Abstract
The Neurospora VS ribozyme is a small nucleolytic ribozyme with unique primary, secondary and global tertiary structures, which displays mechanistic similarities to the hairpin ribozyme. Here, we determined the high-resolution NMR structure of a stem–loop VI fragment containing the A730 internal loop, which forms part of the active site. In the presence of magnesium ions, the A730 loop adopts a structure that is consistent with existing biochemical data and most likely reflects its conformation in the VS ribozyme prior to docking with the cleavage site internal loop. Interestingly, the A730 loop adopts an S-turn motif that is also present in loop B within the hairpin ribozyme active site. The S-turn appears necessary to expose the Watson–Crick edge of a catalytically important residue (A756) so that it can fulfill its role in catalysis. The A730 loop and the cleavage site loop of the VS ribozyme display structural similarities to internal loops found in the active site of the hairpin ribozyme. These similarities provided a rationale to build a model of the VS ribozyme active site based on the crystal structure of the hairpin ribozyme.
INTRODUCTION
The Neurospora Varkud Satellite (VS) ribozyme is part of a family of small nucleolytic RNA enzymes identified from biological sources that also includes the hammerhead, hairpin, hepatitis delta virus (HDV) and glmS ribozymes [for recent reviews see refs (1–4)]. With exception of the glmS ribozyme, these ribozymes control the replication cycle of their parental genome via self-cleavage and the reverse ligation reaction. They all catalyze scission of the RNA backbone by a transesterification reaction, which involves the initial attack of a specific 2′-oxygen on the adjacent 3′-phosphorus and yields two products with 2′-3′ cyclic phosphate and 5′-OH termini. Their common chemical mechanism is equivalent to that of RNase A, and, although they use different tertiary architectures and chemical groups, several share basic mechanistic strategies: activation of the nucleophile, stabilization of an in-line geometry between reactive groups and leaving group stabilization (3). Furthermore, although ribozymes were initially viewed as strict metalloenzymes (5), there is now evidence that several ribozymes employ an acid–base mechanism for the cleavage reaction, and use nucleobases as proton donors and/or acceptors (3).
The Neurospora VS ribozyme was first identified as a satellite RNA in the Varkud-1c strain and a few other isolates of Neurospora (6). A contiguous sequence of 154 nt was shown to be minimally required for self-cleavage in vitro (7). Its secondary structure contains six helical domains: stem–loop I (SLI) forms the substrate and stem–loops II–VI (SLII–SLVI) define the catalytic domain [Figure 1A; (8)]. The VS ribozyme is most active in the presence of divalent ions (9,10). Mg2+ ions allow tertiary-structure formation (8,11,12) and partly contribute to the chemistry of the cleavage reaction (10). Monovalent cations also support cleavage, albeit at a lower rate (10). The self-cleavage reaction takes place when the SLI substrate is located either at the 5′- or 3′-end of the catalytic domain (7,13), and, alternatively, trans-cleavage occurs when the substrate is synthesized separately from the catalytic domain (14). Substrate recognition involves formation of a kissing-loop interaction between loop I of the substrate and loop V of the catalytic domain (11). Formation of this I/V kissing-loop interaction is accompanied by a conformational change in SLI from an unshifted inactive conformation to a shifted active conformation (15). Our present understanding is that, in order for cleavage to occur, the cleavage site internal loop of SLI must dock in a cleft formed by SLII and SLVI to allow its interaction with the A730 loop of SLVI (4,12,16–25).
Figure 1.
The Neurospora VS ribozyme and its A730 internal loop. (A) Primary and secondary structures of the VS ribozyme (wild-type sequence nucleotides 617–783). The site of self-cleavage is indicated by an arrow, and circled nucleotides in loops I and V form the I/V kissing-loop interaction. (B) Schematic of the VS ribozyme (left) and hairpin ribozyme (right) illustrating similarities at the active site (see text). The residues highlighted with white circles are key players of proposed general acid-based mechanisms (4). (C) Primary and predicted secondary structures of the 26-nt SLVI RNA fragment, which includes the A730 loop domain (gray box). Phosphate groups that display inhibitory effect when substituted by Rp phosphorothioate are indicated by an arrow (20,63), and the arrow is filled in those cases where the inhibition could be suppressed by addition of manganese ions (63).
To date, the VS ribozyme is the only nucleolytic ribozyme for which there is no high-resolution structure of the complete RNA. High-resolution NMR structures of stem–loops I and V derived from the VS ribozyme have been obtained (26–31) and provide insights about substrate recognition and activation. The NMR structure of an SLVI fragment containing A730 loop residues was also previously determined, but this structure was found incompatible with previous biochemical data and thus, likely represents an inactive conformation of the A730 loop (29). In addition, low-resolution models have been derived from biochemical data (12), fluorescence resonance energy transfer (32,33) and small-angle X-ray scattering in solution (34). These models help define a global architecture for the VS ribozyme that is unique among nucleolytic ribozymes, but provide no detailed insights into the active site.
Despite the lack of high-resolution structural information, several mechanistic insights have been uncovered for the VS ribozyme (10,21–25) that reveal striking similarities with the hairpin ribozyme [Figure 1B; (4)]. Although the VS and hairpin ribozymes adopt significantly different secondary and tertiary structures, in both cases formation of the active site results from the association of two internal loops (Figure 1B) and similar general acid–base cleavage mechanisms have been proposed (4). Both ribozymes cleave at a purine–purine step within a purine-rich internal loop, between G620 and A621 in the VS ribozyme and between A−1 and G+1 in the hairpin ribozyme (Figure 1B). The cleavage site loops both contain a catalytically important residue proposed to act as the general base, G638 in the SLI internal loop of the VS ribozyme (22,24,25) and G8 in loop A of the hairpin ribozyme (4). Interestingly, additional sequence similarities at the cleavage site internal loops have been reported (35), including a common GCR (R = A or G) sequence just downstream of the cleavage site and a common CGA(A)GCGG sequence [the (A) indicate that an A is found only in the hairpin ribozyme] on the opposite strand that comprises the proposed general base (Figure 1A). The other internal loops forming the active site contain a catalytically important residue proposed to act as the general acid, A756 in the A730 loop of the VS ribozyme (21,23,25) and A38 in loop B of the hairpin ribozyme (4). Association of these two internal loops likely occurs with similar overall topology in both systems (4). In summary, there are considerable similarities between the VS and hairpin ribozymes, which suggest comparable active site architectures to achieve cleavage chemistry.
In order to gain insights into the formation of the VS ribozyme active site, we determined the high-resolution NMR structure of an SLVI fragment containing the A730 loop domain (Figure 1C). The three-dimensional (3D) structure of this A730 internal loop is consistent with a large number of biochemical data and likely represents a ground-state structure prior to its docking with the cleavage site loop. Based on this NMR structure and the previously determined NMR structure of the active conformation of the SLI internal loop, we identify additional similarities between the hairpin and VS ribozymes. We also discuss the structural changes that would be necessary in the SLI and SLVI internal loops for formation of a VS ribozyme active site that mimics the hairpin ribozyme.
