/ASO treatment
Tissue culture medium, trypsin and lipofectamine 2000 and T-REX-293 cells were purchased from Invitrogen (Carlsbad, CA, USA). HeLa cells were obtained from the American Type Tissue Collection (Manassas, VA, USA). HeLa cells were cultured in DMEM supplemented with 10% fetal calf serum, streptomycin (0.1 µg/ml) and penicillin (100 U/ml). The same media was used for T-REX-293 cells with the addition of 5 µg/ml blasticidin. Cells were seeded in 96-well plates at ∼50% confluency then treated the following day with the indicated concentrations of ASO/U1 adaptor in Opti-MEM media (Invitrogen) containing 4–5 µg/ml lipofectamine 2000 for 4–5 h, as described previously (10). Following, transfection cells were washed 1× with PBS, then fed with fresh growth media. The next day, total RNA was purified using an RNeasy 3000 BioRobot (Qiagen, Valencia, CA, USA) and mRNA levels assessed by qRT/PCR performed essentially as described elsewhere (11). Briefly, 10 µl of total RNA was analyzed in a final volume of 50 µl containing 200 nM gene-specific PCR primers, 0.2 mM of each dNTP, 75 nM fluorescently labeled oligonucleotide probe, 5 µl RT–PCR buffer, 5 mM MgCl2, 2 U of Platinum Taq DNA Polymerase (Invitrogen) and 8 U of RNAse inhibitor. Reverse transcription was performed for 30 min at 48°C followed by PCR: 40 thermal cycles of 30 s at 94°C and 1 min at 60°C using an ABI Prism 7700 Sequence Detector (Applied Biosystems). To avoid artifacts based upon well to well variation in cell number, mRNA levels were normalized to the total amount of RNA present in each reaction as determined by Ribogreen assay (12) (Invitrogen). The sequences of the primer/probe sets are listed in Supplementary Materials.
Northern analysis of U1 snRNA was carried out as follows. Cells plated in 10 cm dishes were treated with U1 snRNA directed ASOs at 50–100 nM using lipofectamine 2000 as described above. Total RNA was prepared the following day using RNeasy mini columns according to the manufacturer's protocol. Total of 5 µg RNA was separated in 8% ployacrylamide–7 M urea gels, then transferred to GeneScreen Plus Hybridization Transfer Membranes (PerkinElmer) using a semi-dry transfer apparatus. Northern hybridization was performed using 32P 5′-end labeled oligonucleotide probes as described previously (13). U1-1: agtcgagtttcccacatttg; U1-2: CTCCCCTGCCAGGTAAGTAT; U6: tggaacgcttcacgaatttgcg; U3: accactcagaccgcgttctctcc.
RNAse H protection assays
RNAse H protection assays were performed using a modification of a protocol described by Kaida et al. (8). Briefly, 250 000 cells were transfected with U1 adaptor or ASO for 6 h as detailed above. Cells were washed with PBS and harvested following trypsinzation. The cell pellet was resuspended in 50 µl of RSB-100 buffer (10 nM Tris–HCl, pH 7.4, 100 mM NaCl, 2.5 mM MgCl, 0.1% NP-40 and 1 mM DTT). Cells were rotated at 4°C for 20 min, then centrifuged at 10 000 rpm for 5 min. A 40 µl of cleared cell extract was incubated with 2 µM DNA ASO complimentary to the U1 snRNA 5′-end (5′-CAGGTAAGTAT-3′), 2U Escherichia coli RNAse H (New England Biolabs) for 30 min at 30°C. Total RNA was then purified using an RNeasy Mini Kit (Qiagen). U1 snRNA was analyzed by northern blot as described above.
For cellular protection assays, 293 cells were transfected with U1 adaptor or ASO for 3 h. Following a 1 h recovery, cells were transfected with ASO 469508 at 50 nM for 4 h. Isolation of total RNA and northern hybridization were then carried out as described above.
TET-inducible minigene system
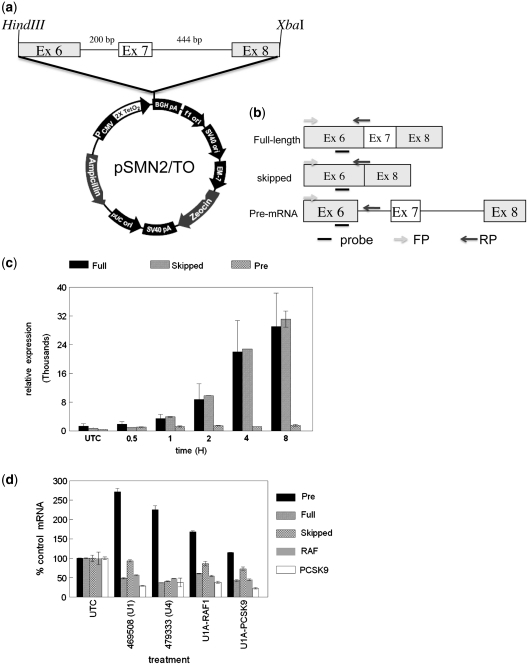
An SMN2 mini gene, comprising the 111-nt long exon 6, a 200-nt shortened intron 6, the 54-nt exon 7, the 444-nt intron 7, the first 75 nt of exon 8, under the control of the CMV and T7 RNA polymerase promoter has been previously described (14). We amplified the minigene with primer set SMN2–Hind3F (AAG CTT aag gct aga gta ctt aat acg act cac) and SMN2–Xba1R (TCT AGA TAA CGC TTC ACA TTC CAG ATC TG), and inserted them into vector into the vector pcDNA 4/TO using HindIII and XbaI restriction sites. The forward primer is complementary to the T7 promoter from pCI-SMN2 and incorporates a HindIII site, while the reverse primer was complementary to the 3′-end of the truncated exon 8 and included a XbaI site. The resultant plasmid, pcSMN2/TO, was transfected into T-REx-293 cells (Invitrogen, Carlsbad, CA, USA), using Effectene transfection reagent according to the manufacturer's protocol (Qiagen, Valencia CA, USA). Cell lines stably integrating the mini gene were selected in DMEM media containing 250 ug/ml Zeocin. Zeocin-resistant colonies were expanded then tested for tetracycline-inducible expression.