MATERIALS AND METHODS
RNA sample preparation
Unlabeled, 15N- and 13C/15N-labeled SLVI RNAs were synthesized in vitro with T7 RNA polymerase and purified as described previously (30). The purified RNAs were first exchanged in a low-salt buffer (10 mM sodium cacodylate pH 6.5, 50 mM KCl, 0.05 mM NaN3 in 90% H2O/10% D2O) with Centricon-3 ultracentrifugation devices (Millipore, MA). They were heated to 95°C for 2 min and then cooled in iced water for 5 min before transferring to the standard NMR buffer [low-salt buffer containing 5 mM MgCl2 99.995% (Sigma-Aldrich, MO, USA)] or other MgCl2-supplemented buffers (low-salt buffer containing either 2.5 mM or 10 mM MgCl2 99.995%) using ultracentrifugation devices. For NMR studies in D2O, the samples were lyophilized several times and resuspended in 99.996% D2O. The concentration of SLVI RNA for NMR studies ranged between 0.4 and 1.5 mM.
Determination of adenine pKa’s in the SLVI fragment
All pH values were measured in D2O with a glass electrode (Radiometer America Inc., OH, USA) placed directly in the NMR sample. No correction was made for the D2O present in the sample. The pH was adjusted with 0.01–0.1 M DCl and NaOD. The reported pH values are the average of the pH values measured before and after the NMR experiments. At each different pH value, 1D 1H and 2D 1H–13C HMQC spectra were collected at 25°C. The 1D 1H spectra were used to verify the integrity of the sample and the 2D 1H–13C HMQC spectra were analyzed to determine the values of adenine C2 chemical shifts. In order to determine adenine pKa of A8 and A20, the log of [(ΔT − Δ)/Δ] was plotted as a function of pH, and the pKa values were derived by linear regression based on the Henderson–Hasselbalch-type equation pH = log[(ΔT − Δ)/Δ] + pKa (36), where Δ is the change in C2 chemical shift at a given pH relative to the unprotonated state (pH 8.6) and ΔT is the total change in chemical shift between the protonated and unprotonated states. Given that full protonation of the A8 (A756) and A20 (A730) residues could not be observed within the pH range studied (pH 4.7–8.6), the values of ΔT were therefore estimated between 6 and 8 ppm in agreement with reported values for folded RNAs (27,29,36–39). Values of pKa derived with ΔT = 6, 7 and 8 ppm were used to calculate an average pKa value and the error was derived from the maximum difference with the mean.
NMR spectroscopy
All NMR data were acquired with 500, 600 and 800 MHz Varian INOVA spectrometers equipped with a pulsed field gradient unit and an actively shielded z-gradient probe, either a 1H{13C/15N} triple-resonance probe (standard or cold probe) or a 1H{15N–31P} indirect detection probe. Exchangeable protons and their attached nitrogens were assigned at 15°C, whereas non-exchangeable protons and their attached carbons were obtained at 25°C in D2O, as previously described (30). In addition, 2D 1H–15N HMQC optimized for transfers via J = 7.0 Hz and 21 Hz (40) were collected at 25°C in D2O for assignment of uridine N1, pyrimidine N3, as well as N7 and N9 of purines. 1H, 13C and 15N chemical shifts were referenced at 25°C to an external standard of 2,2-dimethyl-2-silapentane-5-sulfonic acid at 0.00 ppm. NMR data were processed using the NMRPipe/NMRDraw package (41) and analyzed with NMRView (42).
Structural restraints
An HNN-COSY spectrum was collected in H2O at 15°C to detect 2JNN coupling across Watson–Crick base pairs (43). Distance restraints for exchangeable protons were extracted from 2D flip-back watergate NOESY [mixing time (τm) = 160 ms; (44,45)], 3D amino-optimized 15N-edited NOESY-HSQC [τm of 80 and 160 ms; (46)] and 2D 1H–15N CPMG-NOESY [τm of 80 and 160 ms; (47)] spectra collected at 15°C in H2O. Distance restraints for non-exchangeable protons were extracted from 3D 13C-edited HMQC-NOESY spectra [τm of 80 and 160 ms; (48)] collected at 25°C in D2O. The NOE-derived distance restraints were separated in four classes: strong (1.8–3.3 Å), medium (1.8–4.2 Å), weak (1.8–5.5 Å) and very weak (2.8–7.5 Å) based on NOE crosspeak intensities. Based on NMR evidence for specific base pairing obtained from NOESY and 2D HNN-COSY spectra, canonical distance restraints were employed to define the hydrogen-bond pattern and planarity of the following base pairs: G1–C26, A2–U25, G3–C24, C4–G23, U5–G22, G6–C21, C10–G19, A11–U18, C12–G17 and G13–A16. For the G9–A20 base pair, only two hydrogen-bond distance restraints were employed (G9 H1 to A20 N1 and G9 O6 to A20 H6). Dihedral angle restraints for the sugar puckers (δ) were obtained from analysis of 2D DQF-COSY (49) and 3D HCCH-E.COSY (50) spectra. The backbone dihedral angles α, γ, χ and ζ were restrained using distance restraints derived from comparative NOE analyses (51). Based on NMR evidence, backbone torsion angles of residues in helical regions (1–5, 11–12, 17–18, 22–26) were restrained to A-form values (±15°).
Liquid crystal NMR spectroscopy
For liquid crystal NMR studies, 15N- and 13C/15N-labeled SLVI RNAs (at 0.4–0.5 mM) were aligned by adding a concentrated Pf1 filamentous phage solution (ASLA biotech.) at a final concentration of ∼17 mg/ml (52). Prior to addition of the Pf1 phages, these were exchanged at least two times with the NMR buffer containing 5 mM MgCl2. The 2H NMR splitting for D2O observed at 25°C in these samples was 15.53 Hz for the 15N-labeled SLVI and 15.24 Hz for the 13C/15N-labeled SLVI. Two-dimensional 1H–15N HSQC spectra collected with active JHN coupling in t1 were used to extract imino 1H–15N couplings with the isotropic and Pf1-aligned 15N-labeled SLVI samples. Spin-state selective experiments (53,54) were used to extract one-bond 1H–13C couplings (C1′–H1′, C2–H2, C5–H5, C6–H6 and C8–H8) in isotropic and Pf1-aligned 13C/15N-labeled SLVI samples. One-bond 1H–15N and 1H–13C couplings were measured from fitted peaks using NMRWish in the NMRPipe/NMRDraw package. Extracted residual dipolar coupling (RDC) values were scaled linearly with respect to the observed D2O splitting, to take into account the small difference in the magnitude of the alignment from the different samples. The RDC restraints were separated in two classes to account for overlap of the peaks in the spectra, with error bars estimated at 1 Hz (well resolved) and 2 Hz (partial overlap). RDC restraints for A730 loop residues 6–9 and 20–21 were not included in the structure calculation because of evidence for local dynamics in the internal loop.
Structure calculation
Three-dimensional structures were calculated with restrained molecular dynamics and simulated annealing in X-PLOR-NIH version 2.1.9 (55) using a two-stage protocol. At stage one, an initial set of 50 structures was calculated from structures with randomized backbone angles, as previously described (31). Structures obtained at stage one satisfy all distance and dihedral experimental restraints (no distance violation of >0.2 Å and no torsion angle violation of >5°). At stage two, these structures were refined with the same set of restraints supplemented by RDC restraints and using a single alignment tensor (56). The magnitude of the axial and rhombic components of the alignment tensor were initially obtained using MODULE 1.0 (57), but were subsequently refined by using a grid search to obtain reliable Da and R values (58). The grid search procedure was repeated, using RDCs from the 5′–3′ stem (residues 2–5 and 22–25) and the hairpin stem (residues 11–18), independently. All grid search calculations converged on similar values of Da and R [values of Da(CH) = 16.6 Hz and R = 0.40 were used], suggesting that the relative orientation of the two stems is well defined despite evidence of dynamic residues in the A730 loop. For RDC refinement, restrained molecular dynamics and simulating annealing were used followed by a final energy minimization. During simulated annealing, the force constants for RDC, NOE, impropers, angles and Van der Waals restraints were slowly increased. A final set of 500 structures was calculated, from which the 20 lowest-energy structures that satisfied the experimental restraints (no distance violation >0.2 Å, no torsion angle violation >5° and no RDC violation >5 Hz) were selected for analysis. These 20 lowest-energy structures were used to calculate an average structure that was minimized against NOE and dihedral restraints. All structures were visualized with PyMol (Schrödinger) and analyzed with PyMol and Curves+ (59).
Active site modeling
A subset of three residues (G8, A12 and G13) was extracted from the X-ray structure of the hairpin ribozyme determined with vanadate [pdb entry code 1M5O; (60)]. Residues A12 and G13 in this subset were modified using Coot (61) to match the base type of their equivalents in the VS ribozyme (G620 and A621). The bases of residues G8 and G12 of this modified model were then aligned on the model of the active conformation of SLI of the VS ribozyme (28). A hybrid SLI model was generated in which residues G620, A621, G638 of SLI were replaced by their equivalents in the modified hairpin model. Using Coot, residue A756 of the SLVI structure was moved by hand to allow stacking with A621. Finally, the model of the SLI/SLVI complex was energy minimized with Amber (62).
RESULTS
Magnesium-dependent stabilization of the A730 internal loop
To explore the 3D structure of the A730 loop and gain insights into the active site of the VS ribozyme, the 26-nt SLVI RNA shown in Figure 1C was investigated by NMR spectroscopy. This SLVI RNA is designed to reproduce the secondary-structure context of the A730 internal loop from the VS ribozyme (8), including two sets of two adjacent closing base pairs on each side of this loop (Figure 1). A GAAA tetraloop was also included in the SLVI RNA to help stabilize the hairpin conformation and prevent possible duplex formation at the high RNA concentrations used for NMR studies. We initially verified that the SLVI RNA adopts a single hairpin conformation by native gel electrophoresis (not shown), and this was later confirmed from the ensemble of NMR data.
Given that the VS ribozyme is active in the presence of Mg2+ ions (9) and that Mg2+-ion binding sites have been identified in the A730 loop domain (63), we first examined the effect of MgCl2 concentration on the folding of the SLVI RNA by 1D imino 1H NMR spectra. The 1D imino 1H NMR spectrum of the SLVI RNA recorded in the absence of Mg2+ ions shows signals of variable intensities characteristic of folded RNAs containing dynamic regions (Figure 2, bottom spectrum). These imino proton signals were assigned from a 2D NOESY spectrum collected under the same conditions. It was found that all imino protons of the SLVI RNA give an intense signals in this 1D spectrum, except for the G6 and G9 imino protons, which are not observed, and the U18 and G19 imino protons, which yield signals of weak intensities (Figure 2, bottom spectrum). These absent and weak imino proton signals indicate that the A730 loop domain is unstable in the absence of Mg2+ ions. Upon addition of Mg2+ ions, the imino signals of U18 and G19 become significantly more intense (Figure 2, top spectra) and a weak imino proton signal from G6 appears in the spectrum. Analysis of 2D NOESY and 2D HNN-COSY spectra collected at 5 mM MgCl2 confirms formation of all base pairs from the predicted secondary structure of the SLVI RNA (Figure 1C). Although the imino NMR data do not provide evidence for stabilization of non-canonical base pairs within the A730 loop, they indicate that the A730 loop domain is stabilized by Mg2+ ions, in agreement with previous biochemical studies. A MgCl2 concentration of 5 mM was found sufficient to produce the characteristic spectral changes (Figure 2) and, therefore, was selected for future NMR studies.
Figure 2.
Stabilization of the A730 loop by Mg2+ ions. Imino regions of 1D flip-back watergate (44,45) 1H spectra of SLVI collected at 15°C in NMR buffer containing different concentrations of free MgCl2. Imino proton assignments were derived from 2D NOESY spectra collected in NMR buffer at 0, 5 and 10 mM MgCl2.
The overall 3D structure of the SLVI RNA
The structure of the SLVI RNA was obtained by multidimensional heteronuclear NMR methods. Complete resonance assignments were obtained for all observable 1H, 13C and 15N atoms of SLVI (Supplementary Table S1). The structure determination included NOE-derived distance restraints, dihedral angle restraints and RDC restraints (Table 1). The SLVI structure is well defined by the NMR data with a heavy atom rmsd of 0.67 ± 0.17 Å for the 20 lowest-energy structures (residues 2–25; Table 1 and Figure 3A). The SLVI RNA forms a hairpin structure containing a 5′–3′ stem (residues 2–5 and 22–25; rmsd of 0.53 ± 0.07 Å), a hairpin stem (residues 11–18; rmsd of 0.31 ± 0.01 Å), and the A730 loop (residues 6–10 and 19–21; rmsd of 0.65 ± 0.24 Å). Stem regions form regular A-form helices and the GAAA tetraloop adopts the typical GNRA fold with its characteristic sheared G–A base pair and 3′-purine stack (64,65). The A730 loop domain imparts a ∼150° interhelical angle between the two stems of SLVI and contains two structural characteristics that were not previously identified: a cis-WC/WC G9–A20 base pair and an S-turn motif that protrudes C7 and A8 in the minor groove.
Table 1.
Structural statistics of the SLVI RNA
| Distance restraints | 1017 |
| Number of NOE-derived distance restraints | 965 |
| Inter-nucleotide | 591 |
| Intra-nucleotide | 355 |
| Ambiguous | 19 |
| Hydrogen-bond restraints | 52 |
| Dihedral angle restraints | 88 |
| Residual dipolar coupling restraints | 30 |
| Total number of restraints | 1135 |
| Rmsd from experimental restraints | |
| NOE (Å) (none >0.2) | 0.0075 ± 0.004 |
| Dihedral (°) (none >5) | 0.089 ± 0.009 |
| Residual dipolar couplings (none >5 Hz) | 0.24 ± 0.02 |
| Rmsd from idealized geometry | |
| Bonds (Å) | 0.00555 ± 0.00002 |
| Angles (°) | 1.1968 ± 0.0006 |
| Impropers (°) | 0.422 ± 0.005 |
| Heavy-atom rmsd to the minimized average structure (Å) | |
| Overall (residues 2–25) | 0.67 ± 0.17 |
| 5′–3′ stem (residues 2–5 and 22–25) | 0.53 ± 0.07 |
| Hairpin stem (residues 11–18) | 0.31 ± 0.01 |
| A730 loop (residues 6–10 and 19–21) | 0.65 ± 0.24 |
Figure 3.
NMR solution structure of the SLVI RNA fragment. (A) Stereoview of the 20 lowest-energy structures. The superposition was made on the minimized average structure (not shown) over heavy atoms of residues 2–25. The view is into the minor groove of the A730 active site internal loop. (B and C) Stick representations of the lowest-energy structure of SLVI. For simplicity only heavy atoms are shown and the ribbon replacing the phosphorus and non-bonded oxygen atoms is used to indicate the backbone topology. SLVI nucleotides are color-coded: the loop closing base pairs (G6–C21 and C10–G19) are dark gray, C7 (C755) is magenta, A8 (A756) is green, G9 (G757) is gold and A20 (A730) is blue.