Following TET induction, total RNA was purified using RNeasy mini columns according to the manufacturer's protocol (Qiagen). Expression of pcSMN2/TO full-length, skipped and pre-mRNA was determined by qRT/PCR as detailed above. To avoid amplification of endogenous SMN1/2, all mRNA isoforms shared the same forward primer, T7F2 (TAC TTA ATA CGA CTC ACT ATA GGC TAG CCT CG) which is complimentary to the T7 promoter region (lower case) on pcSMN2/TO. To specifically amplify full-length message a reverse primer bridging the exon 6/7 junction, SMN-E6/7R, was employed (TTT TGT CTA AAA CCC ATA TAA TAG CC). Exon 7 skipped mRNA was amplified with a reverse primer bridging the exon 6/8 junction, SMN-E6/8R (ATG CCA GCA TTT CCA TAT AAT AGC). For pre-mRNA amplification the reverse primer, SMN-I6R, was designed with complementarity to intron 6 (TGT CAG GAA AAG ATG CTG AGT G). A common probe, SMN-E6P, complementary to sequence in exon 6 was used in all reactions (6FAM-CAG ATT CTC TTG ATG ATG CTG ATG CTT TGG- IABkFQ).
Affymetrix exon arrays
The Affymetrix GeneChip Human Exon 1.0 ST Array platform was used for the microarray analysis. The determination of significant differential expression was performed using functions from the ‘Bioconductor’ (15) suite of packages including ‘exonmap’ (16,17) and ‘limma’ (18). The detection of probes showing differential expression was determined by ANOVA using the limma package. P-values were adjusted for multiple testing using the method of Benjamini and Hochberg (19). Probes with adjusted P < 0.01 were called differentially expressed. Differentially expressed probes were divided into exonic or intronic probes via exonmap.
RESULTS
Evaluation of U1 adaptor specificity
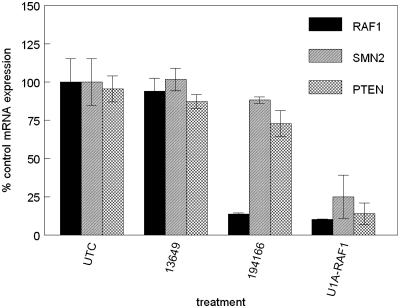
To discriminate potential off-target effects due to sequestration of U1 snRNP by U1 adaptors from those resulting from binding to the mRNA target alone, we evaluated the activity of several U1 adaptors along with ASOs targeted to the same site on the mRNA. An ASO would be expected to bind the targeted mRNA transcript with an affinity similar to that of the U1 adaptor target domain without recruitment of the U1 snRNP, allowing a direct comparison with the alternate degradation mechanism proposed for U1 adaptors. HeLa cells were transfected at 100 nM with a U1 adaptor, a fully 2′-O-methoxyethyl ribose (2′MOE) ASO, or a chimeric 2′MOE/DNA ASO targeted to the same site on RAF1 mRNA. Target mRNA reduction was evaluated by qRT/PCR the following day (Figure 1). The RNAse H-dependent ASO (194166) reduced RAF1 mRNA (solid bars) by >85%, with little reduction of levels of SMN2 (striped bars) or PTEN (hatched bars) non-target messages observed. Treatment with the RAF1 U1 adaptor (U1A-RAF1) at the same concentration resulted in a ∼90% reduction in RAF1 mRNA. In addition, the level of PTEN mRNA was reduced by approximately the same amount, while SMN2 mRNA was reduced by ∼75%. Treatment with ASO 13 496, which does not support cleavage by RNAse H (20), resulted in no significant reduction of either RAF1 or non-target mRNAs. The lack of activity observed with ASO 13496 suggests that binding alone cannot be responsible for specific or non-specific mRNA reduction resulting from treatment with the U1 adaptor or RNase H-dependent ASO.
Figure 1.
U1 adaptor downregulates multiple mRNAs. A U1 adaptor targeted to RAF1 (U1A-RAF1) was administered to Hela cells at a single-dose of 100 nM. A fully modified 2′MOE ASO (13649) and a chimeric 2′MOE/DNA RNAse H-dependent ASO (194166) complementary to the same target sequence were also administered at the same concentration. After 24 h, cells were harvested, RNA was purified and mRNA reduction assayed by qRT/PCR using primers specific for 3 mRNAs; RAF1, PTEN and SMN2. The bars represent per cent mean untreated control for three replicates and the standard errors. UTC = mock-treated control.
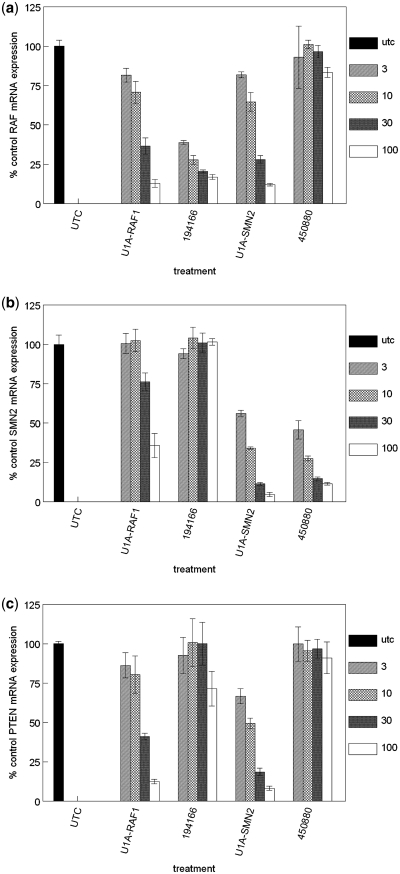
Since it has been reported that IC50's of <5 nM can be achieved with U1 adaptors (1), we wanted to determine if a window of specific target reduction could be observed at lower treatment concentrations. HeLa cells were transfected with U1A-RAF1, a second U1 adaptor targeted to SMN2 (U1A-SMN2) or the corresponding RNAse H-dependent ASOs at multiple concentrations. Expression of RAF1 mRNA was reduced by U1A-RAF1 with an IC50 of ∼20 nM (Figure 2a). U1A-SMN2 also effectively reduced RAF1 mRNA with approximately the same IC50. Reduction of SMN2 mRNA was also evaluated (Figure 2b). While U1A-SMN2 reduced expression of the targeted RNA with an IC50 of ∼3 nM, off-target reduction of SMN2 by U1A-RAF1 was less robust with an IC50 of ∼75 nM. Both U1 adaptors also effectively reduced expression of PTEN, an unrelated, non-target mRNA, with an IC50 of ∼25 nM for U1A-RAF1 and ∼10 nM for U1A-SMN (Figure 2c). ASO 194166 (RAF1) was ∼7-fold more potent than the corresponding U1 adaptor, while ASO 450880 (SMN2) and the corresponding U1 adaptor were equipotent. No significant reduction in non-targeted messages was observed for either ASO. Similar levels of reduction in non-targeted transcripts were observed in primary human umbilical vein endothelial cells (HUVEC) and in 293T cells treated with U1 adaptors (Supplementary Figure S1). Together, these data suggest that transcripts may be differentially sensitive to U1 adaptor-mediated off-target activity and that off-target activity can vary by site and target.