Formation of a cis-WC/WC G–A base pair in the A730 loop
The evidence for a cis-WC/WC G–A base pair in the A730 loop was obtained from 2D 1H–15N CMPG-NOESY and 3D 13C-edited HMQC-NOESY spectra. A strong NOE signal was observed between the A20 H2 and the G9 NH2 protons, which is typical of the cis-WC/WC G–A base pair geometry (Figure 4A). Several NOE signals were also observed that indicate stacking of the G9–A20 base pair on the C10–G19 base pair, including NOEs between A20 H2 and G19 NH and between A20 H1′ and G19 NH (Figure 4A). Initial structural calculations performed without specific hydrogen-bond restraints for the G9–A20 base pair revealed a cis-WC/WC G–A base pair geometry, thus, hydrogen-bond restraints defining this geometry were included in subsequent rounds of structural calculations. The superposition of the 20 lowest-energy structures show that this G–A base pair is well defined by the NMR data, although two conformations with different propeller twists and buckles are observed in the ensemble of structure (Figure 4B). These two alternative conformations observed in the NMR structures may reflect insufficient NMR restraints and/or conformational dynamics for this base pair. The absence of a detectable imino proton signal for G9 (Figure 2) supports conformational dynamics for the G9–A20 base pair.
Figure 4.
Formation of a cis WC/WC G9-A20 base pair in the A730 loop. (A) Selected regions from a 2D 1H–15N CPMG-NOESY spectrum showing NOEs that define the geometry of the G9–A20 base pair. The spectrum was collected at 15°C with a mixing time of 160 ms. (B) The G9–A20 base pair in the 20 lowest-energy structures. The superposition is from Figure 3a. (C) Stacking of the G9–A20 base pair onto the C10–G19 base pair in the lowest-energy structure of SLVI. Dashed lines connect protons for which a NOE is observed in (A).
An S-turn motif in the A730 loop domain
The structure of the A730 loop domain is defined by a large number of NOEs, including several unusual sequential NOEs in the G6–G9 stretch and non-sequential NOEs between nucleotides G6 and G9 (Figure 5A). These NOEs are consistent with the unusual ribose-phosphate backbone of the A730 loop domain, which adopts an S-turn between nucleotides G6 and C10. The S-turn is a common RNA motif, first structurally identified in the loop E of eukaryotic 5S rRNA (66) and the sarcin–ricin loop of 28S rRNA (67), but since found in other structural contexts (68–72). In the A730 loop, the S-turn is created by ribose reversal at A8, with its 2′-OH group pointing in a direction opposite to the 2′-OH groups of adjacent nucleotides (Figure 5B). In the majority of the lowest-energy structures (17/20), the ribose of A8 adopts a 2′-endo conformation, a characteristic of an S-turn, which is in agreement with the intense H1′–H2′ signal in the DQF-COSY spectrum (not shown). The S-turn of the A730 loop leads to bulging out of both the C7 and A8 residues with their Watson–Crick edges exposed in the minor groove. The adjacent G9–A20 base pair possesses a larger C1′–C1′ distance than standard Watson–Crick base pairs that likely helps stabilize the S-turn. Three hydrogen bonds involving G9 and the protruded C7 and A8 bases are found in the ensemble of 20 lowest-energy structures and connect: (i) G9 O4′ and A8 O2′ (2.6 ± 0.4 Å); (ii) G9 O2′ and A8 N3 (3.3 ± 1.0 Å) and (iii) G9 N3 and C7 N4 (3.4 ± 0.8 Å; Figure 5C). Interestingly, an S-turn motif has also been previously found in loop B of the hairpin ribozyme (70), suggesting that it may be important for catalysis by the VS ribozyme (see discussion).
Figure 5.
S-turn motif in the A730 loop of the SLVI RNA. (A) Schematic summarizing the inter-residue NOEs for the A730 internal loop of the SLVI RNA. Black lines indicate NOEs between nucleotides that are adjacent in the sequence, pink lines indicate NOEs between base-pairing residues, blue lines indicate NOEs between G6 and G9, and orange lines refer to NOEs between C7 and G9. For simplicity, all ribose protons (H1′, H2′, H3′, H4′, H5′ and H5″) were grouped under the ribose denomination. (B and C) Close up views of the S-turn motif in the lowest-energy structure showing (B) the ribose reversal at A8 and nearby phosphates and (C) stacking of C7 and A8 in the minor groove and stabilizing hydrogen bonds. In (B) the pro–Rp oxygens are shown in blue and the 2′-oxygens in red. In (C) three hydrogen bonds are shown (A8 N3: G9 2′–OH, C7 NH2: G9 N3 and A8 2′–OH: G9 O4′) that likely stabilize the S-turn motif. For simplicity only heavy atoms are shown and the ribbon replacing the phosphorus and non-bonded oxygen atoms is used to indicate the backbone topology.
Shifted pKa values for adenines of the A730 loop
Given the predicted role of A756 as a general acid in the cleavage reaction, we were interested to determine if the structure of the A730 loop imparts a shifted pKa value for A756. Hence, we determined the pKa of adenines in the A730 loop of SLVI by 13C NMR methods. It has been previously shown that the C2 chemical shift of AMP undergoes an 8-ppm upfield displacement upon protonation at its N1 position (73), and pH-dependent change in C2 chemical shifts have been used to determine adenine pKa’s in folded RNAs (27,29,36–39,73). In SLVI, only two C2 resonances are significantly affected by pH; the A8 and A20 C2 resonances are shifted upfield by 2.4 and 3.3 ppm, respectively, when the pH is decreased from 8.6 to 4.7 (Figure 6A). For both A8 and A20, a single C2–H2 crosspeak is observed in this pH range, indicating fast exchange dynamics on the NMR chemical shift timescale. In such cases, the change in C2 chemical shift at a given pH value can be used to derive pKa values. We obtained a pKa of 4.44 ± 0.10 for A8 and a pKa of 4.74 ± 0.12 for A20 (Figure 6B and Supplementary Figure S1). These pKa values are slightly higher than the pKa value of adenine in single-stranded RNA [∼3.7; (74)] and are compatible with the accessibility of the N1 positions of A8 and A20 in the SLVI structure. Local electronic effects, such as sequence context, base stacking and Mg2+ binding could be responsible for these small pKa shifts. However, there is a discrepancy of about one pH unit between the pKa of A8 and the catalytic pKa [between 5.2 and 5.8; (23,25)]. Such discrepancy is not surprising, since it is expected that formation of the active site will change the chemical environment of the A756 base and thus, affect its pKa.
Figure 6.
Determination of adenine pKa’s in SLVI. (A) Superposition of the aromatic C2–H2 regions of 2D 1H–13C HMQC spectra collected at 25°C and at pH 4.7 (beige), pH 5.1 (black), pH 5.5 (pale blue) and pH 8.6 (grayish blue). Arrows point to significant pH-dependent changes in 13C chemical shift for A8 and A20. (B) Summary of the adenine pKa values in the A730 internal loop.