Figure 2.
Potency and specificity of U1 adaptors and RNAse H ASOs. A second U1 adaptor was designed to target SMN2 (U1A-SMN2). HeLa cells were treated at concentrations of 3 nM, 10 nM, 30 nM, or 100 nM with U1A-SMN2, U1A-RAF1, ASO 194166 (RAF1) or ASO 450880 (SMN2). After 24 h, cells were harvested and RNA purified. Messenger RNA reduction was analyzed by qRT/PCR. The percent mean mock-treated control (UTC) for three to four replicates and the standard error is shown for each concentration. (a) Percent control RAF1 mRNA expression. (b) Percent control SMN2 mRNA expression. (c) Percent control PTEN mRNA expression.
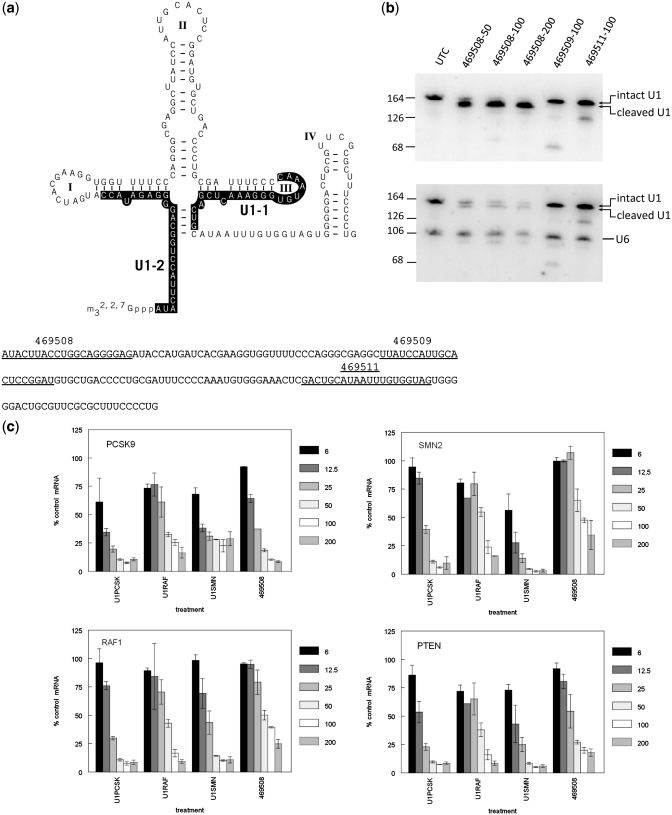
ASO-mediated knockdown of U1 snRNP
To directly evaluate and compare the contribution of U1 snRNP reduction with U1 adaptor off-target activity, we sought to decrease the amount of functional U1 snRNP directly, using RNAse H ASOs designed with complementarity to U1 snRNA (Figure 3a). HeLa cells were transfected with the ASOs at concentrations ranging from 50 nM to 200 nM. The following day, total RNA was purified and U1snRNA reduction analyzed by northern blot. Blots hybridized with the internal U1 northern probe (U1-1) showed a significant shift in the migration of U1snRNA following treatment with 469508 consistent with cleavage of the 5′ U1 snRNA binding sequence, even at the lowest concentration evaluated (Figure 3a and b, upper panel). However, the lack of reduction in signal intensity suggests that there is little degradation of the U1 snRNA following RNaseH cleavage, likely due to the highly structured nature of the RNA target. To confirm this, the same blot was hybridized with a second Northern probe, U1-2, complimentary to the cleaved U1 snRNA 5′-end (Figure 3b, lower panel). As observed with probe U1-1, treatment with ASO 469508 resulted in a shift in mobility consistent with the expected U1 snRNA cleavage product. In addition, a clear reduction in U1 snRNA was observed using probe U1-2, with >75% reduction of U1 snRNA at 50 nM and >90% reduction at 200 nM (normalized to the U6 snRNA loading control). Together, these data confirm that treatment with 469508 efficiently directs RNAse H cleavage of the U1 leader sequence previously shown to be required for U1 splicing activity (8). The other ASOs evaluated, 469509 and 469511, each produced cleavage fragments consistent with their localization on the target, however, the potency of these ASOs was much less than that of 469508.
Figure 3.
Reduction of U1 snRNA with RNAse H ASOs. (a) Chimeric 2′MOE/DNA RNAse H-dependent ASOs were designed to target U1snRNA. The location of 3 RNAse H ASOs is underlined on the U1 snRNA sequence. 469508 (1–20), 469509 (59–78), 469511 (117–138). Northern probe location is highlighted on the U1 snRNA structure. (b) HeLa cells were treated with ASOs at the indicated concentrations (nM). The following day total RNA was purified and U1snRNA reduction analyzed by northern blot with the internal U1 probe (U1-1, upper panel), or with a second probe, U1-2, targeted to the U1 5′-end and the non-target control and U6 snRNA probe (lower panel). Band intensity was quantitated with a Storm 850 Phosphor-Imager and U1 snRNA normalized to U6 snRNA signal for each lane. (c) Effect of U1 adaptor treatment and U1 snRNA reduction on mRNA expression. HeLa cells seeded in 96-well plates were treated with U1A-RAF1, U1A-SMN2, U1A-PCSK9 or anti-U1 ASO 469508, in triplicate at concentrations ranging from 6 to 200 nM. The following day total RNA was purified and mRNA levels of each targeted gene assessed by qRT/PCR. Results are normalized to the total amount of RNA present in each reaction as determined by Ribogreen assay and plotted as percent control compared to mock-treated cells for three to four replicates with the standard error.