DISCUSSION
We are using a modular approach to structurally characterize the Neurospora VS ribozyme, which essentially consists of determining the NMR structure of small fragments of the VS ribozyme that are relevant to its function. We previously determined structures of an activated SLI internal loop and of an SLV fragment by NMR spectroscopy to provide novel information on substrate recognition and activation in the VS ribozyme (28,30,31). Here, we performed NMR studies of an SLVI fragment containing the A730 loop in order to gain structural insights into the formation of the VS ribozyme active site. Below, we demonstrate that our structure is compatible with a large amount of biochemical data on the VS ribozyme and likely represents the conformation of the A730 loop in the folded ribozyme prior to docking with the cleavage site internal loop. We also discuss the resemblance of the A730 loop with loop B of the hairpin ribozyme and its relevance to formation of the active site.
The A730 loop adopts a undocked conformation compatible with chemical probing data
Chemical probing data were previously obtained for a self-cleaving ribozyme in both the absence and presence of Mg2+ ions (8). As observed in our NMR studies of SLVI, the chemical probing data demonstrated that addition of Mg2+ ions stabilize the structure of the A730 loop domain, including several loop closing base pairs that were unstable in the absence of Mg2+ ions (8). The Watson–Crick edges of C755 and A756 were still strongly modified under native conditions (8) in agreement with the structure of SLVI in which the Watson–Crick edges of these two bases are exposed in the minor groove. This suggests that under the conditions used for chemical probing, the A730 loop is not stably docked with the cleavage site internal loop, but can also exist in an undocked state that is compatible with the NMR structure.
Compatibility of the A730 loop structure with mutational and chemical modification data
Thorough mutational analyses of the A730 loop domain were previously carried out and revealed the importance of this loop for catalysis (16,18). In agreement with our structure of the A730 loop, the reversal of the four Watson–Crick base pairs within this domain (U753–G732, G754–C731, C758–G729 and A759–U728) did not significantly affect cleavage activity, but mutations of loop residues had more important effects (16). Although all base substitutions at G757 (G9) and A730 (A20) led to reduced activity, replacing the G9–A20 combination by a G–C, a G–U or an A–A was less detrimental than other substitutions (16,18). Thus, these mutational data indicate the importance of a purine at position 757 and of cis-WC/WC base pairing (75) between residues 757 and 730. This is in agreement with the role of the G9–A20 base pair in stabilizing the S-turn motif. Mutation of A756 (A8) by U, C or G caused the largest reduction in cleavage activity [∼400–800-fold; (16,18)], which is consistent with its proposed mechanistic role as a general acid (21,23,25). Functional group modifications of A756 revealed that the Watson–Crick edge of A756 is particularly important for cleavage (17). In the structure of SLVI, the Watson–Crick edge of A8 is exposed in the minor groove and accessible for docking with the cleavage site internal loop and performing its catalytic role.
Mg2+ binding to the S-turn
The backbone conformation of the S-turn creates two phosphate clusters, suggesting that specific binding of Mg2+ ions may be associated with at least one of these clusters. There is also evidence that other S-turn motifs contain a divalent metal binding site (71,76–79). Evidence for Mg2+ binding to the S-turn of the A730 loop is available from phosphorothioate substitution interference, which was previously employed to identify pro-Rp oxygens that are important for activity. Three inhibitory phosphorothioates, those of C755 (C7), G757 (G9) and C758 (C10), were found in the A730 loop domain [Figure 1C; (20,63)]. Phosphorothioate inhibition at G757 and C758 was suppressed in the presence of thiophilic manganese ions, indicating that the pro-Rp oxygens of G757 and C758 directly coordinate a divalent metal ion [Figure 1C; (63)]. In the SLVI structure, the pro-Rp oxygens of C7 (C755), G9 (G757) and C10 (C758) are part of the S-turn motif, and the proximity of the C7 and G9 pro-Rp oxygens suggest that they may coordinate the same Mg2+ ion (Figure 5B).
Importance of the S-turn motif for catalysis
The A730 loop adopts an S-turn motif that exposes the Watson–Crick edge of A8 (A756), the proposed general acid of the VS cleavage reaction, in an enlarged minor groove. Interestingly, in the X-ray crystal structures of the hairpin ribozyme (60,78), an S-turn was also found in loop B where it allows A38, the proposed general acid of the hairpin cleavage reaction, to protrude in a broad minor groove and make several hydrogen bonds with either a transition state analog or the 2′-cyclic phosphate and 5′-OH of the cleaved form (60,78). Thus, the structure of SLVI reveals additional similarities between the VS and hairpin ribozymes. These structural similarities between the A730 loop of the VS ribozyme and loop B of the hairpin ribozyme could not be predicted from the sequence of these internal loops, which are very different.
Certain non-VS sequences were previously found to partially substitute for the natural A730 loop in a cleavage assay (18). In concordance with protrusion of A756 in the S-turn, it was concluded that a minimal requirement for self-cleavage is the presence of unpaired or non-Watson–Crick paired nucleotides at the location of the A730 loop (18). The most active substitution mutant contained the A-rich bulge of the Tetrahymena group I intron (80). Interestingly, this bulge adopts a structure in which the phosphate backbone makes a corkscrew turn that exposes the Watson–Crick faces of several adenines (80). Similarly, in the VS and hairpin ribozymes, one essential role of the S-turn motif may be to protrude a catalytically important adenine in the active site. In the hairpin ribozyme, however, the catalytically-important adenine (A38), is protruded in the docked state (78), but not in the undocked state (70). This may explain why substitution of the A730 loop domain by loop B of the hairpin ribozyme did not produce a functional VS ribozyme (18).
Insights into the formation of the active site
There is substantial evidence that the cleavage site loop and A730 loop of the VS ribozyme intimately associate to form the active site (4,12,16–25). Given that the NMR structures of both the active conformation of the cleavage site loop (28) and the A730 loop are available, it is interesting to consider how these two loops might interact to form the active site. As described below, these two internal loops of the VS ribozyme display structural similarities to the two internal loops found in the active site of the hairpin ribozyme, and such similarities can be used to build a simple model of the VS ribozyme active site.
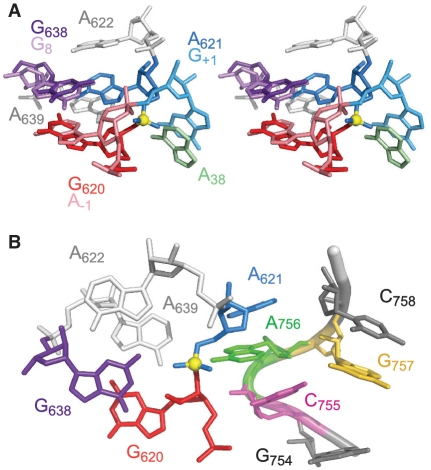
In the crystal structure of a transition state mimic of the hairpin ribozyme, the N1 of G8 is within hydrogen-bonding distance of the 2′-OH nucleophile, and its exocyclic amine forms a hydrogen bond with the scissile phosphate (60). A comparison between this X-ray structure of the hairpin ribozyme and the NMR structure of the cleavage site loop of the VS ribozyme (28) reveals that the G residue proposed to serve as the general base (G638 in the VS ribozyme and G8 in the hairpin ribozyme) and the N − 1 residue (G620 in the VS ribozyme and A − 1 in the hairpin ribozyme) form similar cross-strand purine stacks. The heavy atom superposition between these two sets of bases illustrates the similarity of these purine stacks (Figure 7A; rmsd of 0.8 Å). Thus, only small changes in the relative positioning of G638 with respect to G620 would be necessary to bring G638 in the proper orientation with respect to the cleavage site. For modeling purposes, we modified the positions of the G621 and G638 bases in the cleavage site internal loop structure to mimic the position of functionally equivalent residues in the hairpin ribozyme (Figure 7B).
Figure 7.
Homology modeling of the VS ribozyme active site. (A) Heavy atom superposition of the G638 and G620 nucleotides of the VS ribozyme with the G8 and A − 1 nucleotides of the hairpin ribozyme [pdb entry 1M5O; (60)]. (B) Modeling of the active site by association of the substrate internal loop and the A730 internal loop (see text). For simplicity only heavy atoms are shown and the ribbon replacing the phosphorus and non-bonded oxygen atoms is used to indicate the backbone topology. The yellow sphere represents the scissile phosphate.
The superposition of Figure 7A highlights an important difference between the structures of the cleavage site loops from the hairpin and VS ribozymes; whereas the N − 1 and N + 1 nucleotide of the hairpin ribozyme (A − 1 and G + 1) adopt a splayed conformation, those of the VS ribozyme (G620 and A621) do not. The splayed conformation at the cleavage site is characteristic of nucleolytic ribozymes and likely important for stabilizing the in-line geometry between the 2′-OH nucleophile, the scissile phosphate and the 5′-oxygen leaving group that is typical of SN2 reactions (3). Thus, it is highly likely that A621 must undergo a substantial conformation change to adopt the characteristic splayed conformation at the active site. For modeling purposes, the conformation of A621 was modified such that it mimics the position of the functionally equivalent G + 1 in the hairpin ribozyme (Figure 7B). This brings A621 out of the helix and leaves G–A tandem base pairs in the cleavage site loop, which are known to stabilize internal loops when flanked with G–C base pairs, as found here (81).
Different strategies are adopted by nucleolytic ribozymes to stabilize the splayed conformation of the 5′- and 3′-nucleotides flanking the scissile phosphate, but they all rely on hydrogen bonding and stacking interactions (3). In the hairpin ribozyme, an important base stacking interaction involves G + 1 of loop A and A38 of loop B, which orients this catalytically important base near the 5′-oxygen of the scissile phosphate (Figure 7A). As a last step in this modeling exercise, we attempted to reproduce this latter interaction by stacking A756 (A8) of SLVI on the protruded A621 of our SLI model. Such stacking is compatible with the production of a UV-dependent crosslink between a 4-thio-uridine at position 621 and A756, as previously observed under conditions that are compatible with catalytic activity (19). Interestingly, given the protruded conformation of A756, it was possible to stack A756 on A621 and prevent atomic clashes between the two internal loops with only small displacement of the A756 base (Figure 7B). This stacking also brings the Watson–Crick edge of A756 near the negatively charged scissile phosphate. Such environment would likely stabilize the protonated form of A756 and shifts its pKa towards neutrality (23,25). Furthermore, in the resulting model the minor grooves of the two internal loops interact with one another with a similar overall topology as in the hairpin ribozyme (not shown). Thus, by using the NMR structures of the A730 loop and the active conformation of the SLI internal loop, it is straightforward to model the active site of the VS ribozyme by homology with the active site of the hairpin ribozyme. The resulting model preserves the relative orientation of N − 1, N + 1 and the two key catalytic residues (G620, A621, A756 and G638 in the VS ribozyme) while maintaining the overall topology between the two helical domains. The coherence of the model with existing structural data clearly reinforces the idea that the VS and hairpin ribozymes may adopt similar active site architectures. Nevertheless, a high-resolution structure of the full VS ribozyme is needed to reveal structural details of the active site that are beyond the specific findings of the present study.
ACCESSION CODES
The NMR chemical shifts, NMR restraints and atomic coordinates of the SLVI RNA have been deposited to the RCSB Protein Data Bank with BMRB entry 17292, RCBS entry rcsb101999 and PDB entry 2L5Z, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes for Health Research (CIHR) to P.L. (MOP-86502); Fonds Québécois de la Recherche sur la Nature et les Technologies, Luigi-Liberatore Foundation and Université de Montréal (UdeM) M.Sc. scholarships to G.D. UdeM M.Sc. scholarship to E.B. P.L. holds a Canada Research Chair in Structural Biology and Engineering of RNA. Funding for open access charges: CIHR.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Richter for computer support and D.L. Bryce for providing his structure refinement protocol with RDC restraints. We also thank R.A. Collins and J.G. Omichinski for critical reading of the article. We acknowledge the IBS platform of the Partnership for Structural Biology and the Institut de Biologie Structurale in Grenoble (PSB/IBS), for the NMR access.
REFERENCES
- 1.Collins RA. The Neurospora Varkud satellite ribozyme. Bioch. Soc. Trans. Rev. 2002;30:1122–1126. doi: 10.1042/bst0301122. [DOI] [PubMed] [Google Scholar]
- 2.Lilley DM. The Varkud satellite ribozyme. RNA. 2004;10:151–158. doi: 10.1261/rna.5217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cochrane JC, Strobel SA. Catalytic strategies of self-cleaving ribozymes. Acc. Chem. Res. 2008;41:1027–1035. doi: 10.1021/ar800050c. [DOI] [PubMed] [Google Scholar]
- 4.Lilley DMJ. In: Ribozymes and RNA catalysis. Lilley DMJ, Eckstein F, editors. Cambridge: Royal Society of Chemistry; 2008. pp. 66–91. [Google Scholar]
- 5.Pyle AM. Ribozymes: a distinct class of metalloenzymes. Science. 1993;261:709–714. doi: 10.1126/science.7688142. [DOI] [PubMed] [Google Scholar]
- 6.Saville BJ, Collins RA. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell. 1990;61:685–696. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- 7.Guo HCT, De Abreu DM, Tillier ERM, Saville BJ, Olive JE, Collins RA. Nucleotide sequence requirements for self-cleavage of Neurospora VS RNA. J. Mol. Biol. 1993;232:351–361. doi: 10.1006/jmbi.1993.1395. [DOI] [PubMed] [Google Scholar]
- 8.Beattie TL, Olive JE, Collins RA. A secondary-structure model for the self-cleaving region of Neurospora VS RNA. Proc. Natl Acad. Sci. USA. 1995;92:4686–4690. doi: 10.1073/pnas.92.10.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins RA, Olive JE. Reaction conditions and kinetics of self-cleavage of a ribozyme derived from Neurospora VS RNA. Biochemistry. 1993;32:2795–2799. doi: 10.1021/bi00062a009. [DOI] [PubMed] [Google Scholar]
- 10.Smith MD, Mehdizadeh R, Olive JE, Collins RA. The ionic environment determines ribozyme cleavage rate by modulation of nucleobase pKa. RNA. 2008;14:1942–1949. doi: 10.1261/rna.1102308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rastogi T, Beattie TL, Olive JE, Collins RA. A long-range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J. 1996;15:2820–2825. [PMC free article] [PubMed] [Google Scholar]