We next evaluated the effects of U1 snRNA inactivation with ASO 469508 as compared to U1 adaptor treatment. HeLa cells seeded in 96-well plates were treated in triplicate with anti-U1 ASO 469508 or with U1A-RAF1, U1A-SMN2 or U1A-PCSK9 at concentrations ranging from 6 nM to 200 nM. After 16 h, levels of targeted and non-targeted mRNA were evaluated by qRT/PCR. U1A-PCSK9 and U1A-SMN2 both effectively reduced target mRNAs with IC50's <10nM (Figure 3c). U1A-PCSK9 and U1A-SMN2 also reduced non-target messages with IC50's between 10nM and 25 nM. U1A-RAF1 was ∼5-fold less potent, with an IC50 of ∼50 nM for both target and non-target mRNAs. Levels of each mRNA were also decreased as a result of U1 snRNA reduction with ASO 469508. PCSK9, RAF1 and PTEN mRNA were all reduced to levels comparable to the U1 adaptor off-target reduction for each of these messages with IC50's of 20–50 nM. SMN2 RNA was also reduced following treatment with ASO 469508; however, the IC50 of ∼100 nM was significantly greater than that observed for the specific or non-target U1 adaptors.
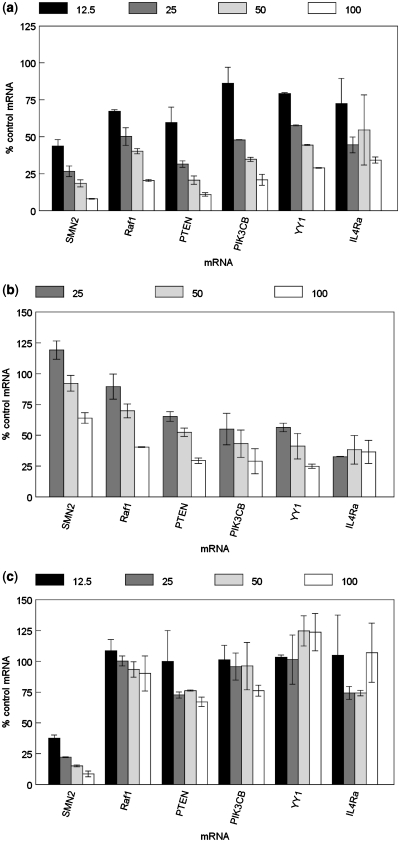
To further evaluate off-target effects, HeLa cells were treated with U1A-SMN2, ASO 450880 and ASO 469508 at concentrations from 12.5 to 100 nM. The following day, expression of a panel of mRNAs was evaluated by qRT/PCR. Treatment with U1A-SMN2 resulted in an IC50 for SMN2 mRNA expression of ∼10 nM (Figure 4a). Non-target mRNAs were also effectively reduced by treatment with U1A-SMN2 with IC50's between 20 and 100 nM, and even at the lowest concentration, significant reduction was observed for several of the non-target transcripts. Treatment with anti-U1 snRNA ASO 469508 also resulted in reduction of the same set of transcripts with similar IC50's (Figure 4b). Note that, as in Figure 3c, SMN2 was the target least affected by disabling of U1 snRNA, suggesting that for this mRNA, a significant proportion of the observed activity for the U1 adaptor is specific and mediated by the proposed mechanism. For other targets, such as PCSK9 (Figure 3c) and PIK3CB (compare Figure 4a and b), the similarity of the potency of mRNA reduction between the U1 adaptor and U1 snRNA reduction suggests that the activity is more related to the effects U1snRNP sequestration by the U1 adaptor rather than the proposed specific deadenylation/degradation of the message mediated by the U1 adaptor. SMN2 ASO 450880 had approximately the same IC50 as the corresponding U1 adaptor, but with no significant activity observed at 10 nM for the other mRNA targets and only slight reduction observed for some transcripts at the highest concentration (Figure 4c), again indicating that the observed off-target activity is specific to treatment with U1 adaptors. These results were confirmed and expanded, as an even larger number of overlapping transcripts were identified by PCR array as being reduced by treatment with U1A-RAF1 and the anti-U1 ASO 469508, but not by the corresponding RNAse H-dependent ASO (Supplementary Figure S2).
Figure 4.
U1 adaptors and anti-U1 snRNA ASOs promote reduction of multiple non-targeted mRNAs. HeLa cells were treated with U1A-SMN2, SMN2 ASO 450880, or U1 ASO 469508 at concentrations from 12.5 to 100 nM. The following day cells were harvested and RNA purified. Messenger RNA reduction was analyzed by qRT/PCR. Percent mock-treated control expression is shown for the six transcripts identified by Entrez Gene symbol at the bottom of the figure. (a) Cells treated with U1A-SMN2. (b) Cells treated with anti-U1 RNAse H ASO 469508. (c) Cells treated with SMN2 RNAse H ASO 450880.
U1 adaptors sequester U1 snRNP
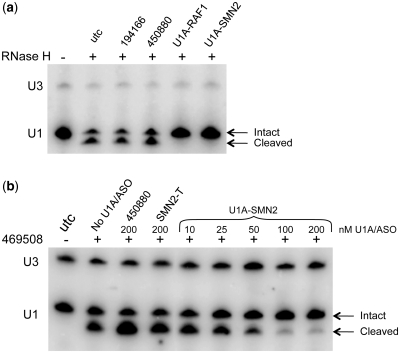
To verify that U1 adaptors sequester U1 snRNP, we performed RNAse H protection assays (Figure 5a). Extracts from cells transfected with U1A-RAF1 and U1A-SMN2 or with the corresponding RNAse H ASOs were incubated with E. coli RNAse H and a DNA ASO probe complimentary to the U1 snRNA 5′ leader sequence. In extracts prepared from untreated cells or cells treated with ASOs, a fragment corresponding to the size expected for RNAse H cleavage was observed. However, in extracts from cells treated with either U1 adaptor, no cleavage product was observed, indicating that the U1 adaptor prevented binding of the DNA ASO probe and subsequent RNAse H digestion.
Figure 5.
U1 adaptors bind the 5′ sequence of U1 snRNA and inhibit antisense hybridization. (a) 293-T cells were transfected with 100 nM U1 adaptor/ASO for 6 h. RNAse H protection assays were performed with whole-cell extracts ± 2U E. coli RNAse H and 2 µM DNA oligonucleotide antisense to the 5′ sequence of U1. Cleavage of U1 snRNA was detected by northern blot using internal U1 probe (U1-1), and normalized using a U3 probe (U3). (b) Cellular RNAse protection assay. Cells were treated with U1A_SMN2 or ASO at concentrations ranging from 10 to 200 nM as shown. Cells were then transfected with anti U1 RNAse H ASO 469508 at 50 nM and U1 snRNA cleavage assessed by northern blot using internal U1 probe (U1-1). Band intensity was quantitated with a Storm 850 Phosphor-Imager and U1 snRNA signal normalized to U3 for each lane.