- 12.Hiley SL, Collins RA. Rapid formation of a solvent-inaccessible core in the Neurospora Varkud satellite ribozyme. EMBO J. 2001;20:5461–5469. doi: 10.1093/emboj/20.19.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rastogi T, Collins RA. Smaller, faster ribozymes reveal the catalytic core of Neurospora VS RNA. J. Mol. Biol. 1998;277:215–224. doi: 10.1006/jmbi.1997.1623. [DOI] [PubMed] [Google Scholar]
- 14.Guo HCT, Collins RA. Efficient trans-cleavage of a stem-loop RNA substrate by a ribozyme derived from Neurospora VS RNA. EMBO J. 1995;14:368–376. doi: 10.1002/j.1460-2075.1995.tb07011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen A, Collins RA. Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol. Cell. 2000;5:469–478. doi: 10.1016/s1097-2765(00)80441-4. [DOI] [PubMed] [Google Scholar]
- 16.Lafontaine DA, Wilson TJ, Norman DG, Lilley DM. The A730 loop is an important component of the active site of the VS ribozyme. J. Mol. Biol. 2001;312:663–674. doi: 10.1006/jmbi.2001.4996. [DOI] [PubMed] [Google Scholar]
- 17.Lafontaine DA, Wilson TJ, Zhao Z-Y, Lilley DMJ. Functional group requirements in the probable active site of the VS ribozyme. J. Mol. Biol. 2002;323:23–34. doi: 10.1016/s0022-2836(02)00910-5. [DOI] [PubMed] [Google Scholar]
- 18.Sood VD, Collins RA. Identification of the catalytic subdomain of the VS ribozyme and evidence for remarkable sequence tolerance in the active site loop. J. Mol. Biol. 2002;320:443–454. doi: 10.1016/s0022-2836(02)00521-1. [DOI] [PubMed] [Google Scholar]
- 19.Hiley SL, Sood VD, Fan J, Collins RA. 4-thio-U cross-linking identifies the active site of the VS ribozyme. EMBO J. 2002;21:4691–4698. doi: 10.1093/emboj/cdf462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones FD, Strobel SA. Ionization of a critical adenosine residue in the Neurospora Varkud satellite ribozyme active site. Biochemistry. 2003;42:4265–4276. doi: 10.1021/bi020707t. [DOI] [PubMed] [Google Scholar]
- 21.Zhao ZY, McLeod A, Harusawa S, Araki L, Yamaguchi M, Kurihara T, Lilley DM. Nucleobase participation in ribozyme catalysis. J. Am. Chem. Soc. 2005;127:5026–5027. doi: 10.1021/ja0502775. [DOI] [PubMed] [Google Scholar]
- 22.Wilson TJ, McLeod AC, Lilley DM. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J. 2007;26:2489–2500. doi: 10.1038/sj.emboj.7601698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith MD, Collins RA. Evidence for proton transfer in the rate-limiting step of a fast-cleaving Varkud satellite ribozyme. Proc. Natl Acad. Sci. USA. 2007;104:5818–5823. doi: 10.1073/pnas.0608864104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaikaran D, Smith MD, Mehdizadeh R, Olive J, Collins RA. An important role of G638 in the cis-cleavage reaction of the Neurospora VS ribozyme revealed by a novel nucleotide analog incorporation method. RNA. 2008;14:938–949. doi: 10.1261/rna.936508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson TJ, Li NS, Lu J, Frederiksen JK, Piccirilli JA, Lilley DM. Nucleobase-mediated general acid-base catalysis in the Varkud satellite ribozyme. Proc. Natl Acad. Sci. USA. 2010;107:11751–11756. doi: 10.1073/pnas.1004255107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michiels PJA, Schouten CHJ, Hilbers CW, Heus HA. Structure of the ribozyme substrate hairpin of Neurospora VS RNA: a close look at the cleavage site. RNA. 2000;6:1821–1832. doi: 10.1017/s1355838200001394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flinders J, Dieckmann T. A pH controlled conformational switch in the cleavage site of the VS ribozyme substrate RNA. J. Mol. Biol. 2001;308:665–679. doi: 10.1006/jmbi.2001.4627. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann B, Mitchell GT, Gendron P, Major F, Andersen AA, Collins RA, Legault P. NMR structure of the active conformation of the Varkud satellite ribozyme cleavage site. Proc. Natl Acad. Sci. USA. 2003;100:7003–7008. doi: 10.1073/pnas.0832440100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flinders J, Dieckmann T. The solution structure of the VS ribozyme active site loop reveals a dynamic “hot-spot”. J. Mol. Biol. 2004;341:935–949. doi: 10.1016/j.jmb.2004.06.084. [DOI] [PubMed] [Google Scholar]
- 30.Campbell DO, Legault P. NMR structure of the Varkud satellite ribozyme stem-loop V RNA and magnesium-ion binding from chemical-shift mapping. Biochemistry. 2005;44:4157–4170. doi: 10.1021/bi047963l. [DOI] [PubMed] [Google Scholar]
- 31.Campbell DO, Bouchard P, Desjardins G, Legault P. NMR structure of Varkud satellite ribozyme stem-loop V in the presence of magnesium ions and localization of metal-binding sites. Biochemistry. 2006;45:10591–10605. doi: 10.1021/bi0607150. [DOI] [PubMed] [Google Scholar]
- 32.Lafontaine DA, Norman DG, Lilley DM. Structure, folding and activity of the VS ribozyme: importance of the 2-3-6 helical junction. EMBO J. 2001;20:1415–1424. doi: 10.1093/emboj/20.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lafontaine DA, Norman DG, Lilley DM. The global structure of the VS ribozyme. EMBO J. 2002;21:2461–2471. doi: 10.1093/emboj/21.10.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipfert J, Ouellet J, Norman DG, Doniach S, Lilley DM. The complete VS ribozyme in solution studied by small-angle X-ray scattering. Structure. 2008;16:1357–1367. doi: 10.1016/j.str.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elder D, Harris RJ. Ribozymes: the hairpin and Varkud ribozymes are related. Riv. Biol. 2003;96:433–439. [PubMed] [Google Scholar]
- 36.Legault P, Pardi A. Unusual dynamics and pKa shifts at the active site of a lead-dependent ribozyme. J. Am. Chem. Soc. 1997;119:6621–6628. [Google Scholar]
- 37.Cai Z, Tinoco IJ. Solution structure of loop A from the hairpin ribozyme from Tobacco ringspot virus satellite. Biochemistry. 1996;35:6026–6036. doi: 10.1021/bi952985g. [DOI] [PubMed] [Google Scholar]
- 38.Smith JS, Nikonowicz EP. NMR structure and dynamics of an RNA motif common to the spliceosome branch-point helix and the RNA-binding site for phage GA coat protein. Biochemistry. 1998;37:13486–13498. doi: 10.1021/bi981558a. [DOI] [PubMed] [Google Scholar]
- 39.Ravindranathan S, Butcher SE, Feigon J. Adenine protonation in domain B of the hairpin ribozyme. Biochemistry. 2000;39:16026–16032. doi: 10.1021/bi001976r. [DOI] [PubMed] [Google Scholar]
- 40.Legault P. Ph.D. thesis. Boulder: University of Colorado at Boulder; 1995. Thesis, structural studies of ribozymes by heteronuclear NMR spectroscopy. [Google Scholar]
- 41.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 42.Johnson BA, Blevins RA. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 43.Dingley AJ, Grzesiek S. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2JNN couplings. J. Am. Chem. Soc. 1998;120:8293–8297. [Google Scholar]
- 44.Piotto M, Saudek V, Skleñár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 45.Grzesiek S, Bax A. The importance of not saturating water in protein NMR. Application to sensitivity enhancement of NOE measurements. J. Am. Chem. Soc. 1993;115:12593–12594. [Google Scholar]