Cellular RNAse H protection assays were also performed. 293T cells were treated with U1A-SMN2 at concentrations ranging from 10 to 200 nM or with the corresponding target ASOs at 200 nM. Cells were then transfected with anti-U1 RNAse H ASO 469508 and U1 snRNA cleavage assessed by northern blot. In the absence of 469508 no cleavage of U1 snRNA was observed (Figure 5b, Lane 1). Approximately 50% cleavage was observed in the absence of the U1 adaptor (Lane 2); however, cleavage was reduced in the presence of U1A-SMN2 with nearly complete blocking of RNAse H cleavage by 469508 at 100 nM and an IC50 of ∼35 nM. Treatment with The RNAse H ASO 450880 or with the target portion of the U1 adaptor only (SMN2-T) had no effect on RNAse H cleavage of U1 snRNA. U1A-RAF1 and U1A-PCSK9 were also evaluated in this assay and found to block RNAse H cleavage of U1 snRNA with IC50's of 50 and 20 nM, respectively (data not shown). These data strongly suggest that the off-target effects of U1 adaptors are related to sequestration of the U1 snRNP.
U1 adaptors alter splicing of pre-mRNA
Since reduction of U1 snRNP has been demonstrated to result in alteration of splicing (7), we utilized an SMN2 minigene system to compare the effects of U1 adaptor treatment and U1 snRNP reduction on alternative splicing. The skipping of exon 7 of the SMN2 gene has been well characterized (14). We subcloned the previously reported SMN2 mini gene (21) into the tetracycline inducible expression plasmid pcDNA 4/TO for stable integration into 293 cells (Figure 6a). SMN2/TO-293 cells were induced with tetracycline at 0.25 ug/ml for 30 min to 6 h. Expression of SMN/TO mRNA assessed using the splice-form specific qRT/PCR primer/probe sets shown in Figure 6b, demonstrated tightly regulated TET induction of the spliced full-length and exon 7 skipped mini gene mRNA (Figure 6c). Reduction of U1 snRNA by ASO 469508, as well as reduction of U4 and U6 snRNAs, was found to result in an increase in SMN2/TO pre-mRNA levels (Supplementary Figure S3A). In addition, reduction of U1, but not U4 or U6 snRNA resulted in an increase in the ratio of skipped relative to full-length spliced mRNA (Supplementary Figure S3B).
Figure 6.
U1 adaptors and anti-U1 snRNA ASOs alter mini gene splicing and expression. (a) A tetracycline inducible SMN2 mini gene, comprised of the 11-nt long exon 6, a 200-nt shortened intron 6, the 54-nt exon 7, the 444-nt intron 7, the first 75 nt of exon 8, from pCI-SMN2 was subcloned into the vector pcDNA 4/TO using PCR generated HindIII and XbaI sites as detailed in the ‘Materials and Methods’ section. Stable TREX 293 cell transformants from transfected cells were obtained by zeocin selection. (b) Location of qRT/PCR primers and probe used to amplify SMN/TO full-length, exon 7 skipped, and pre-mRNA. (c) Kinetics of SMN/TO tetracycline induction. SMN2/TO-293 cells were treated with TET at 0.25 ug/ml for 30 min to 6 h. Expression of SMN/TO mRNA assessed using the splice-form specific qRT/PCR primer/probe sets shown in Figure 5B and as detailed in ‘Materials and Methods’ section. (d) SMN/TO 293 cells were treated with U1A-RAF, U1A-PCSK9, anti-U1 ASO 469508, or anti-U4 ASO 479333 at 50 nM. The following day SMN/TO expression was induced for 4 h, then expression of SMN/TO and target mRNA assessed. Results are presented as percent mock-treated control (UTC) for each mRNA assayed. Pre-mRNA, filled bars; full-length SMN/TO mRNA, striped bars; exon 7-skipped SMN/TO mRNA, hatched bars; RAF1 mRNA, gray bars; PCSK9 mRNA, open bars.
The effects of U1 adaptors on splicing in the SMN minigene system were next evaluated. SMN/TO 293 cells were treated with U1A-RAF, U1A-PCSK9, ASO 469508 (U1 snRNA) or ASO 479333 (U4 snRNA) at 50 nM. The following day SMN/TO expression was induced for 4 h, then expression of SMN/TO and target mRNA assessed. Again, reduction of either U1 or U4 snRNA resulted in an increase in the amount of SMN/TO pre-mRNA (Figure 6d, solid bars). U4 snRNA reduction also led to reduction in expression of both the spliced forms of SMN/TO and of the PCSK9 and RAF1 mRNAs. As previously observed, U1 snRNA reduction resulted in a change in the ratio SMN/TO spliced mRNA, causing an increase in the skipped (hatched bars) relative to the full-length (striped bars) form, although overall expression of spliced SMN/TO message was reduced relative to the untreated control. Both U1 adaptors behaved similarly, with increases in pre-SMN/TO levels and an increase in the skipped relative to the full-length spliced message. Note that treatment with U1A-RAF1 or U1A-PCSK9, as well as ASO 469508 or ASO 479333, reduced RAF1 to similar levels as compared to control cells (gray bars), again demonstrating virtually no selectivity for these U1 adaptors and activities that certainly must be related to sequestration of U1 snRNP. A relative increase in skipping of the endogenous SMN2 exon 7 was also observed following treatment with both U1A-RAF and ASO 469508, while the RAF1 ASO, 194166 again had no effect (Supplementary Figure S4).
U1 adaptor treatment and U1 snRNA knockdown have similar effects on pre-mRNA processing
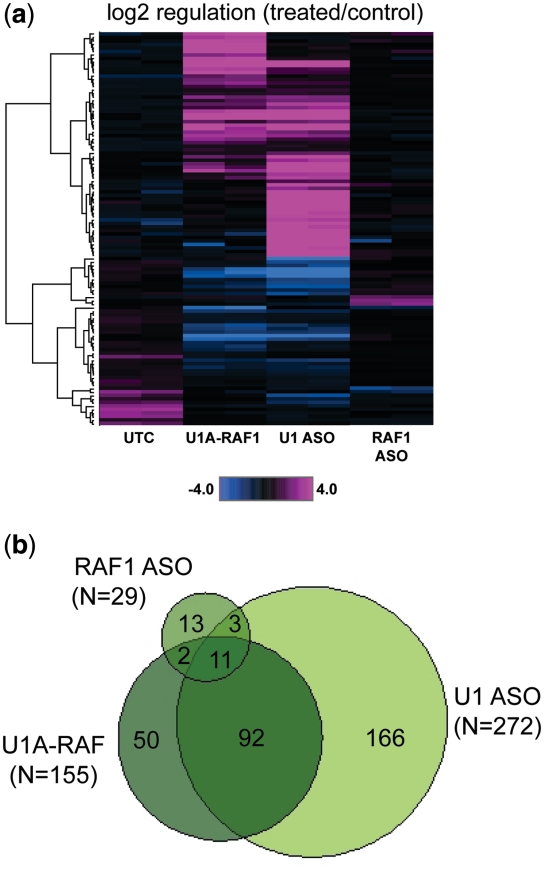
We next sought to evaluate and compare transcriptome-wide effects on expression and splicing following treatment with U1A-RAF1 or direct inactivation of U1 snRNP with ASO 469508. HeLa cells were transfected with U1A-RAF1, ASO 469508, or ASO 194166, at a concentration of 50 nM. After 16 h, target reduction was confirmed by qRT/PCR (Supplementary Figure S5A) and affymetrix exon arrays were performed using total mRNA purified from the U1 adaptor/ASO-treated cells. ANOVA was used to detect probes showing significant changes in either direction among the groups. P-values were then corrected for multiple testing and probes having adjusted P < 0.01 were selected. Compared to untreated cells, log2-fold expression changes were found to be highly significant for U1A-RAF1 and U1 ASO 469508 treated, but not for cells treated with RAF1 ASO 194166 (Figure 7a). Comparison of total signatures for each treatment group versus mock-treated (P < 1e-3 uncorrected) revealed a highly significant overlap (P = 2.5e-160) between genes regulated by U1A-RAF1 and ASO 469508, with ∼60% of the U1A-regulated genes in common with the anti-U1 regulated genes (Figure 7b). ASO 194166 was found to alter the expression of far fewer genes, which overlap with those transcripts altered by U1A-RAF1 (P = 2.7e-22) or with those regulated by the anti-U1 ASO, 469 508 (P = 1.2e-20).
Figure 7.
Global effects of RAF1 U1 adaptor and anti-U1 ASO treatment. HeLa cells were transfected with U1A-RAF, anti-U1 ASO 469508, or ASO 194166 at 50 nM. Total RNA was purified the following day, and mRNA reduction evaluated by qRT/PCR. (a) Affymetrix exon arrays were performed using total RNA. Data were clustered at the transcript level using ANOVA-selected genes (N = 112, treated versus control, P < 0.1). Shown is log2 regulation for U1 adaptor/ASO treated cells versus control. (b) Venn diagrams comparing total signatures for U1 adaptor/ASO versus control. U1A-RAF1, dark green; ASO 469508, light green; ASO 194 166, green.
It has recently been demonstrated that, distinct from its role in splicing, U1 snRNP also functions to protect pre-mRNAs from premature cleavage and polyadenylation (PCPA) (8). Therefore, regulation of intron and exon probe intensity was also analyzed in an attempt to identify changes that may be affecting pre-mRNA processing. Treatment with U1A-RAF1 or with anti-U1 ASO 469508 resulted in significant increases in intron probe levels suggesting disruption of pre-mRNA splicing (Supplementary Figure S5B). Similarly, exon probe expression changes were significant for U1 adaptor and anti-U1 ASO-treated cells, but not cells treated with the RNAse H ASO 194166 (Supplementary Figure S5C). Expression changes for a portion of these transcripts were confirmed by RT/PCR using nuclear or cytoplasmic fractions of total RNA treated with U1A-RAF1 (Supplementary Figure S5D). Most transcripts from U1A-RAF1-treated cells were reduced in the cytoplasmic RNA fraction. In some cases, the transcripts were also reduced in the nuclear fraction. However, many of the transcripts were increased in the nuclear RNA fraction. This increase of unprocessed mRNA in the nucleus and corresponding decrease of the spliced transcript in the cytoplasm is consistent with a model in which U1 adaptors sequester U1 snRNA, effectively lowering the concentration of U1 snRNP in the cell, leading to PCPA and/or disruption in mRNA splicing.
DISSCUSSION
U1 adaptors are designed to promote the degradation of target RNA by tethering the U1 snRNP to the final exon of the mRNA via antisense hybridization (1). Although U1 adaptors have been reported to inhibit both endogenous and reporter genes in a sequence-specific manner, a recent report demonstrating that sequestration of U1 snRNP by RNA decoys can result in the alteration of splicing and subsequent expression of reporter pre-mRNAs (7), suggests the possibility that U1 adaptors may have unintended effects on gene expression. While it has been postulated that sequestration of U1 snRNP by low nanomolar amounts of U1 adaptors would have little effect on the overall splicing given the abundance of the U1 snRNP in the cell, in light of the high concentrations that oligonucleotides are known to achieve in vitro and in vivo (22), this may not be the case. Further, it recently been suggested that cellular levels of U1 in excess of what is required for splicing may be required to suppress premature cleavage and polyadenylation in introns (8), so even small changes in levels of U1 may have deleterious effects on pre-mRNA processing.
Our initial experiments compared the activity of U1 adaptors and ASOs targeting the same sequence. While the activity of U1 adaptors has been compared with siRNAs targeting the same sequence, no such comparison has been made with ASOs designed to utilize an RNAse H-dependent mechanism. We chose to use RNAse H ASOs as a specificity control rather than siRNAs, as the ASO chemistry is more similar to that of a typical U1 adaptor and we were concerned about making comparisons with compounds known to promote degradation of off-target transcripts (23). While the RNase H ASOs targeting RAF1 and SMN2 had similar potency for reduction of the targeted transcript and little significant off-target effect, the target-specific as well as off-target activity of the corresponding U1 adaptors was more variable (Figure 2). The target-specific activity observed for U1A-RAF1 was in close agreement the previously published activity for this compound (1). However, we observed significantly more off-target reduction which, in the case of PTEN mRNA, was nearly equivalent to reduction of the intended target (Figure 2c). This may simply be a consequence of the difference in non-targeted transcripts which we chose to evaluate. The potency of off-target activity may also be related to specific sequence of each U1 adaptor. For instance, U1A-SMN2 was clearly more potent than U1A-RAF1; however, the potency of off-target mRNA reduction, though significant, was less, relative to target-specific reduction, than that of U1A-RAF (Figure 2a and b). Interestingly, U1A-PCSK9 had potency similar to that of U1A-SMN2; however, there was almost no difference between target-specific and off-target activity for this compound (Figure 3c). While there does appear to be a window of specificity for some U1 adaptors, none of the U1 adaptors that we evaluated were free of off-target activity. Even for the most specific, the ratio of on- to off-target activity was ∼7-fold. In contrast, off-target IC50's were never reached for RNAse H-dependent ASOs, despite similarities in target-specific potency.