- 46.Zhang O, Kay LE, Olivier JP, Forman-Kay JD. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J. Biomol. NMR. 1994;4:845–858. doi: 10.1007/BF00398413. [DOI] [PubMed] [Google Scholar]
- 47.Mueller L, Legault P, Pardi A. Improved RNA structure determination by detection of NOE contacts to exchange-broadened amino groups. J. Am. Chem. Soc. 1995;117:11043–11048. [Google Scholar]
- 48.Ikura M, Kay LE, Tschudin R, Bax A. Three-dimensional NOESY-HMQC spectroscopy of a 13C-labeled protein. J. Magn. Reson. 1990;86:204–209. [Google Scholar]
- 49.Rance M, Sorensen OW, Bodenhausen G, Wagner G, Ernst RR, Wuthrich K. Improved spectral resolution in cosy 1H NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun. 1983;117:479–485. doi: 10.1016/0006-291x(83)91225-1. [DOI] [PubMed] [Google Scholar]
- 50.Schwalbe H, Marino JP, King GC, Wechselberger P, Bermel W, Griesinger C. Determination of a complete set of coupling constants in 13C-labeled oligonucleotides. J. Biomol. NMR. 1994;4:631–644. doi: 10.1007/BF00404274. [DOI] [PubMed] [Google Scholar]
- 51.Wijmenga SS, Mooren MMW, Hilbers CW. In: NMR of Macromolecules: A Practical Approach. Roberts GCK, editor. Vol. 134. New York: Oxford University Press; 1993. pp. 217–288. [Google Scholar]
- 52.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nature Struct. Biol. 1998;5:1064–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 53.Brutscher B, Boisbouvier J, Pardi A, Marion D, Simorre JP. Improved sensitivity and resolution in H-1-C-13 NMR experiments of RNA. J. Am. Chem. Soc. 1998;120:11845–11851. [Google Scholar]
- 54.Boisbouvier J, Bryce DL, O'Neil-Cabello E, Nikonowicz EP, Bax A. Resolution-optimized NMR measurement of (1)D(CH), (1)D(CC) and (2)D(CH) residual dipolar couplings in nucleic acid bases. J. Biomol. NMR. 2004;30:287–301. doi: 10.1007/s10858-005-1846-5. [DOI] [PubMed] [Google Scholar]
- 55.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:66–74. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 56.Van Melckebeke H, Devany M, Di Primo C, Beaurain F, Toulme JJ, Bryce DL, Boisbouvier J. Liquid-crystal NMR structure of HIV TAR RNA bound to its SELEX RNA aptamer reveals the origins of the high stability of the complex. Proc. Natl Acad. Sci. USA. 2008;105:9210–9215. doi: 10.1073/pnas.0712121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dosset P, Hus JC, Marion D, Blackledge M. A novel interactive tool for rigid-body modeling of multi-domain macromolecules using residual dipolar couplings. J. Biomol. NMR. 2001;20:223–231. doi: 10.1023/a:1011206132740. [DOI] [PubMed] [Google Scholar]
- 58.Clore GM, Gronenborn AM, Tjandra N. Direct structure refinement against residual dipolar couplings in the presence of rhombicity of unknown magnitude. J. Magn. Reson. 1998;131:159–162. doi: 10.1006/jmre.1997.1345. [DOI] [PubMed] [Google Scholar]
- 59.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rupert PB, Massey AP, Sigurdsson ST, Ferre-D'Amare AR. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 61.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 62.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, Debolt S, Ferguson D, Seibel G, Kollman P. Amber, a package of computer-programs for applying molecular mechanics, normal-mode analysis, molecular-dynamics and free-energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995;91:1–41. [Google Scholar]
- 63.Sood VD, Beattie TL, Collins RA. Identification of phosphate groups involved in metal binding and tertiary interactions in the core of the Neurospora VS ribozyme. J. Mol. Biol. 1998;282:741–750. doi: 10.1006/jmbi.1998.2049. [DOI] [PubMed] [Google Scholar]
- 64.Heus HA, Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA tetraloops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 65.Jucker FM, Heus HA, Yip PF, Moors EH, Pardi A. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 1996;264:968–980. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- 66.Wimberly B, Varani G, Tinoco I., Jr The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry. 1993;32:1078–1087. doi: 10.1021/bi00055a013. [DOI] [PubMed] [Google Scholar]
- 67.Szewczak AA, Moore PB, Chang YL, Wool IG. The conformation of the sarcin/ricin loop from 28S ribosomal RNA. Proc. Natl Acad. Sci. USA. 1993;90:9581–9585. doi: 10.1073/pnas.90.20.9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Correll CC, Munishkin A, Chan YL, Ren Z, Wool IG, Steitz TA. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc. Natl Acad. Sci. USA. 1998;95:13436–13441. doi: 10.1073/pnas.95.23.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leontis NB, Westhof E. A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J. Mol. Biol. 1998;283:571–583. doi: 10.1006/jmbi.1998.2106. [DOI] [PubMed] [Google Scholar]
- 70.Butcher SE, Allain FH, Feigon J. Solution structure of the loop B domain from the hairpin ribozyme. Nat. Struct. Biol. 1999;6:212–216. doi: 10.1038/6651. [DOI] [PubMed] [Google Scholar]
- 71.Zimmermann GR, Wick CL, Shields TP, Jenison RD, Pardi A. Molecular interactions and metal binding in the theophylline-binding core of an RNA aptamer. RNA. 2000;6:659–667. doi: 10.1017/s1355838200000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Allain FH, Gilbert DE, Bouvet P, Feigon J. Solution structure of the two N-terminal RNA-binding domains of nucleolin and NMR study of the interaction with its RNA target. J. Mol. Biol. 2000;303:227–241. doi: 10.1006/jmbi.2000.4118. [DOI] [PubMed] [Google Scholar]
- 73.Legault P, Pardi A. In situ probing of adenine protonation in RNA by 13C NMR. J. Am. Chem. Soc. 1994;116:8390–8391. [Google Scholar]
- 74.Moody EM, Lecomte JT, Bevilacqua PC. Linkage between proton binding and folding in RNA: a thermodynamic framework and its experimental application for investigating pKa shifting. RNA. 2005;11:157–172. doi: 10.1261/rna.7177505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hermann T, Westhof E. Exploration of metal ion binding sites in RNA folds by Brownian-dynamics simulations. Structure. 1998;6:1303–1314. doi: 10.1016/s0969-2126(98)00130-0. [DOI] [PubMed] [Google Scholar]
- 77.Butcher SE, Allain FH-T, Feigon J. Determination of metal ion binding sites within the hairpin ribozyme domains by NMR. Biochemistry. 2000;39:2174–2182. doi: 10.1021/bi9923454. [DOI] [PubMed] [Google Scholar]
- 78.Rupert PB, Ferré-D'Amaré AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 79.Alam S, Grum-Tokars V, Krucinska J, Kundracik ML, Wedekind JE. Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn. Biochemistry. 2005;44:14396–14408. doi: 10.1021/bi051550i. [DOI] [PubMed] [Google Scholar]
- 80.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 81.Walter AE, Wu M, Turner DH. The stability and structure of tandem GA mismatches in RNA depend on closing base pairs. Biochemistry. 1994;33:11349–11354. doi: 10.1021/bi00203a033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.