Treatment of cells with the RNAse H-dependent U1 snRNA ASO, 469508, resulted in cleavage of the 5′-leader sequence responsible for 5′-splice site recognition by the U1 snRNP (Figure 3b) (24). For some transcripts, such as PCSK9 and YY1, reduction of U1 snRNA using this ASO resulted in activity nearly indistinguishable from the off-target activity promoted by the U1 adaptor (Figures 3c, 4a and b), while for other transcripts, such as SMN2 and RAF1, the off-target effect of the U1 adaptor was greater than that observed by directly targeting U1 snRNA. These data suggest that at least a portion of the observed U1 adaptor off-target activity may be related to U1 snRNP sequestration; however, certain transcripts appear to be more susceptible to the effects of U1 reduction than others. It has recently been shown that when levels of U1 were reduced, pre-mRNAs containing intronic polyadenylation signals are prematurely cleaved and polyadenylated (PCPA) within the intron (8). Our RNAse H protection assays clearly showed that U1 adaptors effectively reduced the levels of functional U1 snRNP at concentrations consistent with those at which off-target transcript reduction was observed (compare Figures 3c and 5b). Therefore, it is likely that differences in the presence of intronic poly(A) signals or in the ability of a particular U1 adaptor to sequester U1 snRNA, may account for differences in off-target activity between transcripts. There did appear to be some correlation of off-target activity with the affinity of U1 adaptors for U1 snRNA, as U1A-SMN2 and U1A-PCSK9 blocked U1 snRNA more effectively than did U1A-RAF1 (Figure 5). It has been suggested that vulnerability to PCPA would be expected to increase with increasing intron size if U1 snRNP is not available to suppress utilization of cryptic polyadenylation signals. Indeed, SMN2 which was relatively resistant to the non-specific effects of U1 adaptor treatment and U1 snRNA reduction, has fewer, and on average, shorter introns, than either RAF1 or PTEN.
To further explore the mechanism of U1 adaptor-mediated off-target mRNA reduction, we compared the effect of U1 adaptor and ASO treatment on splicing and expression using a tetracycline inducible SMN2 minigene construct (Figure 6) (25). In agreement with Roca and Krainer (7), who used U1-specific RNA decoys to sequester U1 snRNP, we found that ASO 469508-mediated cleavage of the 5′-end of U1 snRNA resulted in reduced SMN2 exon 7 inclusion in the TET-inducible mini gene (Figure 6d). In addition, a significant increase in the level of the unspliced pre-mRNA for the mini gene was observed. It is likely that U1 reduction differentially affects splicing by favoring interactions of exons with the more favorable splice site consensus sequences (26,27) when U1snRNP is present in limiting quantities. Note that reduction of U4 snRNA resulted in an equal reduction of the skipped and full-length messages with a corresponding increase in the level of the pre-mRNA (see also Supplementary Figure S3). U2 and U6 snRNA were also reduced in this system with no effect on the ratios of the skipped to full-length mRNA (data not shown). Treatment with U1 adaptors resulted in an expression profile similar to that observed with U1 reduction; i.e. an increase in pre-mRNA levels and the ratio of skipped to full-length spliced mRNA. Together these data suggest that U1 adaptors may have unintended effects on mRNA splicing as the result of sequestration of U1 snRNP. Our results differ from those previously reported by Goraczniak et al. (1) who observed no effects of treatment with U1 adaptors on splicing of a reporter gene or of four endogenous genes. We chose to evaluate effects on splicing near the IC80 for the U1 adaptor rather than the IC50. As a result, we treated cells with ∼10-fold more U1 adaptor. It is also possible that our tetracycline-inducible splicing reporter system is more sensitive to the effects of U1 reduction. However, inclusion of exon 7 in the endogenous SMN2 gene was also reduced by U1A-RAF1 or anti-U1 ASO treatment (Supplementary Figure S4). Our data suggest that U1 adaptors vary in their ability to promote off-target mRNA splicing and reduction (Figures 3c and 6d), but that, at least in some cases, they are no more specific than is reducing U1 snRNA.
In a previous study, cells treated with U1 adaptors targeting PCSK9 were assessed by microarray profiling, and compared head-to-head with an siRNA directed against PCSK9 (1). The two methods of transcript reduction were determined share a high degree of overlap, with 93% concordance in gene expression changes and it was concluded that U1 adaptors do not have a significant off-target profile as compared with siRNA. However, it is possible that the high degree of concordance resulted from comparison of two treatments both of which may produce significant off-target effects (28). In fact, significant changes in gene expressions were observed in both U1 adaptor and siRNA-treated groups (1). We used Affymetrix Exon Arrays to evaluate off-target effects of treatment with U1A-RAF1 and anti-U1 ASO 469508 since they had similar potency for mRNA reduction (Figures 3 and 4). Given that significant and widespread off-target activity due to seed region homology has been reported for siRNAs (23,28), while the hybridization of a typical 18–20 nt ASO has been determined to approach nearly theoretical specificity (29,30), we chose employ the corresponding RNAse H-dependent ASO as our experimental control. While treatment with the corresponding ASO produced few significant changes in gene expression relative to mock-treated control cells, treatment with both U1A-RAF1 and ASO 469508 lead to significant changes in gene regulation (Figure 7a). Importantly, a large degree of overlap was observed between the U1A-RAF1 and the anti-U1 ASO 469508 groups, while the RAF1 ASO regulated far fewer genes which overlapped significantly with those regulated by U1A-RAF1 or ASO 469508 (Figure 7b). Further analysis of the array data revealed that specific intron and exon probes were increased or decreased in U1A-RAF and anti-U1 ASO 469508 treated cells relative to control (Supplementary Figure S5B and C). Several of these transcripts were confirmed by qRT/PCR. Consistent with disruption of splicing and PCPA, the pre-mRNA for a significant proportion of the transcripts was increased in the nucleus of cells treated with U1A-RAF1 or ASO 469508 with a corresponding decrease in the spliced mRNA in the cytoplasm (Supplementary Figure S5D).
Our data confirm that U1 adaptors are capable of reducing targeted mRNA transcripts at concentrations comparable to that obtained with chimeric ASOs supporting RNAse H-mediated cleavage of the target. However, quantitative RT/PCR with multiple U1 adaptors, PCR arrays and Affymetrix exon arrays all show considerable reduction of non-target transcripts by U1 adaptors as compared to ASOs at concentrations where phenotypic effects would be expected to occur. It is possible that a portion of the observed non-specific activity may be the consequence off-target hybridization and subsequent deadenylation/degradation of the transcript (31), or the result of RNAse H-mediated cleavage of non-target sequences bound by the LNA/DNA portion the oligonucleotide. All U1 adaptors used in this study had several DNA gaps of three or four bases. It has been shown that DNA gaps of four bases can efficiently direct RNAse H cleavage of a target (32); however, no target or off-target reduction of mRNA was observed when cells were treated with only the target portion of the U1A-RAF1 adaptor (Figures 1 and 5b). Our data suggest that a significant portion of U1 adaptor-mediated off-target mRNA reduction is the result of sequestering of U1 snRNP. For some U1 adaptors such as U1A-SMN2 a narrow window between on- and off-target potency was observed, while for others such as U1A-RAF1 and U1A-PCSK9 there was very little difference. Clearly, U1 adaptor sequences will require careful screening to minimize U1 snRNP sequestration and establish maximal specificity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: ISIS Pharmaceuticals.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Peter Linsley for help in the analysis of exon array data. We also thank Walt Lima, Xue-hai Liang and Hongjiang Wu for stimulating discussions; Frank Bennett and Frank Rigo for critical reading of the manuscript; and Tracy Reigle for help in preparation of the figures.
REFERENCES
- 1.Goraczniak R, Behlke MA, Gunderson SI. Gene silencing by synthetic U1 Adaptors. Nat. Biotechnol. 2009;27:257–263. doi: 10.1038/nbt.1525. [DOI] [PubMed] [Google Scholar]
- 2.Seraphin B, Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- 3.Eperon IC, Makarova OV, Mayeda A, Munroe SH, Caceres JF, Hayward DG, Krainer AR. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol. 2000;20:8303–8318. doi: 10.1128/mcb.20.22.8303-8318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhuang Y, Weiner AM. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 5.Furth PA, Choe WT, Rex JH, Byrne JC, Baker CC. Sequences homologous to 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol. Cell. Biol. 1994;14:5278–5289. doi: 10.1128/mcb.14.8.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gunderson SI, Polycarpou-Schwarz M, Mattaj IW. U1 snRNP Inhibits Pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 7.Roca X, Krainer AR. Recognition of atypical 5′ splice sites by shifted base-pairing to U1 snRNA. Nat. Struct. Mol. Biol. 2009;16:176–182. doi: 10.1038/nsmb.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 468:664–669. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker BF, Lot SS, Condon TP, Cheng-Flournoy S, Lesnik EA, Sasmor HM, Bennett CF. 2′-O-(2-methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J. Biol. Chem. 1997;272:11994–12000. doi: 10.1074/jbc.272.18.11994. [DOI] [PubMed] [Google Scholar]
- 10.Vickers TA, Zhang H, Graham MJ, Lemonidis KM, Zhao C, Dean NM. Modification of MyD88 mRNA Splicing and Inhibition of IL-1beta Signaling in Cell Culture and in Mice with a 2′-O-Methoxyethyl-Modified Oligonucleotide. J. Immunol. 2006;176:3652–3661. doi: 10.4049/jimmunol.176.6.3652. [DOI] [PubMed] [Google Scholar]
- 11.Winer J, Kwang C, Jung S, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase ± polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto JG, Beadles-Bohling AS, Wiren KM. Comparison of RiboGreen and 18S rRNA quantitation for normalizing real-time RT-PCR expression analysis. BioTechniques. 2004;36:58–60. doi: 10.2144/04361BM06. [DOI] [PubMed] [Google Scholar]
- 13.Liang XH, Liu Q, Fournier MJ. rRNA Modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5:e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gentleman R, Carey V, Bates D, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yates T, Okoniewski MJ, Miller CJ. X:Map: annotation and visualization of genome structure for affymetrix exon array analysis. Nucleic Acids Res. 2008;36:D780–D786. doi: 10.1093/nar/gkm779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okoniewski MJ, Miller CJ. Comprehensive analysis of affymetrix exon arrays using BioConductor. PLoS Comput. Biol. 2008;4:e6. doi: 10.1371/journal.pcbi.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smyth GK, Yang YH, Speed T. Statistical issues in microarray data analysis. Methods Mol. Biol. 2006;224:111–136. doi: 10.1385/1-59259-364-X:111. [DOI] [PubMed] [Google Scholar]
- 19.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 20.Monia BP, Johnston JF, Sasmor H, Cummins LL. Nuclease resistance and antisense activity of modified oligonucleotides targeted to Ha-ras. J. Biol. Chem. 1996;271:14533–14540. doi: 10.1074/jbc.271.24.14533. [DOI] [PubMed] [Google Scholar]
- 21.Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 22.Levin AA, Yu RZ, Geary RS. Basic principles of the pharmacokinetics of antisense oligonucleotide drugs. In: Stanely Crooke T, editor. Antisense Drug Discovery: Principles, Strategies and Applications. Boca Raton, FL: 2nd edn, CRC Press, Taylor & Francis Group; 2007. pp. 183–216. [Google Scholar]
- 23.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA ‘off-target’ transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuang Y, Weiner AM. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 25.Cartegni L, Hastings ML, Calarco JA, de Stanchina E, Krainer AR. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am. J. Hum. Genet. 2006;78:63–77. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buratti E, Chivers M, Kralovicova J, Romano M, Baralle M, Krainer AR, Vorechovsky I. Aberrant 5′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2007;35:4250–4263. doi: 10.1093/nar/gkm402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roca X, Sachidanandam R, Krainer AR. Determinants of the inherent strength of human 5′ splice sites. RNA. 2005;11:683–698. doi: 10.1261/rna.2040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 29.Monia BP, Johnston JF, Ecker DJ, Zounes MA, Lima WF, Freier SM. Selective inhibition of mutant Ha-ras mRNA expression by antisense oligonucleotides. J. Biol. Chem. 1992;267:19954–19962. [PubMed] [Google Scholar]
- 30.Vickers TA, Koo S, Bennett CF, Crooke ST, Dean NM, Baker BF. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. J. Biol. Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 31.Abad X, Vera M, Jung SP, Oswald E, Romero I, Amin V, Fortes P, Gunderson SI. Requirements for gene silencing mediated by U1 snRNA binding to a target sequence. Nucleic Acids Res. 2008;36:2338–2352. doi: 10.1093/nar/gkn068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H, Lima WF, Crooke ST. Properties of cloned and expressed human RNase H1. J. Biol. Chem. 1999;274:28270–28278. doi: 10.1074/jbc.274.40.28270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.